Introduction
Hepatitis B virus (HBV) infection remains a
significant public health concern globally. Approximately 350
million individuals worldwide are chronically infected with HBV,
and such infections may lead to the development of liver cirrhosis
or hepatocellular carcinoma (1,2).
Although various types of antiviral drugs, including
nucleotide/nucleotide analogue and interferon, have been used to
eradicate this virus in recent years, no significant progress has
been achieved (3). Increasing
evidence has demonstrated that patients acutely infected with HBV
usually develop marked, multispecific cytotoxic T lymphocyte (CTL)
responses to the virus, whereas chronically infected individuals
exhibit weak responses (4,5). Therefore, the development of
immunotherapeutic strategies to improve weak virus-specific T cell
responses is critical.
Dendritic cells (DCs) are considered the most potent
antigen-presenting cells (APCs); and as such, are able to initiate
immune responses against invading pathogens (6) and have been observed to be
responsible for the cross-presentation of antigens in vitro
and in vivo as well as the stimulation of naïve cluster of
differentiation (CD)8+ T cell proliferation and
maturation (7). A previous study
revealed that defective CTL responses may be attributed to impaired
DC function (8). Therefore,
promoting and improving the functions of DCs may comprise an
efficient treatment strategy for persistent HBV infections.
Ubiquitin (Ub) is a highly conserved small
regulatory protein (9), and a
centrally significant component of the Ub-proteasome system (UPS),
which attaches covalently to numerous cellular proteins through a
highly regulated process (10,11).
Major histocompatibility complex (MHC) class-I antigen presentation
is strictly dependent on the supply of appropriate peptides,
mediated by the UPS, with which to efficiently prime
CD8+ T cells and initiate an adaptive immune response
(12,13). The HBV core antigen (HBcAg) is a
highly immunogenic subviral particle that in natural and
recombinant forms may induce marked immune responses characterized
by acute T-cell activity (14).
These hypotheses prompted the present study to investigate whether
a Ub-modified HBcAg fusion protein was able to enter DCs and be
presented by MHC class-I molecules in order to elicit robust CTL
responses.
However, direct intracellular protein delivery is
inhibited by the lipophilic nature of biological membranes
(15). The cytoplasmic
transduction peptide (CTP), which is derived from the protein
transduction domain (PTD) of the human immunodeficiency virus-1
trans-activator of transcription protein, is a novel and
deliberately designed transduction protein used to efficiently
deliver biomolecules into the cytoplasm (16). Therefore, the properties of CTP
provide an opportunity for antigens to enter DCs and thus be
presented by MHC class I molecules.
In the present study, a prokaryotic expression
vector for Ub-HBcAg-CTP (GGRRARRRRRR) was constructed and purified.
Subsequently, the biological activity of the purified fusion
protein was examined to determine whether it was able to be
presented by MHC class-I molecules and consequently efficiently
enhance HBV-specific CTL responses in vitro.
Materials and methods
Animals and cell lines
The present study was approved by the Ethics
Committee of Shanghai JiaoTong University Affiliated Sixth People’s
Hospital (Shanghai, China). Thirty BALB/c mice (H-2d),
aged 6–8 weeks, were purchased from the Shanghai Experimental
Animal Centre of the Chinese Academy of Sciences (Shanghai, China)
and maintained in the Experimental Animal Center of the Shanghai
Sixth People’s Hospital under specific pathogen-free conditions
(22–24°C; humidity 50–55%; 12 h light/12 h dark cycle). The mice
were pellet fed standard mouse food and given access to sterilized
water. The mice were cared for and treated in accordance with the
guidelines established by the Shanghai Public Health Service Policy
on the Humane Care and Use of Laboratory Animals. After one week,
10 BALB/c mice were sacrificed by cervical dislocation following
anesthesia with 3% pentobarbital sodium (Sigma-Aldrich, St. Louis,
MO, USA) via intramuscular injection (1:5 dilution; dose, 10 g/0.1
ml). The bone marrow was collected from their femurs and tibiae,
which was the source of the bone marrow cells. Briefly, the femurs
and tibiae were removed from the mice and the surrounding muscle
tissue was removed from the bones. The intact bones were placed in
70% ethanol for 5 min, for disinfection, and were then washed with
phosphate-buffered saline (PBS; Keygentec, Nanjing, China). Both
ends were cut with scissors and the marrow was flushed with PBS.
Clusters within the marrow suspension were dispersed by vigorous
pipetting. The experiment was repeated three times.
HEK293T cells (Nanjing Medical University, Nanjing,
China) were cultured in Dulbecco’s modified Eagle’s medium
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 μg/ml streptomycin (Invitrogen Life Technologies,
Carlsbad, CA, USA), under humidified conditions with 5%
CO2 at 37°C. The H-2d mastocytoma cell line
P815/c (expressing the HBV core antigen) (Nanjing Medical
University) was maintained in our lab (Department of Infectious
Disease, Shanghai JiaoTong University Affiliated Sixth People’s
Hospital), and was cultured under the same conditions as the
HEK293T cells.
Vector construction
The plasmid pcDNA3.1 (-)-Ub-HBcAg was constructed
and maintained in our lab. The Ub-HBcAg cDNA sequence was generated
via polymerase chain reaction (PCR) to obtain an 820 bp PCR
product. The paired primer sequences were as follows: Forward,
5-AATGGATCCGGCGGCCGTCGTGCGCGTCGTCGTCGTCGTCGTATGGACAT
TGACCCG-3′ and reverse, 5′-CCCAAGCTTGCCACCTCTCAGG CGAAGG-3′
(Sangon Biotech Co., Ltd., Shanghai, China). The underlined
nucleotides represent the BamHI and HindIII sites,
respectively. The Ub-HBcAg-CTP gene (Sangon Biotech Co., Ltd.) was
inserted into the pMAL-c2X prokaryotic expression vector
(Invitrogen Life Technologies) at the BamHI and
HindIII (New England Biolabs, Ipswich, MA, USA) sites. The
control genes (Ub-HBcAg and HBcAg-CTP) were also amplified via PCR
and cloned separately into pMAL-c2X vectors. The aforementioned
plasmids were further identified via restriction enzyme digestion
and bidirectional DNA sequencing.
