Introduction
Upper tract urothelial carcinoma (UTUC) is an
uncommon, clinically heterogeneous and potentially fatal disease.
UTUC accounts for only 5–10% of urothelial carcinomas (1,2) and
60% of UTUCs are invasive at diagnosis compared with only 15% for
bladder tumors (1,3). The established early diagnostic
methods for UTUC, including multi detector computed tomographic
urography, magnetic resonance imaging, urinary cytology, cystoscopy
and ureteroscopy, are either invasive or lack sensitivity (3). Thus, the majority of UTUC cases are
diagnosed at an advanced stage, resulting in poor prognoses.
Results from studies on bladder cancer, which is more common and
more thoroughly studied, are often extrapolated to UTUC (4); however, molecular markers for the
detection of UTUC are largely absent. Individuals with low risk
UTUC can partially benefit from conservative therapy (3,5), but
radical nephroureterectomy is still considered the gold standard
for the treatment of localized UTUC (3,6).
Therefore, correct early diagnosis is critical for designing
appropriate treatment schedules to increase survival and decrease
morbidity. Early biomarkers that can complement and improve current
strategies for UTUC detection should be identified.
MicroRNAs (miRNAs/miRs) are a class of 17–27
nucleotide (nt) single stranded RNA molecules that can regulate
gene expression by repressing translation or cleaving RNA
transcripts in a sequence specific manner. The biogenesis of miRNA
has been comprehensively reported in numerous reviews (7,8), and
the ability of miRNAs to inhibit the translation of oncogenes and
tumor suppressors suggests their involvement in carcinogenesis
(9,10). miRNAs present in human blood
samples are known as circulating miRNAs. Circulating miRNAs have
been recently demonstrated to be present in a stable and
reproducible state (11). Serum
and plasma samples are easily acquired in a relatively non invasive
manner, and isolated miRNAs can be readily detected by reverse
transcription quantitative polymerase chain reaction (RT qPCR),
which is a widely used clinical and laboratory technique. The
unique expression profile of serum or plasma miRNA has been used as
a fingerprint for various malignancies, including carcinomas of the
urinary tract (12,13). Extracellular miRNAs, either
embedded in microvesicles or protected by RNA binding
proteins/lipids, can provide diagnostic, prognostic or even
therapeutic targets for UTUC.
However, in spite of the aberrant expression of
miRNAs being implicated in the clinical prognosis of human UTUC
(14), the global circulating
miRNA patterns in UTUC patients have not been studied, to the best
of our knowledge. Circulating miRNAs are promising candidates for
non invasive cancer testing due to the stability and tissue
specificity of miRNAs in the blood. Circulating miRNAs have been
recently reported as diagnostic markers for various cancer types
(15–18). The present study used the deep
sequencing platform Illumina HiSeq™ 2000, followed by RT-qPCR, to
systematically and extensively identify circulating miRNAs as a
novel class of potent non invasive biomarkers for UTUC.
Materials and methods
Ethics statement
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Nanjing Medical
University (Nanjing, China). Written informed consent was obtained
from all participants involved in this study.
Study population, sample collection and
serum RNA isolation
The present study collected serum samples from 58
UTUC patients (mean age, 68.0 ± 11.6 years) undergoing radical
nephroureterectomy at the First Affiliated Hospital of Nanjing
Medical University (Nanjing, China), from 2007 to 2013. A total of
42 matched cancer free controls (mean age, 67.9 ± 9.0 years) were
also included. These controls were patients with hematuria who were
further diagnosed with urinary tract infection, benign prostate
hyperplasia and urinary tract calculi at the First Affiliated
Hospital of Nanjing Medical University (Nanjing, China). Clinical
and pathological charac teristics, including age, gender, family
history of cancer, stage and grade, were recorded and are
summarized in Table I. The
diagnosis of UTUC was established according to the results of
histopathological analysis. Tumors were graded and classified
according to the WHO (2004) and TNM classifications of the
International Union Against Cancer (2009) (19,20).
