Introduction
Borna disease virus (BDV) is a non-cytolytic,
neurotropic, non-segmented RNA virus of negative polarity and a
member of the family Bornaviridae within the order Mononegavirales
(1). The BDV genome spans ~8.9 kb
and contains six major open reading frames (ORFs) (2). BDV has been in the focus of interest
on account of its ability to cause severe neurobehavioral diseases
in animals and to introduce cDNA of its RNA transcripts into host
genomes (3,4). Studies initiated in the 1980s using
serological methods have suggested that human BDV infection may be
linked to neuropsychiatric diseases (5,6).
However, subsequent attempts to confirm human BDV infection by
non-serological methods have revealed inconsistent results
(4,7–9).
Therefore, the role of BDV role in human psychiatric disorders (if
any) remains inconclusive.
A previous epidemiological study by our group has
demonstrated that there may be an association between BDV infection
and certain human neuropsychiatric disorders, including viral
encephalitis, schizophrenia and bipolar disorder (10). In addition, a multi-center study by
our group conducted in three western Chinese provinces, which used
non-serological methods, including reverse transcription
quantitative polymerase chain reaction (RT-qPCR), ELISA and western
blot analysis, revealed that BDV may be associated with various
neuropsychiatric disorders, including viral encephalitis,
schizophrenia, multiple sclerosis and Parkinson's disease (11).
MicroRNAs (miRNAs) are a conserved class of
endogenous non-coding RNAs of ~22 nucleotides that modulate the
post-transcriptional expression of specific genes by base-pairing
with specific sequences (miRNA response element) in the
3′-untranslated regions (UTRs) of target mRNAs, thereby causing
mRNA degradation or translational inhibition (12). miRNAs exert pleiotropic effects on
gene expression and regulate diverse biological processes,
including cell proliferation, differentiation and apoptosis
(13). Previous studies have
suggested that certain miRNAs are integral components of host-virus
interactions and affect viral infection and host response processes
by targeting the viral genome or mRNAs (14,15).
For example, Qian et al (16) demonstrated that miR-122 exerts a
direct antiviral function by inhibiting BDV translation and
replication as well as indirectly acting through interferon (IFN)
to increase host innate immunity to modulate the virus-host
interaction. Zhai et al also found that persistent BDV
infection inhibited the expression of type I IFNs through the
suppression of miR-155 (17).
Neonatal rats with intracerebral BDV infection have
been described as excellent models of neonatal Borna disease (NBD)
in which to investigate the effects of BDV on learning, memory,
emotional impairment and social behavior, which are similar to
abnormalities observed in several human mental disorders, including
schizophrenia, affective disorders and autism (18–21).
The aim of the present study was to isolate miRNAs
from the hippocampi of BDV-infected and non-infected neonatal rats
in order to identify the differential miRNA expression profile.
Bioinformatic analysis was then performed to investi gate the
dysregulated miRNA target genes and the signaling pathways
involved, which may help to facilitate further understanding of the
important roles of miRNAs in BDV infection.
Materials and methods
Ethics statement
The Ethics Committee of Chongqing Medical University
approved the present study. All experiments were in accordance with
the National Institutes of Health Guidelines for Animal Research
(22). Special care was taken to
minimize the number and suffering of animals.
Sample collection
Newborn Sprague-Dawley rats (male; n=20; weighing,
10–12 g) purchased from the Chongqing Medical University Animal
Center [Animal Usage License no. SYXK (Chongqing) 20020007] were
monitored twice daily. Within 24 h of birth, rat pups were
intracranially (i.c.) inoculated in their right cerebral
hemispheres using a Hamilton syringe with either 50 µl
(104 FFU/ml) of BDV strain solution (kindly provided by
Professor Hanns Ludwig, AG Borna-Virus Infections, Free University
of Berlin, Germany) as previously described (23) or phosphate-buffered saline (PBS;
Jinmei Biotechnology, Jiangsu, China; control: Sham inoculated).
The virus strain was a human BDV Hu-H1 strain isolated from a
bipolar patient's white blood cells (24). Offspring were housed with their
mothers under biosafety level S2 for up to four weeks and then
gender-matched and separated. They were housed under a 12-h
light/dark rhythm at constant humidity and temperature and provided
with food and water ad libitum. Controls were housed under
standard conditions (25). On
postnatal day 56, anesthesia was performed by administration of 10%
chloral hydrate (100 g/0.4 ml; Jining BaiYi Chemical Co., Ltd.,
Shandong, China) by intraperitoneal injection (i.p.) (26), and the rat was sacrificed by
dislocation of the cervical vertebrae. The whole brains were
excised, the hippocampi were dissected from the brain on ice and
then submerged in liquid nitrogen followed by storage at −80°C
until later analysis.
RNA extraction for microarray and quality
inspection
Total RNA was extracted from the hippocampi of rats
infected intra-cranially with either BDV or sterile PBS using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA)
according to the manufacturer's instructions. The samples were
homog-enized in liquid nitrogen prior to adding TRIzol (1 ml TRIzol
per 50–100 mg tissue sample). RNA samples were dissolved in 25 µl
DNAse/RNase-free H2O and stored at −80°C until later
use. Eight samples (n=4 per group) of RNA were subjected to
GeneChip® miRNA3.0 Array analysis (Affymetrix, Santa
Clara, CA, USA). The concentration, purity and integrity of RNA
were measured by the Thermo Nanodrop 1000 (Thermo Fisher
Scientific, Waltham, MA USA) and an Agilent 2100 Bioanalyzer
(Agilent Technologies, Santa Clara, CA, USA).