Protein expression, purification and
western blotting
The recombinant plasmids were transformed into the
Escherichia coli BL21 (DE3) bacterial strain (Keygentec) to
induce the expression of the recombinant fusion proteins. Following
being lysed by sonication (SM-650D; Shunma, Nanjing, China) and
centrifuged (5415C; Thermo Fisher Scientific, Waltham, MA, USA) at
8,000 × g for 5 min at 4°C, the supernatants containing the
Ub-HBcAg-CTP, Ub-HBcAg and HBcAg-CTP fusion proteins were purified
using an amylose resin column (Polysciences, Inc., Eppelheim,
Germany) according to the manufacturer’s instructions and were
evaluated via western blot analysis with an anti-HBcAg mouse
monoclonal antibody (1:500 dilution; Abcam, Cambridge, UK) at 4°C
overnight, and a horseradish peroxidase-conjugated goat anti-mouse
secondary monoclonal antibody (1:5,000 dilution; Wuhan Boster
Biological Technology, Ltd., Wuhan, China) for 2 h at room
temperature. The maltose binding protein-tag (Sangon Biotech Co.,
Ltd.) was ultimately cleaved by the Tobacco Etch Virus protease
(Invitrogen Life Technologies). All proteins were stored at 4°C
until use.
DC generation
DCs were generated according to a previously
published method with certain modifications (17). Briefly, bone marrow cells from the
femurs and tibiae of the mice were collected and cultured at a
density of 2×106 cells/ml in RPMI 1640 medium (Hyclone
Biocehmical Product Co., Ltd., Beijing, China) supplemented with
10% FBS, 20 ng/ml recombinant mouse granulocyte-monocyte colony
stimulating factor (rmGM-CSF; Peprotech EC Ltd., London, UK) and 10
ng/ml recombinant mouse interleukin 4 (rmIL-4; Peprotech EC Ltd.).
Following a two day incubation, the adherent cells were divided
into five groups, and four of the groups were cultured for an
additional 72 h in the presence of Ub-HBcAg-CTP, Ub-HBcAg,
HBcAg-CTP and HBcAg (Abcam; all concentrations 20 μg/ml),
respectively. All groups were treated with lipopolysaccharide (20
ng/ml; Sigma-Aldrich). On day eight, the non-adherent and loosely
adherent cells were harvested as DCs.
DC morphology, intracellular localization
and western blot analysis
The day five and day eight DCs were observed via
scanning (Quanta 450) and transmission (Tecnai 12) electron
microscopy (FEI Company, Eindhobven, Netherlands). The samples were
treated according to the standard experimental methods (18). The day five DCs were cocultured
with the aforementioned proteins at a concentration of 20
μg/ml for 24 h. Following washing with PBS, the cells were
fixed in 100% pre-chilled methanol (Keygentec). Following an
additional three washes with PBS, the cells were permeabilized with
0.3% Triton X-100 (Keygentec) and blocked with 10% normal goat
serum (Wuhan Boster Biological Technology, Ltd.). The cells were
subsequently incubated overnight with an anti-HBcAg mAb (1:500
dilution) at 4°C. The cells were then further incubated with goat
anti-mouse fluorescein isothiocyanate (FITC)-Immunoglobulin G
(Wuhan Boster Biological Technology, Ltd.) for 1 h at room
temperature. Following washing with PBS and DAPI-staining
(Keygentec), the cells were visualized with a LSM 510 laser
scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) and
analyzed using LSM image examiner software (Carl Zeiss). The mean
fluorescence intensity (MFI) of the entire view was calculated from
five sites (the four corners and center of each section).
Day five DCs, which were treated as described above,
were also cocultured with or without the specific proteasome
inhibitor MG-132 (10 μmol; Sigma-Aldrich). Following 24 h of
incubation, the DCs were harvested to analyze the level of HBcAg
via western blotting.
Western blot analysis
Following a 24 h incubation the DCs were harvested
and washed twice with PBS. The cells were then gently dispersed
into a single-cell suspension and homogenized using
radioimmunoprecipitation assay lysis buffer (Keygentec). Protein
concentrations were determined using Pierce Bicinchoninic Acid
Protein Assay Reagent kit (Pierce Biotechnology, Inc., Rockford,
IL, USA). The homogenates were diluted to the desired protein
concentration using 2X SDS-PAGE loading buffer (Invitrogen Life
Technologies). The samples were then boiled and loaded onto
polyacrylamide mini-gels (Invitrogen Life Technologies) for
electrophoresis. The protein samples were then transferred to
polyvinylidene fluoride membranes (EMD Millipore, Bedford, MA, USA)
using semi-dry apparatus. The membranes were then incubated with
HBcAg monoclonal human anti-mouse antibody at 4°C overnight,
followed by an incubation with horseradish peroxidase-conjugated
goat anti-mouse immunoglobulin G secondary antibody at room
temperature for 2 h. GAPDH was used as the control (GAPDH antibody,
1:1,000, 4°C overnight; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA). Image-Pro Plus (version, 6.0; Media Cybernetics, Inc,
Bethesda, MD, USA) was used to visualize and quantify the blots.
Gray value was used to compare the differences between the groups.
Gray value=gray value of HBcAg/gray value of GAPDH.
IL-12p70 production, DC immunophenotypic
analysis and T cell proliferation
Day 5 DCs were cocultured with the aforementioned
proteins for 72 h, and the IL-12p70 concentrations in the
supernatants were measured using a standard sandwich ELISA kit
(R&D Systems, Minneapolis, MN, USA) according to the
manufacturer’s instructions. The concentrations were calculated and
expressed as pg/ml. The surface molecules of day eight DCs were
analyzed following incubation with phycoerythrin (PE)-labeled
monoclonal antibodies against mouse CD11c (1:50 dilution), CD80
(1:50 dilution), CD83 (1:100 dilution), CD86 (1:100 dilution) and
MHC class-I (1:40 dilution) (eBioscience, San Diego, CA, USA) at
4°C in the dark for 30 min. Fluorescence analyses were performed on
a COULTER EPICS XL Flow Cytometer (Beckman Coulter, Miami, FL, USA)
using Expo32-ADC software (Beckman Coulter).
Day five DCs were cocultured with Ub-HBcAg-CTP,
Ub-HBcAg, HBcAg-CTP and HBcAg (20 μg/ml; Abcam). After 72 h, day
eight DCs were pretreated with 25 μg/ml mitomycin C
(Sigma-Aldrich) for 30 min. T cells were sorted from splenocytes of
allogenenic naïve mice using nylon wool columns (Polysciences.
Inc.) and grown as responder cells in coculture with DCs at various
responder/stimulator (T cell/DC) ratios (5:1, 10:1 and 20:1). The
cells were incubated in a final volume of 100 μl for 96 h,
during which 10 μl Cell Counting Kit-8 solution (Dojindo
Laboratories, Kumamoto, Japan) was added for 4 h. The absorbance
values of the cultures were read at a wavelength of 450 nm
(Multiskan Ascent; Thermo Fisher Scientific).