Blood samples of all patients were drawn within 24 h after
admission. The coagulated blood samples were collected in tubes
containing a separating gel and clot activator and centri fuged at
1,500 × g for 15 min at 4°C. The supernatant was centrifuged at
1,500 × g for 15 min to precipitate cell debris and stored at −80°C
until use. Total RNA of 12 UTUC patients and 12 controls was
isolated from 200 µl serum samples using an miRNeasy
Serum/Plasma kit (Qiagen, Hilden, Germany) according to the
manufacturer's instructions.
 | Table IDemographic and clinical features of
patients with upper tract urothelial carcinoma and controls in the
training and validation sets. |
Table I
Demographic and clinical features of
patients with upper tract urothelial carcinoma and controls in the
training and validation sets.
| Variable | Cases (n=58)
| Controls (n=42)
| P valuea |
|---|
| n | % | n | % |
|---|
| Age (years) | | | | | |
| <68 | 28 | 48.3 | 22 | 52.4 | 0.685 |
| ≥68 | 30 | 51.7 | 20 | 47.6 | |
| Gender | | | | | |
| Male | 41 | 29.3 | 30 | 71.4 | 0.936 |
| Female | 17 | 70.7 | 12 | 28.6 | |
| Tobacco smoking
status | | | | | |
| Never | 32 | 55.2 | 26 | 61.9 | 0.501 |
| Positiveb | 26 | 44.8 | 16 | 38.1 | |
| Alcohol consumption
status | | | | | |
| Never | 40 | 82.8 | 31 | 73.8 | 0.278 |
| Positiveb | 18 | 17.2 | 11 | 26.2 | |
| Family history of
cancer | | | | | |
| No | 42 | 72.4 | 37 | 88.1 | 0.057 |
| Yes | 16 | 27.6 | 5 | 11.9 | |
| Tumor stage | | | | | |
| I | 21 | 36.2 | | | |
| II | 14 | 24.1 | | | |
| III | 20 | 34.5 | | | |
| IV | 3 | 5.2 | | | |
| Tumor grade | | | | | |
| PUNLMP | 1 | 1.7 | | | |
| Low grade | 17 | 29.3 | | | |
| High grade | 40 | 69.0 | | | |
HiSeq 2000 sequencing and bioinformatics
analysis
Small RNAs of 16-30 nt in length were first isolated
from the total RNA by size fractionation in a 15% Tris/borate/EDTA
urea polyacrylamide gel, and these small RNAs were ligated to an
activated 5′ adaptor. A 3′ adaptor was then ligated to the small
RNA 5′ adaptor, and RT using a RT primer was performed to create
cDNA constructs. The RT reaction was performed using primers that
were complementary to the two adaptors. The amplified cDNA
constructs were purified and sequenced using an Illumina HiSeq 2000
platform (Illumina, Inc., San Diego, CA, USA) with the following
parameters: The adaptor molecule and empty sequence were directly
trimmed, and low-quality tags were filtered. Only reads 18–32
bp-reads were selected. All sequences that matched transfer RNA,
ribosomal RNA and DNA repeats were filtered. Thus, two types of
alignment were generated: Reads uniquely mapped to known miRNAs and
reads generating other unknown small RNAs. The expression of miRNAs
was calculated, and DEGseq was used to differentially screen the
miRNAs between the cases and controls by the criterion of
P<0.001. All of the abovementioned procedures were performed by
Shanghai Majorbio Bio-pharm Technology (Shanghai, China).
miRNA quantification by RT-qPCR
A two-phase case-control study was designed to
identify serum miRNAs as potential markers for UTUC. In the
training set, miRNAs were measured in serum samples from 12 UTUC
patients and 12 controls. In the validation set, miRNAs were
measured in serum samples from 46 UTUC patients and 30 controls.
miRNAs were prepared using an All-in-One™ miRNA qRT-PCR Detection
kit (cat. no. AOMD-Q060; GeneCopoeia-FulenGen, Guangzhou, China)
according to the manufacturer's instructions. Extracted RNA was
reverse-transcribed in the presence of poly-A polymerase with an
oligo-dT adaptor. A SYBR green qPCR assay was performed with an
Applied Biosystems Step One Plus System (Applied Biosystems, Foster
City, USA) with forward primer for the mature miRNA sequence and a
universal adaptor reverse primer. The respective validated
miRNA-specific forward primer (GeneCopoeia-FulenGen) was used for
the qPCR assay. Amplification was performed under the following
conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15
sec, 60°C for 30 sec and 72°C for 40 sec. The expression levels of
miRNA were normalized to RNU6-2 as described previously
(21). All reactions, including
controls with no template RNA, were performed in triplicate. The
relative expression of miRNA compared with RNU6-2 was
calculated using the 2−ΔCt method.