MicroRNA microarray
Following RNA isolation, the Flash Tag Biotin HSR
RNA Labeling kit (Affymetrix) was used for miRNA labeling according
to the manufacturer's instructions. Each 0.05–0.5 µg of sample was
3′-end-labeled with a biotin fluorescent label using the Gene Chip
3′IVT Express kit (Affymetrix). After the labeling procedure was
terminated, the total mixture with the biotin-labeled samples and
hybridization buffer were hybridized in a Gene Chip Hybridization
Oven 645 (Affymetrix), which provides an active mixing action and a
constant incubation temperature to improve hybridization uniformity
and enhance the signal, according to the manufacturer's
instructions. Following hybridization, the slides were washed
several times using the GeneChip Hybridization Wash and Stain kit
(Affymetrix) and then dried by centrifugation at 3000 x g for 15
min at 25°C. The slides were then scanned with the GeneChip Scanner
3000 (Affymetrix) as previously described (27).
Array data analysis
The array data were analyzed using miRBase v17
(Affymetrix), which was also used for identifying the significantly
differentially expressed miRNAs. With the use of Gene Set
Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp) and its
Molecular Signatures Database 1.0 (MSigDB 1.0), the genes were
classified according to BioCarta (http://www.biocarta.com/) pathway and gene ontology
(GO) biological analysis to identify pathways actively regulated by
the significantly differentiated miRNAs (28).
RT-qPCR
According to the Qiagen Supplementary Protocol
(Qiagen GmbH, Hilden, Germany) for purification of small RNAs from
the rat hippocampi, total RNA was extracted using a Qiagen miRNeasy
Mini kit (Qiagen) and then eluted in 700 µl QIAzol reagent (Qiagen)
in 1.5 ml microcentrifuge tubes. Following chloroform (Chongquing
ChaunDong Chemical Co., Ltd., Chongqing, China) extraction, the
samples (n=3 per group) were centrifuged at 12,000 xg for 15 min at
4°C. The upper aqueous phase was transferred to a fresh collection
tube, and the RNA was precipitated by adding 1.5 volumes (usually
525 µl) of 100% ethanol (ChengDu KeLong Chemical Co., Ltd.,
Sichuan, China) to the aqueous phase. After being mixed thoroughly
by pipetting, the solution was transferred onto an RNeasy Mini spin
column (every column had an aqueous phase of <700 µl; Qiagen)
and centrifuged at ≥8,000 xg at room temperature for 15 sec in a
2-ml collection tube. Subsequently, 700 µl buffer RWT (Qiagen) was
added onto the RNeasy Mini spin column, which was then centrifuged
at ≥8,000 xg for 15 sec, followed by addition of 500 µl buffer PRE
(Qiagen) onto the column and centrifugation at ≥8,000 xg for 1 min.
Finally, the RNA pellet was re-suspended in 30–50 µl RNase-free
water (Shanxi Chaoying Biotechnology, Shanxi, China). The
concentration, purity and integrity of RNA were measured by the
Thermo Nanodrop 1000 and the Aglient 2100 Bioanalyzer.
Nine miRNAs with greater than or equal to two-fold
differential expression according to the microarray results were
selected for RT-qPCR validation. All the RNA samples were
reverse-transcribed to cDNA using the All-in-One™ miRNA qRT-PCR
Detection kit (GeneCopoeia, Rockville, MD, USA) according to the
manufacturer's instructions. The reaction mixture consisted of 5 µl
5X RT buffer (GeneCopoeia), 1 µl 2.5 U/µl PolyA polymerase, 1 µl
RTase Mix and 2,000 ng total RNA template in a total volume of 25
µl. Reverse transcription was performed in a Gene Amp PCR System
9700 (Applied Biosystems, Life Technologies, Thermo Fisher
Scientific) at 37°C for 60 min and 85°C for 5 min. An RT-negative
control was included in each batch of reactions. The qPCR reactions
were performed with the ABI Prism 7900 system (Applied Biosystems)
using the All-in-One™ miRNA q-PCR kit (GeneCopoeia) according to
the manufacturer's instructions. The reaction mixture consisted of
10 µl 2X All-in-One qPCR mix, 2 µl All-in-One™ miRNAqPCR primer (2
µM), 2 µl Universal Adaptor PCR primer (2 µM), and 2 µl
First-strand cDNA (diluted 1:5) in a total volume of 20 µl. PCR
reactions were initiated by a 10-min incubation at 95°C followed by
40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 10 sec.
A melting curve was performed at the end of the PCR run over a
range of 66−95°C, increasing the temperature stepwise by 0.5°C
every 6 sec. All samples were measured in triplicate, and no
template controls were included on the same plate. The qPCR
reactions were performed as described above and repeated three
times in triplicate.
Statistical analysis for RT-qPCR
The relative abundance of each miRNA was calculated
using the comparative Ct (2™ΔΔCt) method. The results
were assessed by a t-test, χ2 test or Wilcoxon
Mann-Whitney test as appropriate. P<0.05 was considered to
indicate a statistically significant difference between values.
Statistical analysis was performed using the SPSS 21.0 software
package (International Business Machines, Armonk, NY, USA).