Intracellular cytokine analysis of
proliferative T cells
Proliferative T cells were stimulated for 6 h in the
presence of 25 μg/ml phorbol 12-myristate 13-acetate
(Sigma-Aldrich), 1 μg/ml ionomycin (Sigma-Aldrich) and 1.7
μg/ml monensin (Sigma-Aldrich) (19). The cells were subsequently stained
with a PE-Cy5-conjugated anti-CD3 mAb (eBioscience) and a
FITC-conjugated anti-CD8α mAb (eBioscience) for 15 min at room
temperature. Following fixation and permeabilization using the
Fix/Perm reagents A and B (BD Biosciences, San Jose, CA, USA) for
15 and 5 min respectively, the cells were incubated with a
PE-labeled anti-interferon (IFN)-γ mAb (eBioscience) for 30 min.
Fluorescence analyses were performed on a COULTER EPICS XL flow
cytometer (Beckman Coulter) with Expo 32-Advanced Digital
Compensation software (Navios™ Cytometer, version 1.0; Beckman
Coulter).
Cytokine secretion and CTL assay
T cells were cocultured with mature DCs in a
humidified atmosphere containing 5% CO2 at 37°C for four
days at a T cell/DC ratio of 10:1. The concentrations of various
cytokines (IFN-γ and IL-2) in the supernatants were measured using
mouse cytokine ELISA kits (R&D Systems). The concentrations
were expressed as pg/ml.
The P815/c cells were seeded as the target cells,
and previously stimulated T cells were used as the effector cells.
The T cells were cocultured with the P815/c cells at
effector/target ratios of 5:1, 10:1 and 20:1 for 4 h at 37°C in a
humidified atmosphere containing 5% CO2. HBcAg-specific
CTL activity was measured using a lactate dehydrogenase release
assay (CytTox 9 6 ® non-radioactive cytotoxicity kit;
Promega Corporation, Madison, WI, USA), according to the
manufacturer’s instructions. The absorbance values were recorded at
a wavelength of 490 nm (Multiskan Ascent). The percentage
cytotoxicity was calculated as follows: [(experimental release -
effector spontaneous release - target spontaneous release) /
(target maximum release - target spontaneous release)] × 100
(20).
Statistical analysis
Each value in the present study was obtained from a
minimum of three independent experiments and data are expressed as
the mean ± standard deviation. The differences between groups were
determined using one-way analysis of variance. Statistical data
analyses were performed with SPSS 16.0 software (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Fusion protein expression, purification
and western blotting
The Ub-HBcAg-CTP, Ub-HBcAg and HBcAg-CTP fusion gene
constructs were 813, 780 and 585 bp in size, respectively, as
determined by PCR amplification. All the target prokaryotic
expression plasmids were successfully constructed and identified by
restriction endonuclease analysis and bidirectional DNA sequencing
(data not shown). The purified fusion proteins were identified to
be ~80% pure via SDS-PAGE analysis (Fig. 1A) and the products were further
confirmed by western blot analysis (Fig. 1B).
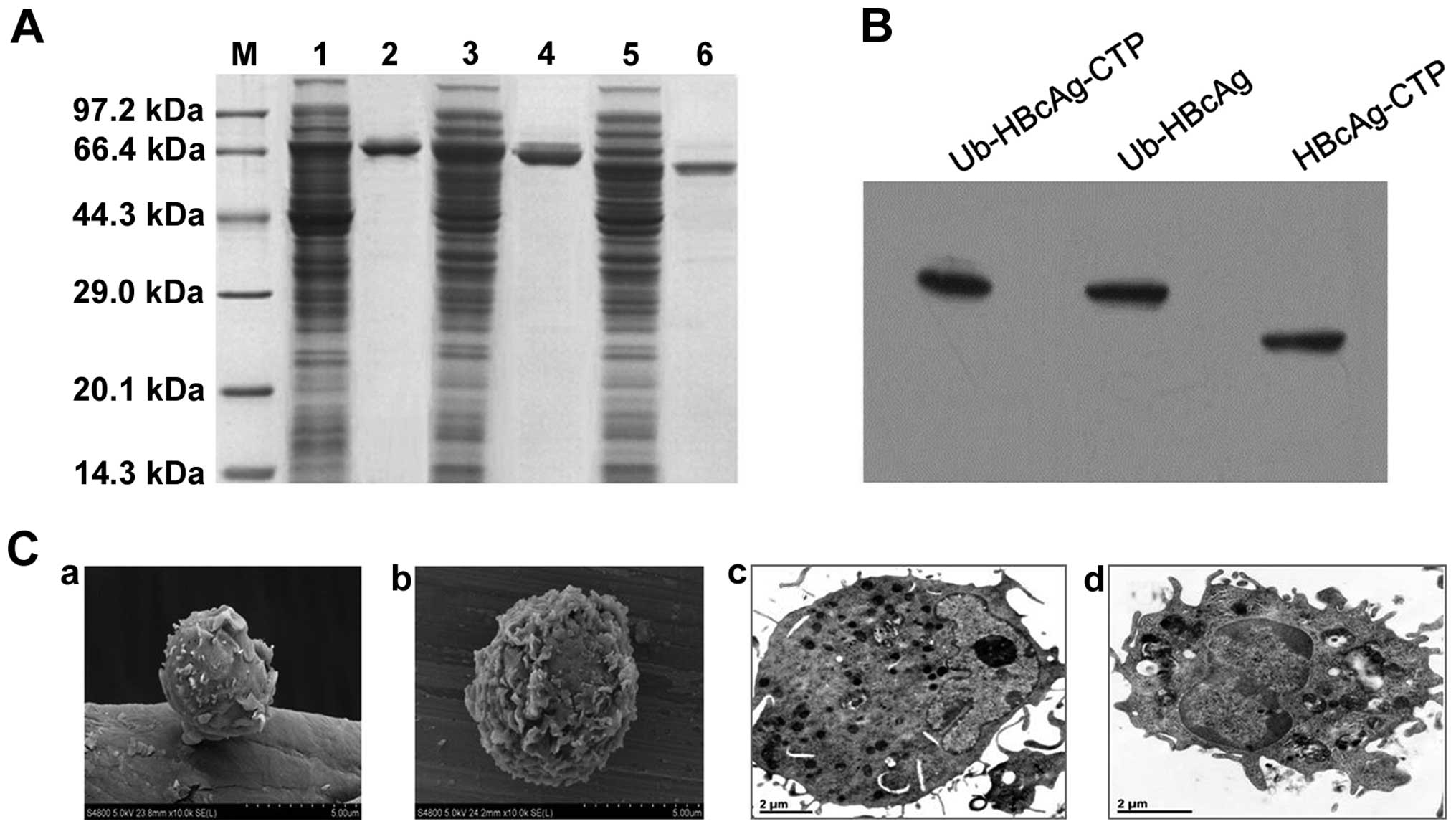 | Figure 1Fusion protein expression,
purification, western blot analysis and DC morphology. (A)
Expression and purification of fusion proteins by SDS-PAGE. Lanes:
M, molecular weight standards; 1 and 2, expression and purification
of MBP-Ub-HBcAg-CTP recombinant protein, respectively; 3 and 4,
expression and purification of MBP-Ub-HBcAg recombinant protein,
respectively; 5 and 6, expression and purification of MBP-HBcAg-CTP
recombinant protein, respectively. (B) Purified fusion proteins
were analyzed via western blotting using anti-HBcAg monoclonal
antibody. (C) Ultrastructures of day five and day eight DCs were
observed using (a and b) scanning electron microscopy
(magnification, ×5,000) and (c and d) transmission electron
microscopy (magnification, ×20,000), respectively. HBcAg, hepatitis
B virus core antigen; DC, dendritic cell; CTP, cytoplasmic
transduction peptide; MBP, maltose binding protein; Ub,
ubiquitin. |
DC maturation, cytoplasmic localization
and western blotting
On the DC surfaces, increased numbers of ruffes and
rough dendrites were observed by scanning electron microscopy.