Statistical analysis
Student's t-test or the χ2 test
were used to determine the differences in the selected demographic
variables between the cases and controls. The relative expres sion
of miRNA was analyzed using the 2−ΔΔCt method. The Mann
Whitney U test was used to compare the expression of serum miRNAs
between the different groups. R software (version 3.0.1; MathSoft,
Seattle, WA, USA) was used to run unsupervised clustering analysis
and principal component analysis (PCA). Sensitivity, specificity
and area under the curve (AUC) for serum miRNA levels were
determined using receiver operator characteristic (ROC) analysis.
All tests were two sided, and P<0.05 (95% CI) was considered to
indicate a statistically significant difference. Statistical
analysis was performed using SPSS 13.0 software (SPSS, Inc.,
Chicago, IL, USA).
Results
Characteristics of the study
population
The frequency distributions of the selected
characteristics of the cases and controls are shown in Table I. No significant differences in
age, gender, tobacco smoking and alcohol consumption were
identified between the UTUC patients and controls (P>0.05). The
family history of malignancies was similar between the cases and
controls. The clinicopathological characteristics are presented in
Table I.
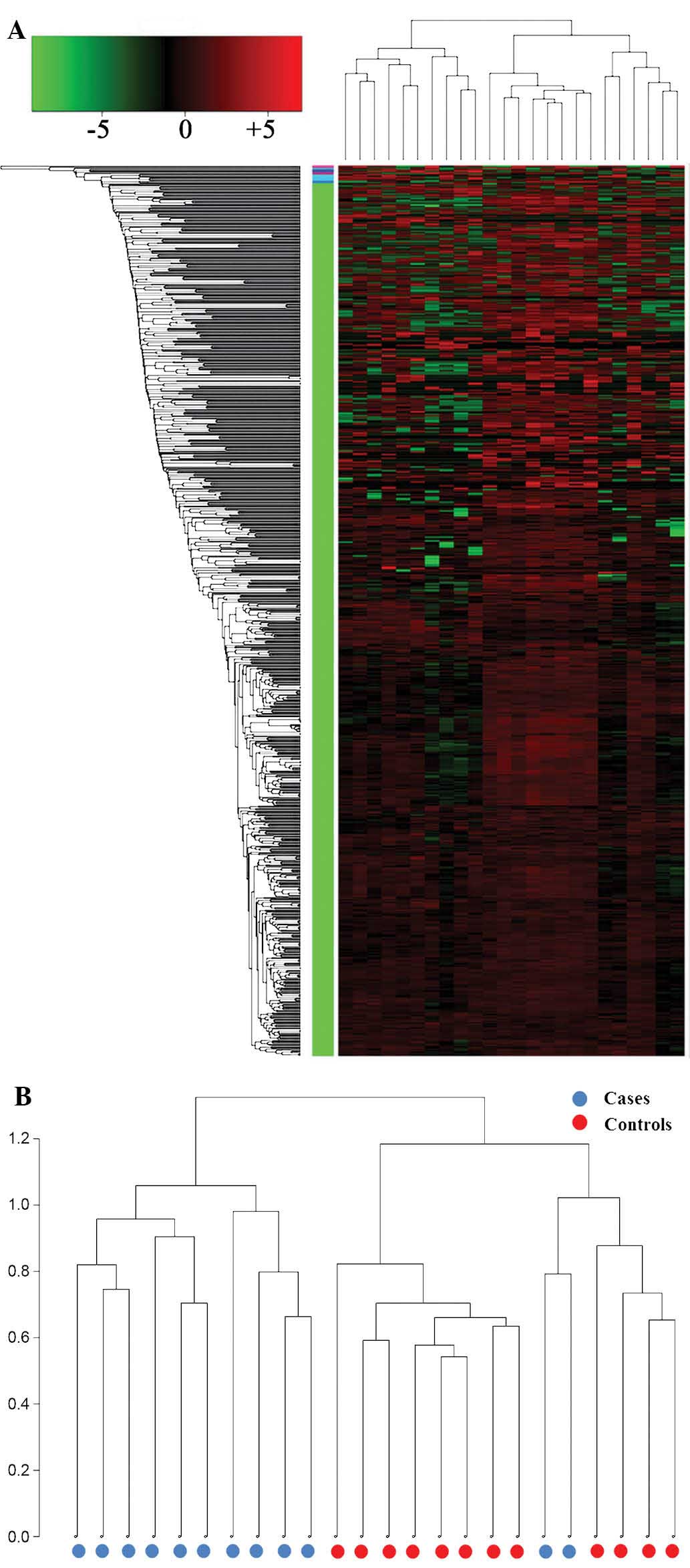
Unsupervised clustering analysis and
PCA
The differential expression of miRNAs between the
UTUC and control serum samples was analyzed by performing
unsupervised clustering that was blinded to the clinical
annotations. The dendrogram generated by cluster analysis showed a
clear distinction between the UTUC and control samples based on the
serum miRNA profile (Fig. 1).