Results
Differentially expressed miRNAs
The GeneChip® miRNA3.0 Array employed in
the present study contained >2,990 capture probes, covering all
human, mouse and rat miRNAs annotated in miRBase v17 as well as all
viral miRNAs associated with these species. Volcano plot filtering
was performed to identify miRNAs with a greater than or equal to
two-fold differential expression between the BDV-infected and
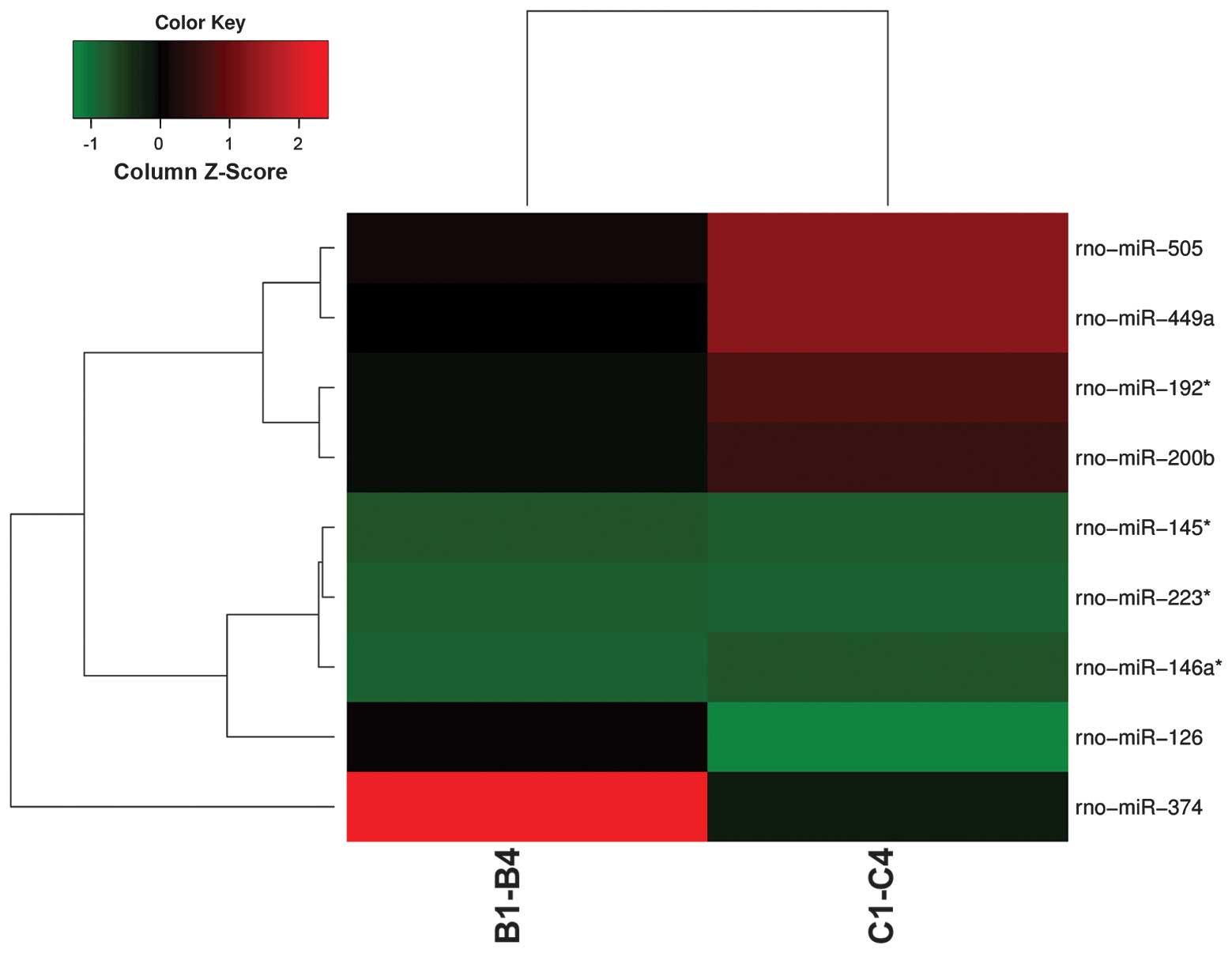
control groups. Fig. 1 displays
the results of a two-way hierarchical clustering of miRNAs and
samples. Seven miRNAs (miR-145*, miR-146a*,
miR-192*, miR-200b, miR-223*, miR-449a and
miR-505) showed increased expres sion, whereas two miRNAs (miR-126
and miR-374) showed decreased expression in the BDV-infected group.
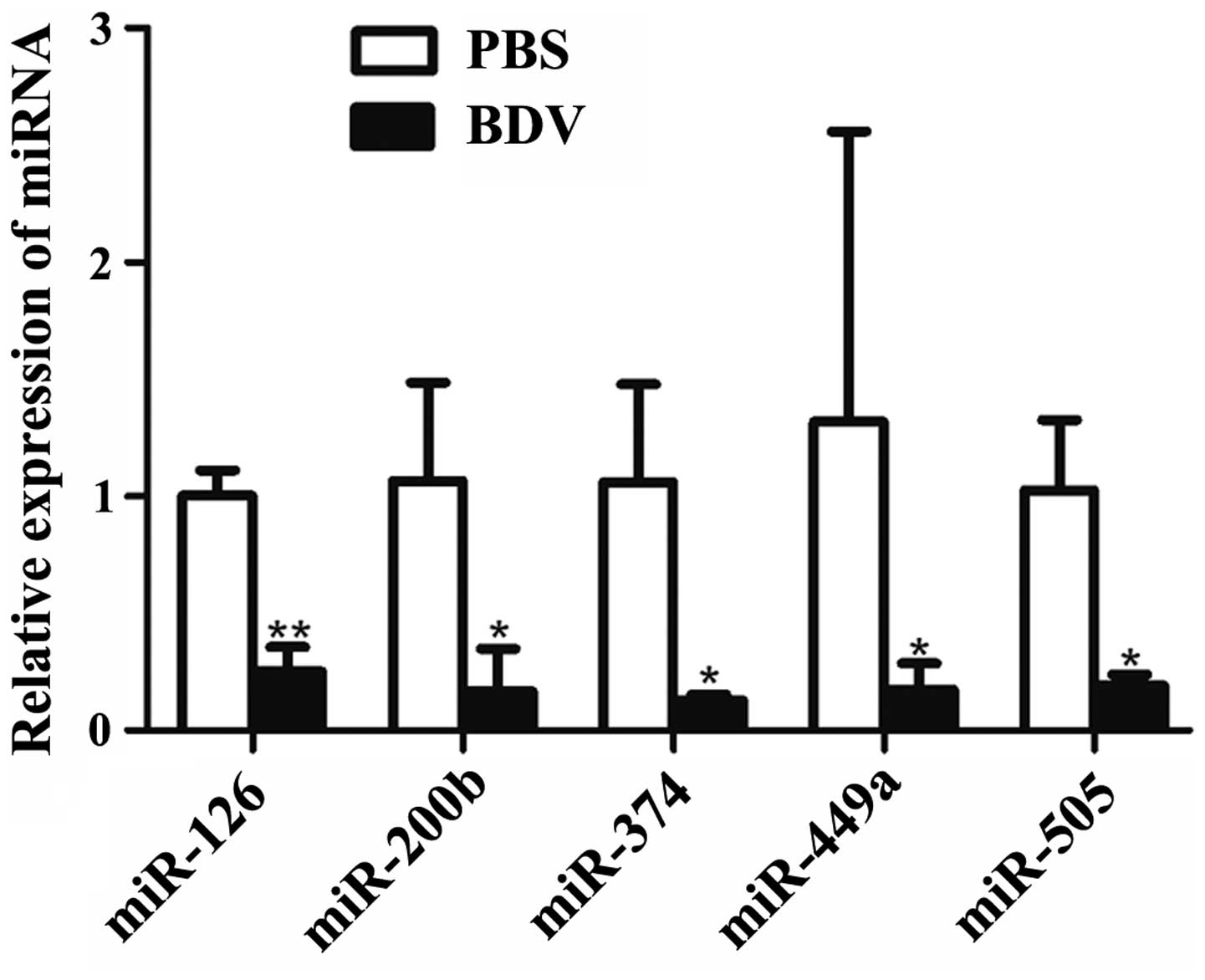
RT-qPCR analysis further indicated that five miRNAs (miR-126,
miR-200b, miR-374, miR-449a and miR-505) showed significantly
decreased expression (P<0.05) in response to BDV infection
(Fig. 2).