Similarly, the morphological characteristics of mature DCs were
detected by transmission electron microscopy, and identified larger
nuclei, as well as increased numbers of mitochondria and ribosomes
(Fig. 1C).
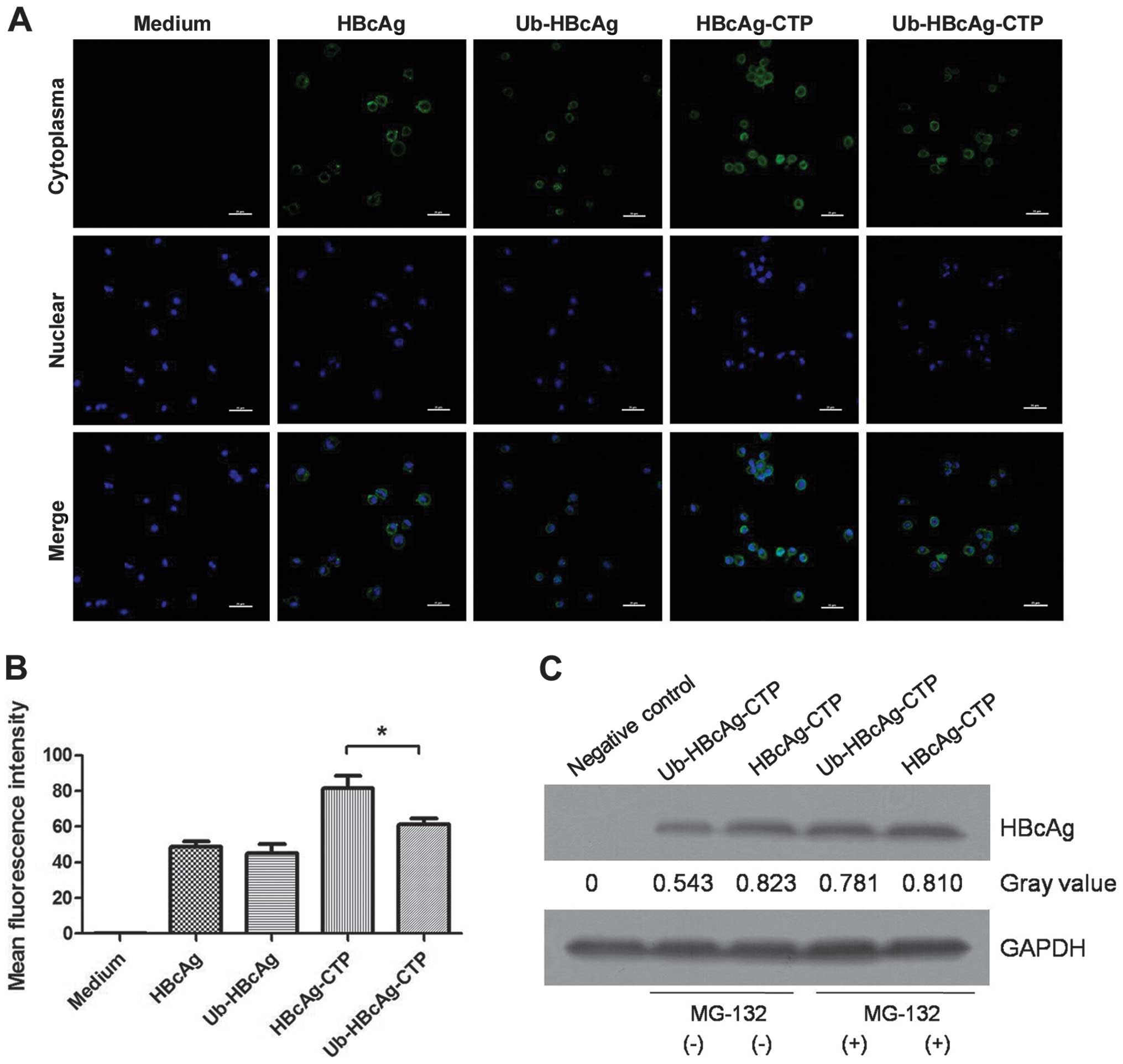
Immunofluorescent staining and confocal microscopy
were used to characterize the intracellular localization of the
various proteins in DCs. The HBcAg protein was identified in the
cytoplasm of DCs treated with Ub-HBcAg-CTP and HBcAg-CTP via the
presence of green fluorescence (FITC-labeled HBcAG) (Fig. 2A). This fluorescence was clearly
distinguished from the nuclear-specific counterstain, DAPI. Green
fluorescence was observed to aggregate primarily on the DC
cytomembranes in the Ub-HBcAg and HBcAg-treated groups (Fig. 2A), demonstrating the potent
transduction potential and cytoplasmic localization mediated by the
CTP peptide. The green fluorescence intensity detected in the
Ub-HBcAg-CTP group was weaker than that in the HBcAg-CTP group,
indirectly demonstrating the lower HBcAg levels in the former group
(Fig. 2B). Furthermore, the levels
of HBcAg were detected in the Ub-HBcAg-CTP and HBcAg-CTP groups
using western blot analysis to determine the rates of intracellular
degradation. As expected, HBcAg levels in Ub-HBcAg-CTP-group were
lower than those in the HBcAg-CTP-group, but recovered to the same
level as those of HBcAg following the addition of MG-132 (Fig. 2C).