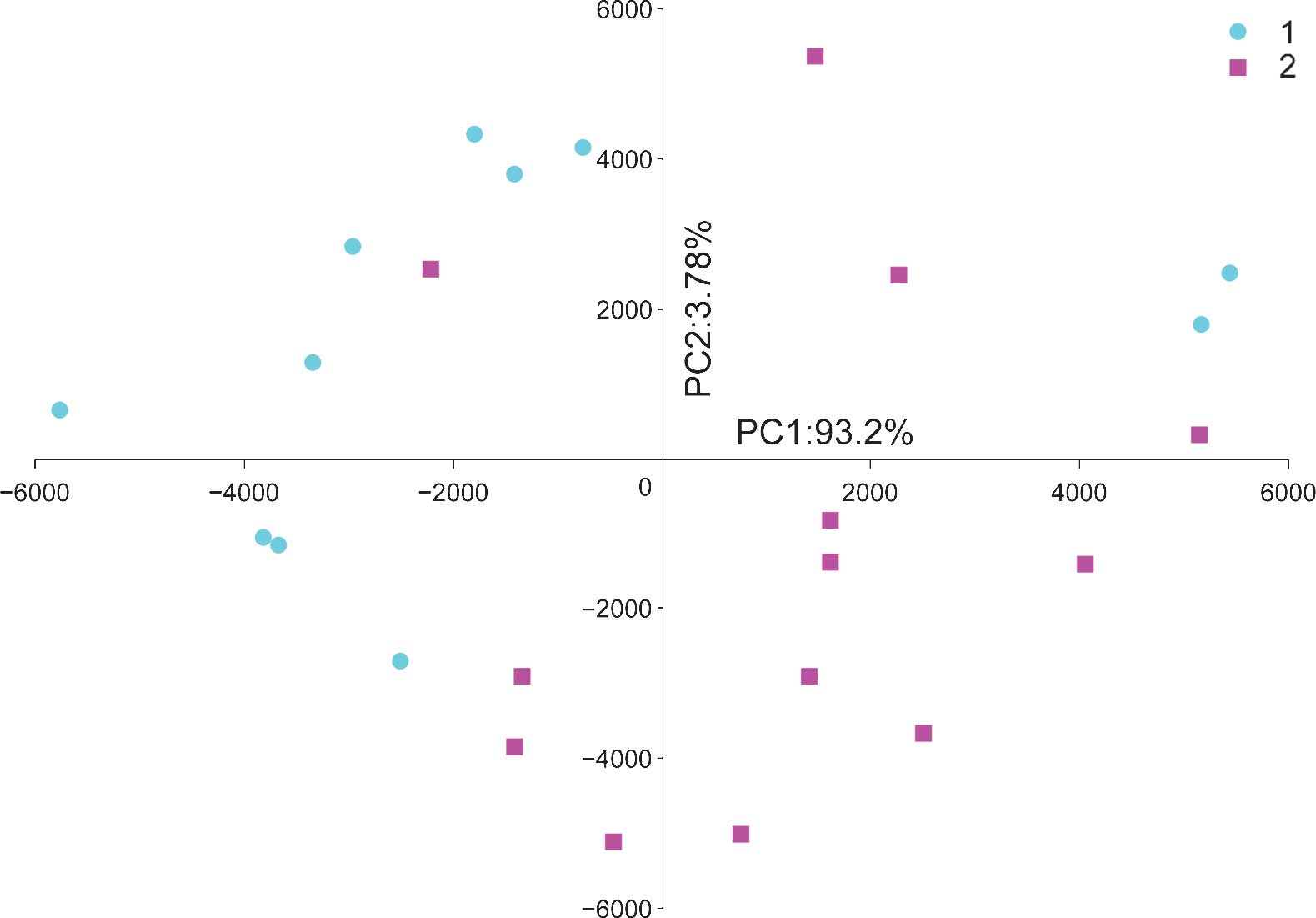
PCA was used to classify 24 samples (12 UTUC and 12
control samples) based on the expression profile of 711 miRNAs
expressed in all samples. The PCA plot illustrates the global
characteristics of these samples (Fig.