Prediction of target genes and functional
bioinformatic analysis
Using the MSigDB 1.0, genes were predicted as
targets for the nine differentially expressed miRNAs by sequencing
of miR-145*, miR-146a*, miR-192*,
miR-200b, miR-223*, miR-449a, miR-505, miR-126, and
miR-374. These genes were submitted to Gene Set Enrichment Analysis
(GSEA), which was used for GO biological process categorization,
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathway analysis, and
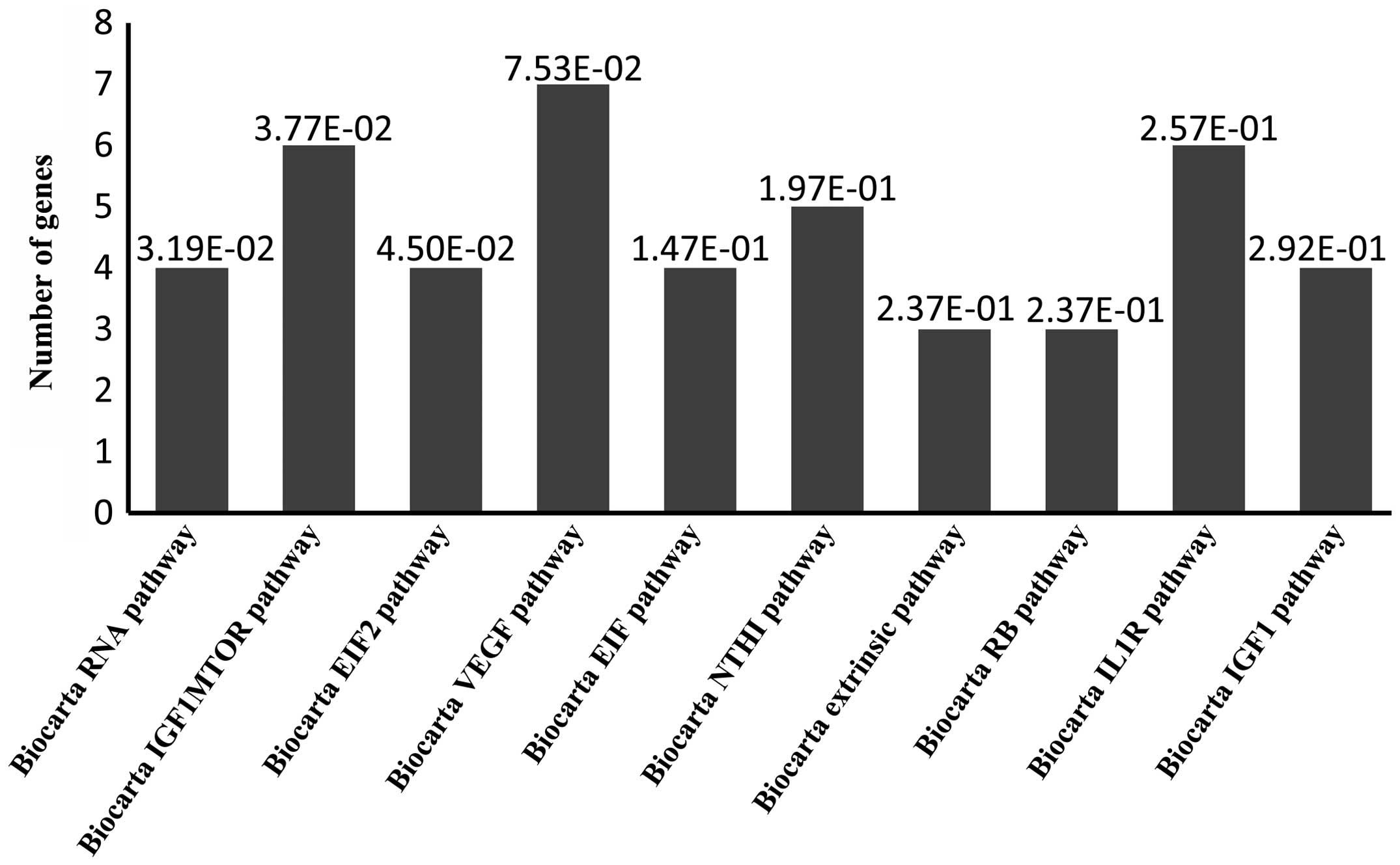
Biocarta pathway analysis. Biocarta pathway anlaysis predicted
target genes associated with 'RNA', 'IGF1mTOR', 'EIF2', 'VEGF',
'EIF', 'NTHI', 'extrinsic', 'RB', 'IL1R' and 'IGF1' pathways
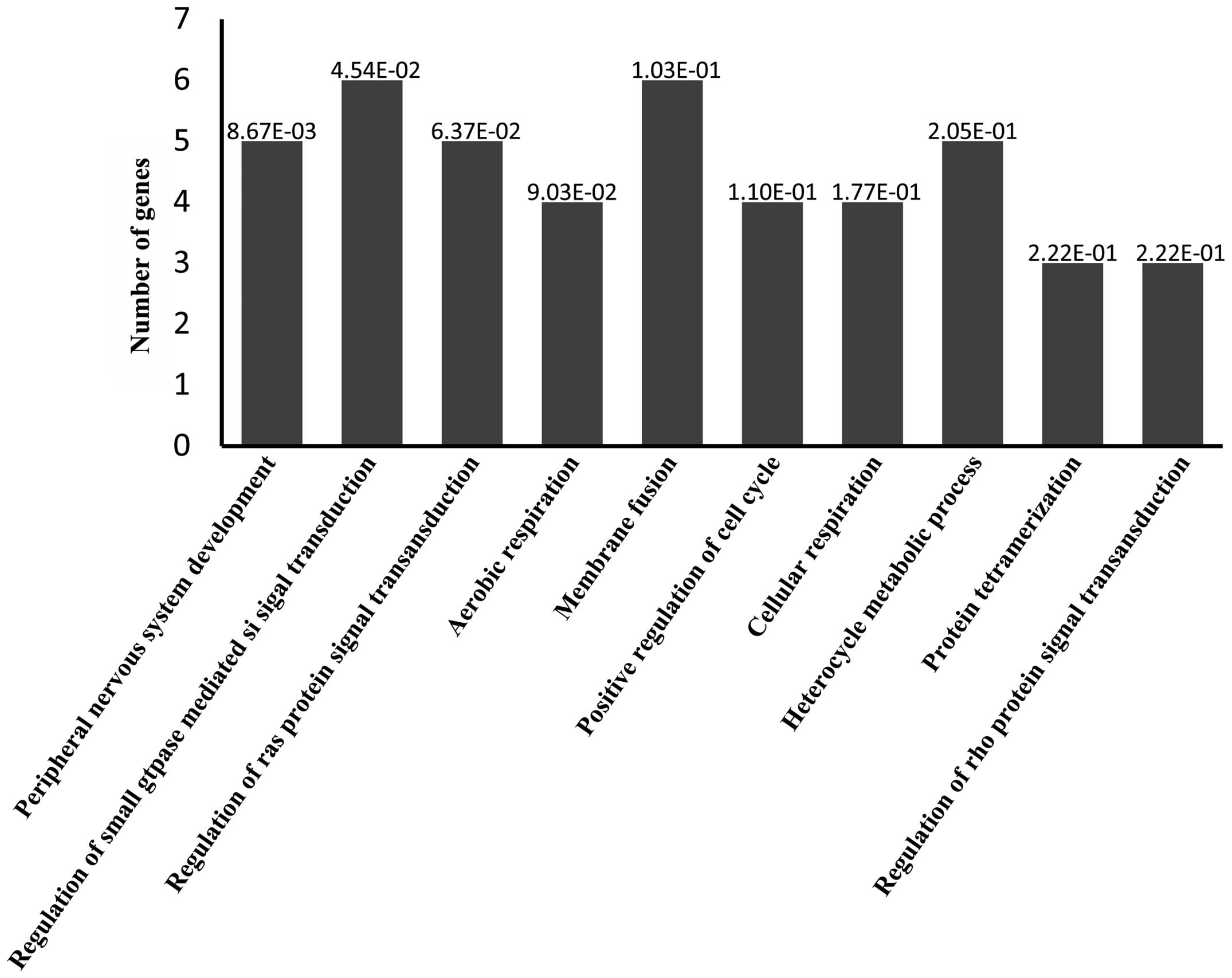
(Fig. 3). GO analysis predicted
target genes associated with 'peripheral nervous system
development', 'regulation of small GTPase-mediated signal
transduction', 'regulation of Ras protein signal transduction',
'aerobic respiration', 'membrane fusion', 'positive regulation of
cell cycle', 'cellular respiration', 'heterocycle metabolic
process', 'protein tetramerization' and 'regulation of Rho protein
signal transduction' processes (Fig.
4). Predicted target genes were classified according to
Biocarta pathway analysis by using GSEA bioinformatics resources
(Table I). Several GO biological
processes were predicted by GO analysis (Table II).
 | Table IBiocarta pathways enriched for
targets of the nine differentiated miRNAs. |
Table I
Biocarta pathways enriched for
targets of the nine differentiated miRNAs.