Ub-HBcAg-CTP leads to increased IL-12
production and surface molecule expression
IL-12 is considered a vital cytokine, produced by
DCs, and is a key cytokine involved in the induction of Type-1
immune responses (21). The
IL-12p70 concentrations were assayed in the various protein-treated
groups. The results demonstrated that DCs cocultured with
Ub-HBcAg-CTP secreted significantly greater quantities of IL-12p70
(84.38±9.625 pg/ml) than those of the other groups (P<0.05;
Fig. 3B). However, no differences
were identified among the other groups. Additionally, following
treatment, the quantity of DCs (CD11c+) in the culture
was ~85% (flow cytometric analysis) and the surface molecules CD80,
CD83, CD86 and MHC class-I were highly expressed on the
Ub-HBcAg-CTP-treated DCs (Fig.
3A).
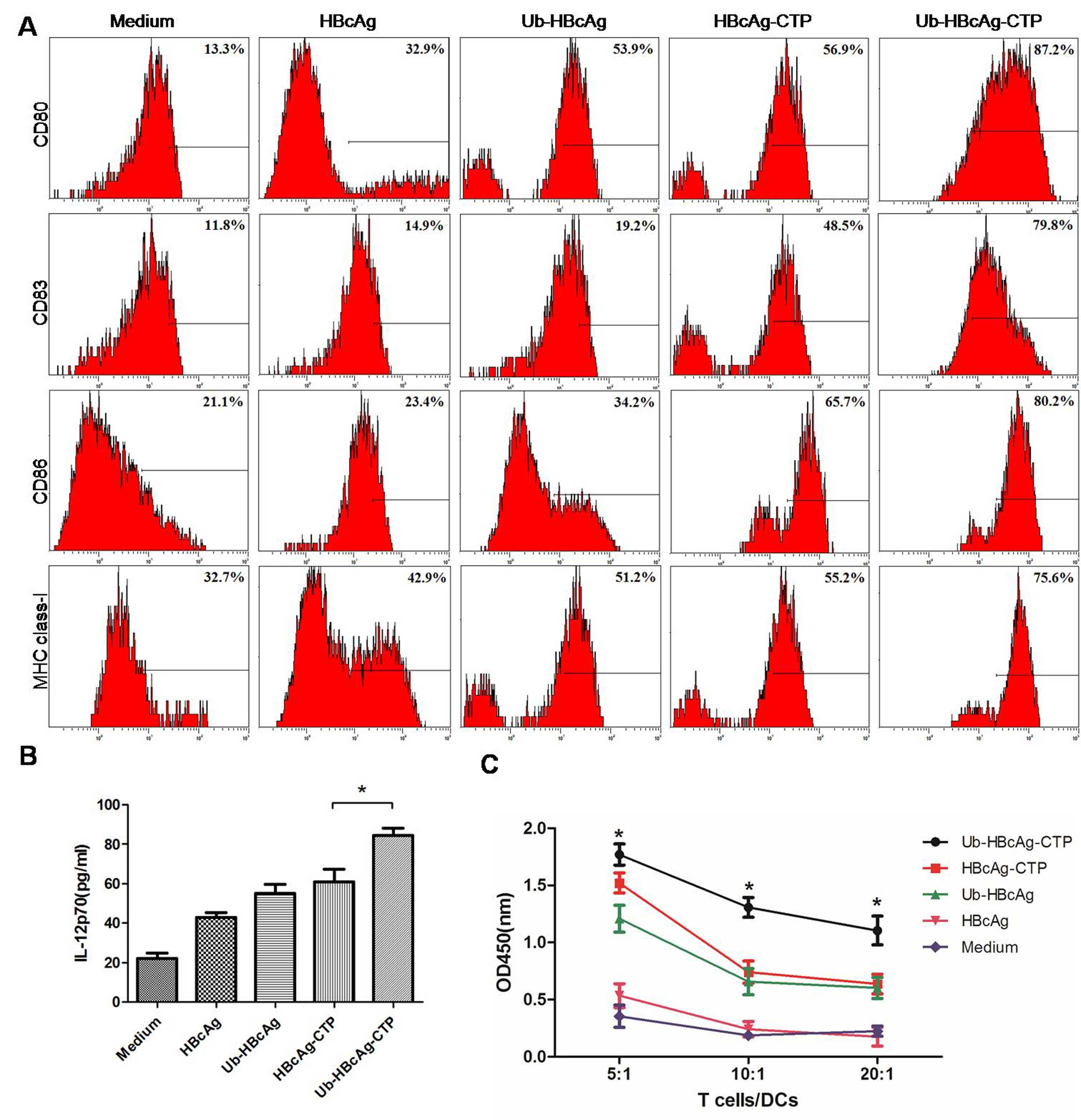 | Figure 3DC immunophenotypic analysis,
detection of IL-12p70 production and T-cell proliferation. (A)
Surface molecules on DCs (CD80, CD83, CD86, and major
histocompatibility complex class-I) were detected using flow
cytometry. Surface molecule expression was significantly higher in
the Ub-HBcAg-CTP group than in the other groups.
*P<0.05, **P<0.01. (B) IL-12p70
concentrations in supernatants of DCs were measured via ELISA. Data
is expressed as the mean ± SD. *P<0.05 vs. HBcAg-CTP
group. (C) T-cell proliferation capacities at various
responder/stimulator (T cell/DC) ratios. Data is expressed as the
mean ± SD. *P<0.05, **P<0.01 vs.
HBcAg-CTP group. HBcAg, hepatitis B virus core antigen; DC,
dendritic cell; CTP, cytoplasmic transduction peptide; MBP, maltose
binding protein; Ub, ubiquitin; SD, standard deviation; IL,
interleukin; CD, cluster of differentiation; OD, optical
density. |
T-cell proliferation
T cells were cocultured with DCs, which had been
pulsed with various proteins in order to analyze their T-cell
proliferation capacities. It was identified that the T-cell
proliferation capacity was markedly higher in the Ub-HBcAg-CTP
group (P<0.05; Fig. 3C), and
was enhanced by lower T cell/DC ratios.