| Gene set | Number of
genes | P-value | Target genes |
|---|
| BioCarta RNA
pathway | 4 |
3.19×10−2 | EIF2S1, EIF2S2,
CHUK, TP53 |
| BioCarta IGF1MTOR
pathway | 6 |
3.77×10−2 | EIF2S1, EIF2S2,
EIF2B5, EIF4E, IGF1R, RPS6KB1 |
| BioCarta EIF2
pathway | 4 |
4.50×10−2 | EIF2S1, EIF2S2,
EIF2B5, EIF2AK3 |
| BioCarta VEGF
pathway | 7 |
7.53×10−2 | EIF2S1, EIF2S2,
EIF2B5, VHL, PLCG1, PXN, FLT1 |
| BioCarta EIF
pathway | 4 |
1.47×10−1 | EIF2S1, EIF2S2,
EIF4E, EIF4A2 |
| BioCarta NTHI
pathway | 5 |
1.97×10−1 | CHUK, MAP3K7,
MAP2K6, DUSP1, NR3C1 |
| BioCarta extrinsic
pathway | 3 |
2.37×10−1 | F2R, TFPI, F3 |
| BioCarta RB
pathway | 3 |
2.37×10−1 | TP53, CDK2,
MYT1 |
| BioCarta IL1R
pathway | 6 |
2.57×10−1 | CHUK, MAP3K7,
MAP2K6, JUN, IL1A, TGFB2 |
| BioCarta IGF1
pathway | 4 |
2.92×10−1 | IGF1R, JUN, MAP2K1,
PTPN11 |
 | Table IIPredicted miRNA target genes by gene
ontology analysis. |
Table II
Predicted miRNA target genes by gene
ontology analysis.
| Gene set | Number of
genes | P-value | Target genes |
|---|
| Peripheral nervous
system development | 5 |
8.67×10−3 | ACCN1, EGR2,
SERPINI1, UGT8, PMP22 |
| Regulation of small
GTPase mediated signal transduction | 6 |
4.54×10−2 | ARF6, RALBP1, FGD4,
PLCE1, MFN2, CDC42BPB |
| Regulation of Ras
protein signal transduction | 5 |
6.37×10−2 | ARF6, RALBP1, FGD4,
PLCE1, MFN2 |
| Aerobic
respiration | 4 |
9.03×10−2 | NNT, SDHD, UQCRH,
SLC25A14 |
| Membrane
fusion | 6 |
1.03×10−1 | VPS4B, GOSR2,
VAMP3, NAPG, VAPA, RABEP1 |
| Positive regulation
of cell cycle | 4 |
1.10×10−1 | TGFB2, EREG,
CITED2, TGFA |
| Cellular
respiration | 4 |
1.77×10−1 | NNT, SDHD, UQCRH,
SLC25A14 |
| Heterocycle
metabolic process | 5 |
2.05×10−1 | HPRT1, MAT2B,
COX15, CPOX, MTHFD2 |
| Protein
tetramerization | 3 |
2.22×10−1 | HPRT1, TP53,
IGF1R |
| Regulation of Rho
protein signal transduction | 3 |
2.22×10−1 | ARF6, RALBP1,
FGD4 |
Discussion
The present study reported the dysregulated miRNAs
in the hippocampi of neonatal rats infected with BDV Hu-H1. Among
the five dysregulated miRNAs identified by RT-qPCR, miR-126,
miR-200b and miR-449a showed a strong association with nervous
system development, cell differentiation, proliferation and
apoptosis, while miR-374 and miR-505 showed no significant
association with these processes.
Numerous previous studies have focused on rats
infected with BDV, usually with BDV strain He/80, strain V and
strain H1766 that originate from horses with Borna disease
(4,18–20,29,30).
By contrast, the present study used rats infected with BDV virus
strain Hu-H1 that originated from a human bipolar patient. In
agreement with the findings of the present study, BDV-infected rats
display neurogen-esis-associated disorders, including impairments
in learning, memory and emotion. The hippocampus is an essential
brain region for cognition and emotion (31), and miRNAs have vital roles in
hippocampal survival, development, function and plasticity
(32–34). Although previous studies on
specific miRNA expression and function in BDV-infected
oligoden-droglial cells have been published (16,17),
the present study was the first to comprehensively assess miRNA
expression in the hippocampi of BDV-infected neonatal rats.
Over-expression of miR-126 leads to nuclear factor
(NF)-κB activation via downregulation of inhibitor of NF-κB
(35). A study has revealed the
interactions of BDV with the NF-κB system (36), which is involved in the regulation
of cellular apoptosis and host defence. Therefore, the finding of
the present study that miR-126 was down-regulated in BDV-infected
rat hippocampi further supported the hypothesis that BDV infection
inhibits NF-κB through downregulating miR-126, finally leading to
disturbances in synaptic plasticity and induction of cellular
apoptosis (37).
miR-200b regulates multiple cellular functions,
including motility, proliferation and apoptosis (38). One study on microglia has
established that miR-200b directly inhibits c-Jun (39), a substrate of the c-jun N-terminal
kinase (JNK)/mitogen-activated protein kinase (MAPK) pathway in the
brain (40), which regulates
microglial activation (41) and
other diverse biological functions, including proliferation,
differentiation and apoptosis in various cell types (42–45).
Impaired microg-lial migration contributes to the pathogenesis of
several brain diseases, including prion disease (46), Parkinson's disease (47) and Alzheimer's disease (48). extracellular signal-regulated
kinase (ERK) is one MAPK family member in mammalian cells (49), and BDV Hu-H1 activates the ERK-RSK
complex downstream of the Raf/MAPK kinase (MEK)/ERK signaling
cascade in human oligodendroglial cells, thereby inhibiting cell
proliferation (50). These
combined findings supported the hypothesis that BDV-induced
downregulation of miR-200b may accentuate neurodegenerative
diseases through activating the ERK-RSK complex of the Raf/MEK/ERK
signaling cascade and targeting the JNK/MAPK signaling pathway,
which regulates microglial activation.
miR-449a modulates cell cycle regulation and
apoptosis through regulating cyclin D1 and B-cell lymphoma 2 (BCL2)
expression in SGC7901 cells (51).
Previous findings by our group demonstrated that BDV Hu-H1 inhibits
cellular proliferation and promotes apoptosis in human
oligodendrocytes via BCL2-associated X protein upregulation and
BCL2 downregulation (52). In
addition, miR-449a downregulation releassd CDK6 kinases, and
miR-449a overexpression significantly inhibits E2F1 expression
(53). CDK6/E2F1 is an important
regulator of cell proliferation and cell cycle progression
(54). Based on these combined
findings, the effect of BDV on miR-449a expression is likely to
have an important role in cell cycle regulation, proliferation and
apoptosis through the BCL2 and CDK6/E2F1 pathways.