Ub-HBcAg-CTP leads to increased IFN-y
production by CD8+ T cells in vitro
The levels of CD8+ T cell-produced IFN-γ
were measured by intracellular staining and fow cytometry. The
cells were stained with an anti-CD3 mAb to identify T cells within
the cell populations, and the samples were subsequently subjected
to intracellular staining. As shown in Fig. 4A, the percentage of specific
IFN-γ+CD8+ T cells was greater in the
Ub-HBcAg-CTP group than in the other groups (P<0.05), suggesting
that Ub-HBcAg-CTP-treated DCs may enhance the generation of
specific CTLs.
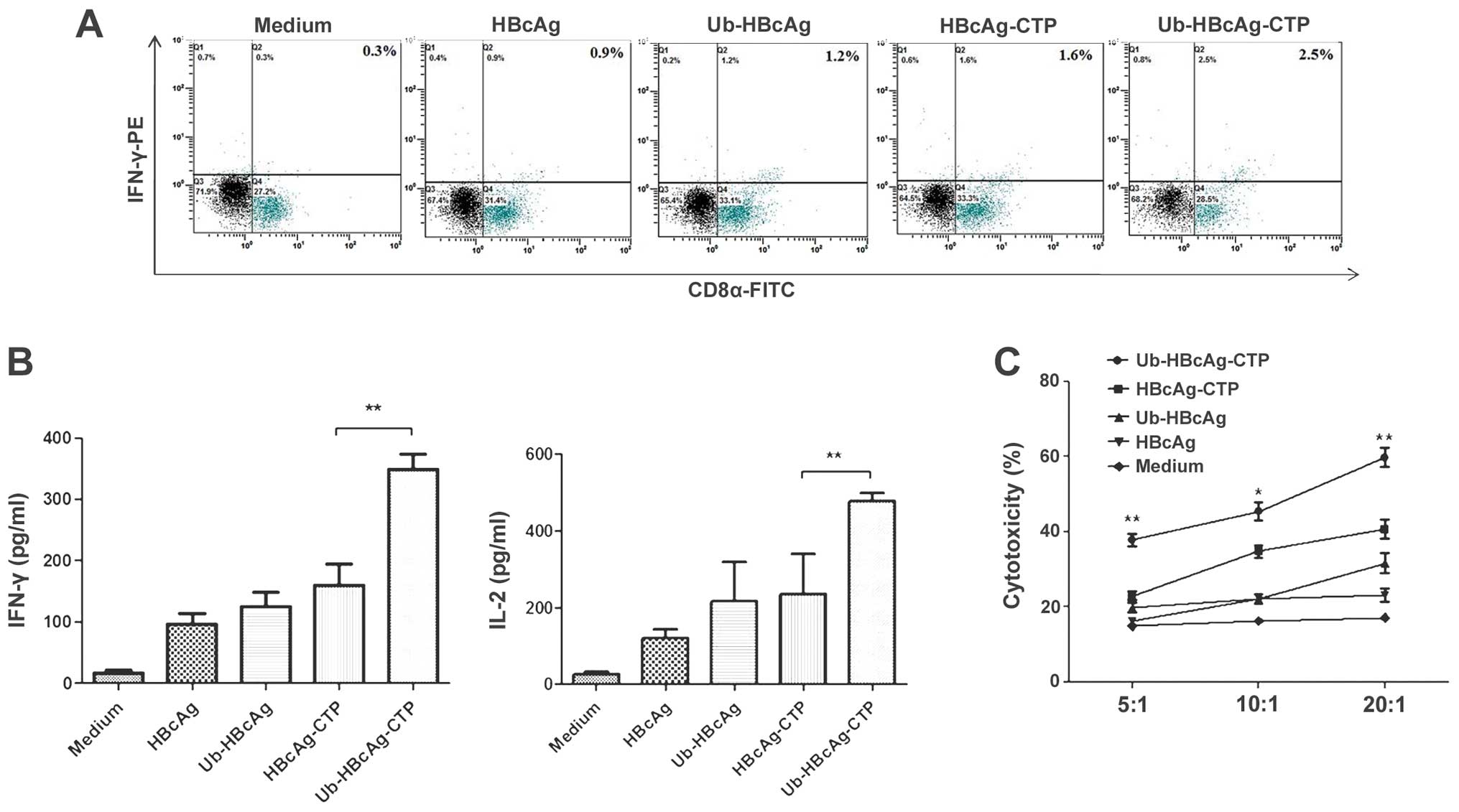 | Figure 4Intracellular cytokine analysis of
proliferative T cells, cytokine production and CTL assay. (A)
Specific IFN-γ+CD8+ T cells were detected by
flow cytometry. (B) Levels of IFN-γ and IL-2 in the supernatants of
proliferative T cells stimulated by various protein-treated DCs.
Data is expressed as the mean ± standard deviation.
*P<0.05. (C) Specific CTL activity was measured by
lactate dehydrogenase release assay. The proliferative T cells were
incubated with target cells at various effector/target ratios (5:1,
10:1 and 20:1). CTL activity was indicated as the mean percentage
of specific lysis (mean ± standard deviation).
*P<0.05, **P<0.01, compared with
control. CTL, cytotoxic T lymphocytes; IFN-γ, interferon-γ; IL,
interleukin; CD, cluster of differentiation; DC, dendritic cell;
HBcAg, hepatitis B virus core antigen; CTP, cytoplasmic
transduction peptide; Ub, ubiquitin; FITC, fluorescein
isothiocyanate. |
Ub-HBcAg-CTP-induces the enhancement of
cytokine production and CTL activity
Subsequently, the levels of IFN-γ and IL-2 secreted
by T cells were measured. The Ub-HBcAg-CTP group produced higher
levels of IFN-γ (348.8±24.78 pg/ml) and IL-2 (476.5±20.81pg/ml)
than those in the other groups (P<0.05, Fig. 4B). Clearer CTL responses were
detected at differing effector/target ratios in the Ub-HBcAg-CTP
group, compared with those in the other groups (Fig. 4C). These results suggested that
Ub-HBcAg-CTP may induce marked specific CTL responses, which was
consistent with the high level of IFN-γ expressed in
CD8+ T cells.
Discussion
Various immunotherapeutic strategies have been
developed to eliminate HBV; however, these strategies have not had
a substantial impact (22). The
host antiviral immune response, particularly CTL activity, to
HBV-antigens has been established as the main determinant in the
processes of viral replication and clearance (23,24).