In the present study, the RT-qPCR results were
inconsistent with the microarray results obtained prior to the
validation in the present study. This is likely due to the
limitations in the sensitivity, quantification and false positives
inherent to microarray technology (27). This phenomenon has also occurred in
previous miRNA expression studies in gastric cancer cells (55) and spinal cord injury (56).
In conclusion, the present study reported the
dysregulation of miRNAs in the hippocampi of neonatal rats infected
with BDV Hu-H1. Among the five dysregulated miRNAs identified by
RT-qPCR, miR-126, miR-200b and miR-449a showed a strong association
with nervous system development, cell differentiation,
proliferation and apoptosis. Further studies on miRNA target gene
identification and their specific biological functions are required
to address the specific regulatory mechanisms of the dysregulated
miRNAs in BDV infection.
Acknowledgments
The authors would like to thank Professor Hanns
Ludwig (AG Borna-Virus Infections at the Free University of Berlin,
Germany) and Dr Liv Bode (Robert Koch Institute, Free University of
Berlin, Berlin, Germany) for providing the BDV Hu-H1 strain. The
authors would also like to thank Dr ND Melgiri for his assistance
in editing and proofreading the manuscript. The present study was
financially supported by the National Natural Science Foundation of
China (grant no. 31300137), the Chongqing Foundation Frontier
Research Plan Project (grant no. cstc2013jcyjA10003) and the
National Basic Research Program of China (973 Program, grant no.
2009CB918300).
References
|
1
|
Zhang L, Wang X, Zhan Q, et al: Evidence
for natural Borna disease virus infection in healthy domestic
animals in three areas of western China. Arch Virol. 159:1941–1949.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Briese T, Schneemann A, Lewis AJ, et al:
Genomic organization of Borna disease virus. Proc Natl Acad Sci
USA. 91:4362–4366. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Horie M, Honda T, Suzuki Y, et al:
Endogenous non-retroviral RNA virus elements in mammalian genomes.
Nature. 463:84–87. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bode L and Ludwig H: Borna disease virus
infection, a human mental-health risk. Clin Microbiol Rev.
16:534–545. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Amsterdam JD, Winokur A, Dyson W, et al:
Borna disease virus: a possible etiologic factor in human affective
disorders? Arch Gen Psychiatry. 42:1093–1096. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rott R, Herzog S, Fleischer B, et al:
Detection of serum antibodies to Borna disease virus in patients
with psychiatric disorders. Science. 228:755–756. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carbone KM: Borna disease virus and human
disease. Clin Microbiol Rev. 14:513–527. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ikuta K, Ibrahim MS, Kobayashi T and
Tomonaga K: Borna disease virus and infection in humans. Front
Biosci. 7:d470–d495. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwemmle M: Borna disease virus infection
in psychiatric patients: are we on the right track? Lancet Infect
Dis. 1:46–52. 2001. View Article : Google Scholar
|
|
10
|
Li Q, Wang Z, Zhu D, et al: Detection and
analysis of Borna disease virus in Chinese patients with
neurological disorders. Eur J Neurol. 16:399–403. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Xu MM, Zeng L, et al: Evidence
for Borna disease virus infection in neuropsychiatric patients in
three western China provinces. Eur J Clin Microbiol Infect Dis.
33:621–627. 2014. View Article : Google Scholar
|
|
12
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ghosh Z, Mallick B and Chakrabarti J:
Cellular versus viral microRNAs in host-virus interaction. Nucleic
Acids Res. 37:1035–1048. 2009. View Article : Google Scholar :
|
|
15
|
Gottwein E and Cullen BR: Viral and
cellular microRNAs as determinants of viral pathogenesis and
immunity. Cell Host Microbe. 3:375–387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qian J, Zhai A, Kao W, et al: Modulation
of miR-122 on persistently Borna disease virus infected human
oligodendroglial cells. Antiviral Res. 87:249–256. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhai A, Qian J, Kao W, et al: Borna
disease virus encoded phos-phoprotein inhibits host innate immunity
by regulating miR-155. Antiviral Res. 98:66–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hornig M, Weissenböck H, Horscroft N and
Lipkin WI: An infection-based model of neurodevelopmental damage.
Proc Natl Acad Sci USA. 96:12102–12107. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pletnikov MV, Rubin SA, Schwartz GJ, Moran
TH, Sobotka TJ and Carbone KM: Persistent neonatal Borna disease
virus (BDV) infection of the brain causes chronic emotional
abnormalities in adult rats. Physiol Behav. 66:823–831. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu YJ, Schulz H, Lin CC, et al: Borna
disease virus-induced neuronal degeneration dependent on host
genetic background and prevented by soluble factors. Proc Natl Acad
Sci USA. 110:1899–1904. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ludwig H and Bode L: Borna disease virus:
new aspects on infection, disease, diagnosis and epidemiology. Rev
Sci Tech Off Int Epiz. 19:259–288. 2000.
|
|
22
|
Clark JD, Gebhart GF, Gonder JC, Keeling
ME and Kohn DF: Special report: the 1996 guide for the care and use
of laboratory animals. ILAR J. 38:41–48. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang R, Gao H, Zhang L, et al: Borna
disease virus infection perturbs energy metabolites and amino acids
in cultured human oligodendroglia cells. PLoS One. 7:e446652012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bode L, Dürrwald R, Rantam FA, Ferszt R
and Ludwig H: First isolates of infectious human Borna disease
virus from patients with mood disorders. Mol Psychiatry. 1:200–212.
1996.PubMed/NCBI
|
|
25
|
Lei Y, Li D, Deng J, et al: Metabolomic
profiling of three brain regions from a postnatal infected Borna
disease virus Hu-H1 rat model. Metabolomics. 10:484–495. 2013.