Therefore, the induction of HBV-specific CD8+ T cells is
a current goal in the development of effective immune-based
therapeutic interventions for the treatment of HBV infection,
intended to enhance antigen presentation in order to induce broad
CTL responses.
C D8+ T cell priming requires the direct
and/or cross-presentation of antigenic peptides on MHC class I
molecules by APCs (25). DCs are
among the most potent APCs and possess a unique capacity to
interact with naïve T cells and consequently induce immune
responses (26). Patients with
chronic HBV infections generally exist in an immune-comprised
state, comprising immune tolerance with impaired DC function
(27,28). Therefore, efforts should be made to
enhance the antigen presenting capacities of DCs.
CTP, which is derived from PTD, was verified to
facilitate specific cytoplasmic localization and possess potent
membrane transduction potential (16). Accordingly, cytoplasmic functional
molecules may be more efficiently targeted by CTP-mediated delivery
(16,29). In the present study, a
Ub-HBcAg-CTP-expressing vector was constructed in order to achieve
increased transduction into cells. Following transduction, the
Ub-HBcAg-CTP fusion protein was detected via confocal microscopy,
in order to characterize the intracellular localization. It was
revealed that CTP-containing fusion proteins remained primarily in
the cytoplasm of DCs, whereas proteins without CTP were detected
primarily on the cell surface. This result confirmed that target
proteins were able to be transported efficiently into the DC
cytoplasm via CTP, thus providing a basis for the further
verification of protein immunogenicity.
Ub is a well-conserved and ubiquitously expressed
protein that is conjugated to target proteins (30). It is best known for its role as a
marker for protein destruction by the UPS (31). In the present study, the MFI of
Ub-HBcAg-CTP-transduced DCs was weaker than that of the HBcAg-CTP
group, thus indirectly demonstrating the degradation of HBcAg.
Furthermore, HBcAg was quantitatively measured by western blot
analysis. It was identified that the Ub-tagged HBcAg protein was at
a lower level in the absence of MG-132 (specific proteasome
inhibitor), indicating rapid recognition of the fusion protein by
the UPS, leading to HBcAg degradation. Ub-mediated antigen
processing has been widely used to enhance immune responses in
infectious disease and cancer studies (32,33).
A previous study confirmed that Ub-fused proteins may improve CTL
activity by promoting the introduction of the encoded protein into
the MHC class I pathway (34).
HBcAg, which possesses unique immunological features, elicits
prominent immune responses during chronic HBV infection (35). Therefore, a Ub-modified HBcAg gene
was amplified.
In the present study, the immunomodulatory effects
of these proteins on bone marrow-derived DC (BMDC) development
in vitro was investigated. It was identified that
Ub-HBcAg-CTP-treated BMDCs expressed higher levels of surface
molecules. Additionally, the associated mature DCs secreted higher
levels of IL-12 in response to various intracellular pathogenic
infections. This response has a key role in the initiation of
specific T cell-mediated immune responses and the promotion of
T-helper (Th)1 cell activation and differentiation (21). Accordingly, IL-12p70 production was
evaluated. As desired, the DC production of IL-12p70 was markedly
increased in response to Ub-HBcAg-CTP treatment, compared with that
in the other treatment groups. The DC morphology in the
Ub-HBcAg-CTP group was also observed to include numerous
mitochondria, as well as increased quantities of ribosomes and
rough endoplasmic reticulum. The results demonstrated that the
Ub-modified HBcAg protein may facilitate DC maturation and increase
MHC class-I expression.
Th1 cells are known to produce large quantities of
Type-1 cytokines, including IFN-γ and IL-2, whereas Th2 cells
produce large quantities of Type-2 cytokines, including IL-4 and
IL-10 (36). In the current study,
the secretion of Type-1 cytokines (IFN-γ and IL-2) was demonstrated
to be significantly increased in the Ub-HBcAg-CTP group, which
indicated that Ub-HBcAg-CTP-pulsed DCs had a tendency to promote
Th1 polarization and activate cell-mediated immunity. In addition,
in order to determine whether this maturation sufficiently
modulated CD8+ T cells, the production of IFN-γ was
measured via intracellular cytokine staining. The cells in the
Ub-HBcAg-CTP group were found to induce a greater quantity of
IFN-γ+CD8+ T cells and more robust specific
CTL activity. The specific cytotoxic response assay confirmed the
enhancing effect of Ub-HBcAg-CTP on specific CTL responses. The
results demonstrated that Ub-HBcAg-CTP fusion proteins may enhance
the capacity of T cells to proliferate, secrete cytokines and
develop into CTLs in vitro. Based on these findings, it is
possible that the role of the Ub-HBcAg-CTP fusion protein in the
induction of specific CTLs may be attributed to the fact that
UPS-degraded antigenic peptides are presented by
cell-surface-expressed MHC class-I molecules and are, therefore,
specifically recognized by CTLs.
In conclusion, the present study demonstrated that
Ub-HBcAg-CTP fusion proteins do not only increase the expression of
MHC class-I molecules, but also enhance the presentation of
antigenic peptides and elicit robust HBcAg-specific CTL immune
responses in vitro. Although the data presented in the
present study may not be fully translatable to studies in humans,
these findings may suggest a potential therapeutic strategy for
chronically infected HBV patients.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81270502)
and the Natural Science Foundation of Shanghai (grant no.
11ZR1427100).
References
|
1
|
Ganem D and Prince AM: Hepatitis B virus
infection - natural history and clinical consequences. New Engl J
Med. 350:1118–1129. 2004. View Article : Google Scholar
|
|
2
|
Shepard CW, Simard EP, Finelli L, Fiore AE
and Bell BP: Hepatitis B virus infection: epidemiology and
vaccination. Epidemiol Rev. 28:112–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Raney AK, Hamatake RK and Hong Z: Agents
in clinical development for the treatment of chronic hepatitis B.
Expert Opin Investig Drugs. 12:1281–1295. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rehermann B: Immune responses in hepatitis
B virus infection. Semin Liver Dis. 23:21–38. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kakimi K, Isogawa M, Chung J, Sette A and
Chisari FV: Immunogenicity and tolerogenicity of hepatitis B virus
structural and nonstructural proteins: implications for
immunotherapy of persistent viral infections. J Virol.