View Article : Google Scholar
|
|
26
|
Xie H, Zhu Y, Jiang W, et al:
Lactoferrin-conjugated superpara-magnetic iron oxide nanoparticles
as a specific MRI contrast agent for detection of brain glioma in
vivo. Biomaterials. 32:495–502. 2011. View Article : Google Scholar
|
|
27
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50.
2014.
|
|
28
|
Subramanian A, Tamayo P, Mootha VK, et al:
Gene set enrichment analysis: a knowledge-based approach for
interpreting genome-wide expression profiles. Proc Natl Acad Sci
USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pletnikov MV, Moran TH and Carbone KM:
Borna disease virus infection of the neonatal rat: developmental
brain injury model of autism spectrum disorders. Front Biosci.
7:d593–d607. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gosztonyi G: Natural and experimental
Borna disease virus infections-neuropathology and pathogenetic
considerations. APMIS. (Suppl 116): 53–57. 2008. View Article : Google Scholar
|
|
31
|
Fanselow MS and Dong HW: Are the dorsal
and ventral hippo-campus functionally distinct structures? Neuron.
65:7–19. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Davis TH, Cuellar TL, Koch SM, et al:
Conditional loss of Dicer disrupts cellular and tissue
morphogenesis in the cortex and hippocampus. J Neurosci.
28:4322–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Konopka W, Kiryk A, Novak M, et al:
MicroRNA loss enhances learning and memory in mice. J Neurosci.
30:14835–14842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hébert SS and De Strooper B: Alterations
of the microRNA network cause neurodegenerative disease. Trends
Neurosci. 32:199–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng X, Wang H, Ye S, et al: Up-regulation
of microRNA-126 may contribute to pathogenesis of ulcerative
colitis via regulating NF-kappaB inhibitor IkappaBalpha. PLoS One.
7:e527822012. View Article : Google Scholar
|
|
36
|
Planz O, Pleschka S and Wolff T: Borna
disease virus: a unique pathogen and its interaction with
intracellular signalling pathways. Cell Microbiol. 11:872–879.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hans A, Bajramovic JJ, Syan S, et al:
Persistent, noncytolytic infection of neurons by Borna disease
virus interferes with ERK 1/2 signaling and abrogates BDNF-induced
synaptogenesis. FASEB J. 18:863–865. 2004.PubMed/NCBI
|
|
38
|
Brabletz S and Brabletz T: The ZEB/miR.200
feedback loop-a motor of cellular plasticity in development and
cancer? EMBO Rep. 11:670–677. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jadhav SP, Kamath SP, Choolani M, Lu J and
Dheen ST: microRNA-200b modulates microglia-mediated
neuroinflam-mation via the cJun/MAPK pathway. J Neurochem.
130:388–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Juhila J, Sipilä T, Icay K, et al:
MicroRNA expression profiling reveals miRNA families regulating
specific biological pathways in mouse frontal cortex and
hippocampus. PloS one. 6:e214952011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dheen ST, Kaur C and Ling EA: Microglial
activation and its implications in the brain diseases. Curr Med
Chem. 14:1189–1197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sabapathy K, Hu Y, Kallunki T, et al: JNK2
is required for efficient T-cell activation and apoptosis but not
for normal lymphocyte development. Curr Biol. 9:116–125. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Graves LM, Guy HI, Kozlowski P, et al:
Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature.
403:328–332. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang SH, Sharrocks AD and Whitmarsh AJ:
Transcriptional regulation by the MAP kinase signaling cascades.
Gene. 320:3–21. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ciesielski-Treska J, Grant NJ, Ulrich G,
et al: Fibrillar prion peptide (106–126) and scrapie prion protein
hamper phagocytosis in microglia. Glia. 46:101–115. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park JY, Paik SR, Jou I and Park SM:
Microglial phagocytosis is enhanced by monomeric α.synuclein, not
aggregated α synuclein: Implications for Parkinson's disease. Glia.
56:1215–1223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mizuno T: The biphasic role of microglia
in Alzheimer's disease. Int J Alzheimer's Dis. 2012:2012.
|
|
49
|
Frödin M and Gammeltoft S: Role and
regulation of 90 kDa ribosomal S6 kinase (RSK) in signal
transduction. Mol Cell Endocrinol. 151:65–77. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu X, Yang Y, Zhao M, et al: Proteomics
reveal energy metabolism and mitogen-activated protein kinase
signal transduction perturbation in human Borna disease virus
Hu-H1-infected oligodendroglial cells. Neuroscience. 268:284–296.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hu J, Fang Y, Cao Y, Qin R and Chen Q:
miR-449a Regulates proliferation and chemosensitivity to cisplatin
by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci.
59:336–345. 2014. View Article : Google Scholar
|
|
52
|
Li D, Lei Y, Deng J, et al: Human but not
laboratory borna disease virus inhibits proliferation and induces
apoptosis in human oligodendrocytes in vitro. PLoS One.
8:e666232013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang EB, Kong R, Yin DD, et al: Long
noncoding RNA ANRIL indicates a poor prognosis of gastric cancer
and promotes tumor growth by epigenetically silencing of
miR-99a/miR-449a. Oncotarget. 5:2276–2292. 2014.PubMed/NCBI
|
|
54
|
Sherr CJ and McCormick F: The RB and p53
pathways in cancer. Cancer Cell. 2:103–112. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yu BQ, Su LP, Li JF, et al: microrna
expression signature of gastric cancer cells relative to normal
gastric mucosa. Mol Med Rep. 6:821–826. 2012.PubMed/NCBI
|
|
56
|
Strickland ER, Hook MA, Balaraman S, Huie
JR, Grau JW and Miranda RC: MicroRNA dysregulation following spinal
cord contusion: implications for neural plasticity and repair.
Neuroscience. 186:146–160. 2011. View Article : Google Scholar : PubMed/NCBI
|