76:8609–8620. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steinman RM and Hemmi H: Dendritic cells:
translating innate to adaptive immunity. Curr Top Microbiol
Immunol. 311:17–58. 2006.PubMed/NCBI
|
|
7
|
Pozzi LA, Maciaszek JW and Rock KL: Both
dendritic cells and macrophages can stimulate naive CD8 T cells in
vivo to proliferate, develop effector function, and differentiate
into memory cells. J Immunol. 175:2071–2081. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Almand B, Resser JR, Lindman B, et al:
Clinical significance of defective dendritic cell differentiation
in cancer. Clin Cancer Res. 6:1755–1766. 2000.PubMed/NCBI
|
|
9
|
Glickman MH and Ciechanover A: The
ubiquitin-proteasome proteolytic pathway: destruction for the sake
of construction. Physiol Rev. 82:373–428. 2002.PubMed/NCBI
|
|
10
|
Shabek N and Ciechanover A: Degradation of
ubiquitin: the fate of the cellular reaper. Cell Cycle. 9:523–530.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hershko A and Ciechanover A: The ubiquitin
system. Annu Rev Biochem. 67:425–479. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Warnatsch A, Bergann T and Krüger E:
Oxidation matters: the ubiquitin proteasome system connects innate
immune mechanisms with MHC class I antigen presentation. Mol
Immunol. 55:106–109. 2013. View Article : Google Scholar
|
|
13
|
Burr ML, Boname JM and Lehner PJ: Studying
ubiquitination of MHC class I molecules. Methods Mol Biol.
960:109–125. 2013.PubMed/NCBI
|
|
14
|
Schödel F, Moriarty AM, Peterson DL, et
al: The position of heterologous epitopes inserted in hepatitis B
virus core particles determines their immunogenicity. J Virol.
66:106–114. 1992.PubMed/NCBI
|
|
15
|
Singh TR, Garland MJ, Cassidy CM, et al:
Microporation techniques for enhanced delivery of therapeutic
agents. Recent Pat Drug Deliv Formul. 4:1–17. 2010. View Article : Google Scholar
|
|
16
|
Kim D, Jeon C, Kim JH, et al: Cytoplasmic
transduction peptide (CTP): new approach for the delivery of
biomolecules into cytoplasm in vitro and in vivo. Exp Cell Res.
312:1277–1288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Y, Chen Z, Jia H, Wu W, Zhong S and
Zhou C: Induction of Tc1 response and enhanced cytotoxic T
lymphocyte activity in mice by dendritic cells transduced with
adenovirus expressing HBsAg. Clin Immunol. 119:280–290. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tatu RF, Anuşca DN, Groza SŞ, et al:
Morphological and functional characterization of femoral head
drilling-derived mesenchymal stem cells. Rom J Morphol Embryol.
55:1415–1422. 2014.
|
|
19
|
Crawford TQ, Ndhlovu LC, Tan A, et al:
HIV-1 infection abrogates CD8+ T cell mitogen-activated protein
kinase signaling responses. J Virol. 85:12343–12350. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen JH, Yu YS, Chen XH, Liu HH, Zang GQ
and Tang ZH: Enhancement of CTLs induced by DCs loaded with
ubiquitinated hepatitis B virus core antigen. World J
Gastroenterol. 18:1319–1327. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Giermasz AS, Urban JA, Nakamura Y, et al:
Type-1 polarized dendritic cells primed for high IL-12 production
show enhanced activity as cancer vaccines. Cancer Immunol
Immunother. 58:1329–1336. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Zou ZQ, Liu CX and Liu XZ:
Immunotherapeutic interventions in chronic hepatitis B virus
infection: a review. J Immunol Methods. 407:1–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsai SL, Sheen IS, Chien RN, et al:
Activation of Th1 immunity is a common immune mechanism for the
successful treatment of hepatitis B and C: tetramer assay and
therapeutic implications. J Biomed Sci. 10:120–135. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Phillips S, Chokshi S, Riva A, Evans A,
Williams R and Naoumov NV: CD8(+) T cell control of hepatitis B
virus replication: direct comparison between cytolytic and
noncytolytic functions. J Immunol. 184:287–295. 2010. View Article : Google Scholar
|
|
25
|
Schliehe C, Bitzer A, van den Broek M and
Groettrup M: Stable antigen is most effective for eliciting
CD8+ T-cell responses after DNA vaccination and
infection with recombinant vaccinia virus in vivo. J Virol.
86:9782–9793. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chiang CL, Balint K, Coukos G and
Kandalaft LE: Potential approaches for more successful dendritic
cell-based immunotherapy. Expert Opin Biol Ther. Jan 2–2015.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tavakoli S, Mederacke I, Herzog-Hauff S,
et al: Peripheral blood dendritic cells are phenotypically and
functionally intact in chronic hepatitis B virus (HBV) infection.
Clin Exp Immunol. 151:61–70. 2008. View Article : Google Scholar
|
|
28
|
Op den Brouw ML, Binda RS, van Roosmalen
MH, et al: Hepatitis B virus surface antigen impairs myeloid
dendritic cell function: a possible immune escape mechanism of
hepatitis B virus. Immunology. 126:280–289. 2009. View Article : Google Scholar :
|
|
29
|
Chen HZ, Wu CP, Chao YC and Liu CY:
Membrane penetrating peptides greatly enhance baculovirus
transduction efficiency into mammalian cells. Biochem Biophys Res
Commun. 405:297–302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schaefer A, Nethe M and Hordijk PL:
Ubiquitin links to cytoskeletal dynamics, cell adhesion and
migration. Biochem J. 442:13–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao G and Luo H: The ubiquitin-proteasome
pathway in viral infections. Can J Physiol Pharmacol. 84:5–14.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang QM, Kang L and Wang XH: Improved
cellular immune response elicited by a ubiquitin-fused ESAT-6 DNA
vaccine against Mycobacterium tuberculosis. Microbiol Immunol.
53:384–390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pal A and Donato NJ: Ubiquitin-specific
proteases as therapeutic targets for the treatment of breast
cancer. Breast Cancer Res. 16:4612014. View Article : Google Scholar
|
|
34
|
Rodriguez F, Zhang J and Whitton JL: DNA
immunization: ubiquitination of a viral protein enhances cytotoxic
T-lymphocyte induction and antiviral protection but abrogates
antibody induction. J Virol. 71:8497–8503. 1997.PubMed/NCBI
|
|
35
|
Cao T, Lazdina U, Desombere I, et al:
Hepatitis B virus core antigen binds and activates naive human B
cells in vivo: studies with a human PBL-NOD/SCID mouse model. J
Virol. 75:6359–6366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen W, Shi M, Shi F, et al: HBcAG-pulsed
dendritic cell vaccine induces Th1 polarization and production of
hepatitis B virus-specific cytotoxic T lymphocytes. Hepatol Res.
39:355–365. 2009. View Article : Google Scholar : PubMed/NCBI
|