2). Despite heterogeneity within the two groups, the first
principal component (PC1) was able to differentiate the UTUC
samples from the control samples, and the difference between the
two groups was >95%.
RT-qPCR analysis of serum miRNAs in
UTUC
To validate the putative markers identified from the
profiling results, the present study confirmed the concentrations
of 21 candidate miRNAs, which were selected following HiSeq 2000
sequencing, with a SYBR green-based RT-qPCR assay. The criteria for
selecting miRNA for RT-qPCR analysis were as follows: i) Counts of
miRNAs >100 in all sequenced samples; ii) fold change >2 or
<0.5 and P<0.001; iii) the absolute value of the coefficient
in PC1 >500; and iv) miRNAs sub-categorized by the unsupervised
clustering method. Consequently, 21 miRNAs that met the inclusion
criteria were selected for further analysis. The results are
summarized in Table II.
 | Table IIDifferentially expressed miRNAs
between serum of patients with upper tract urothelial carcinoma and
controls. |
Table II
Differentially expressed miRNAs
between serum of patients with upper tract urothelial carcinoma and
controls.
| miRNA name | Lowest count in all
samples | Fold change (cases
vs. controls) | P value | Predicted by |
|---|
| has-let-7a-5p | 88495 | 2.08 | <0.001 | PCA |
| has-let-7b-5p | 13358 | 3.18 | <0.001 | PCA |
| has-let-7c | 65799 | 2.44 | <0.001 | PCA |
| has-let-7f-5p | 85201 | 2.05 | <0.001 | PCA |
| has-miR-16-5p | 477766 | 2.43 | <0.001 | PCA |
| has-miR-183-5p | 7393 | 4.24 | <0.001 | PCA |
| has-miR-191-5p | 53946 | 2.57 | <0.001 | PCA |
| has-miR-22-3p | 64832 | 0.46 | <0.001 | PCA |
| has-miR-25-3p | 86758 | 2.78 | <0.001 | PCA |
| hsa miR 26a 5p | 14980 | 2.33 | <0.001 | PCA |
| has-miR-33b-3p | 6636 | 3.98 | <0.001 | CA |
| has-miR-3609 | 127 | 12.68 | <0.001 | CA |
| has-miR-370 | 106 | 0.39 | <0.001 | CA |
| has-miR-423-5p | 9077 | 2.48 | <0.001 | PCA |
| has-miR-431-5p | 169 | 0.23 | <0.001 | CA |
| hsa-miR-451a | 183231 | 2.18 | <0.001 | PCA |
|
hsa-miR-4763-5p | 122 | 10.81 | <0.001 | CA |
| hsa-miR-486-5p | 1537466 | 2.99 | <0.001 | PCA |
| hsa-miR-655 | 148 | 5.18 | <0.001 | CA |
|
hsa-miR-664a-3p | 24258 | 8.12 | <0.001 | CA |
| hsa-miR-92a-3p | 161639 | 0.46 | <0.001 | PCA |
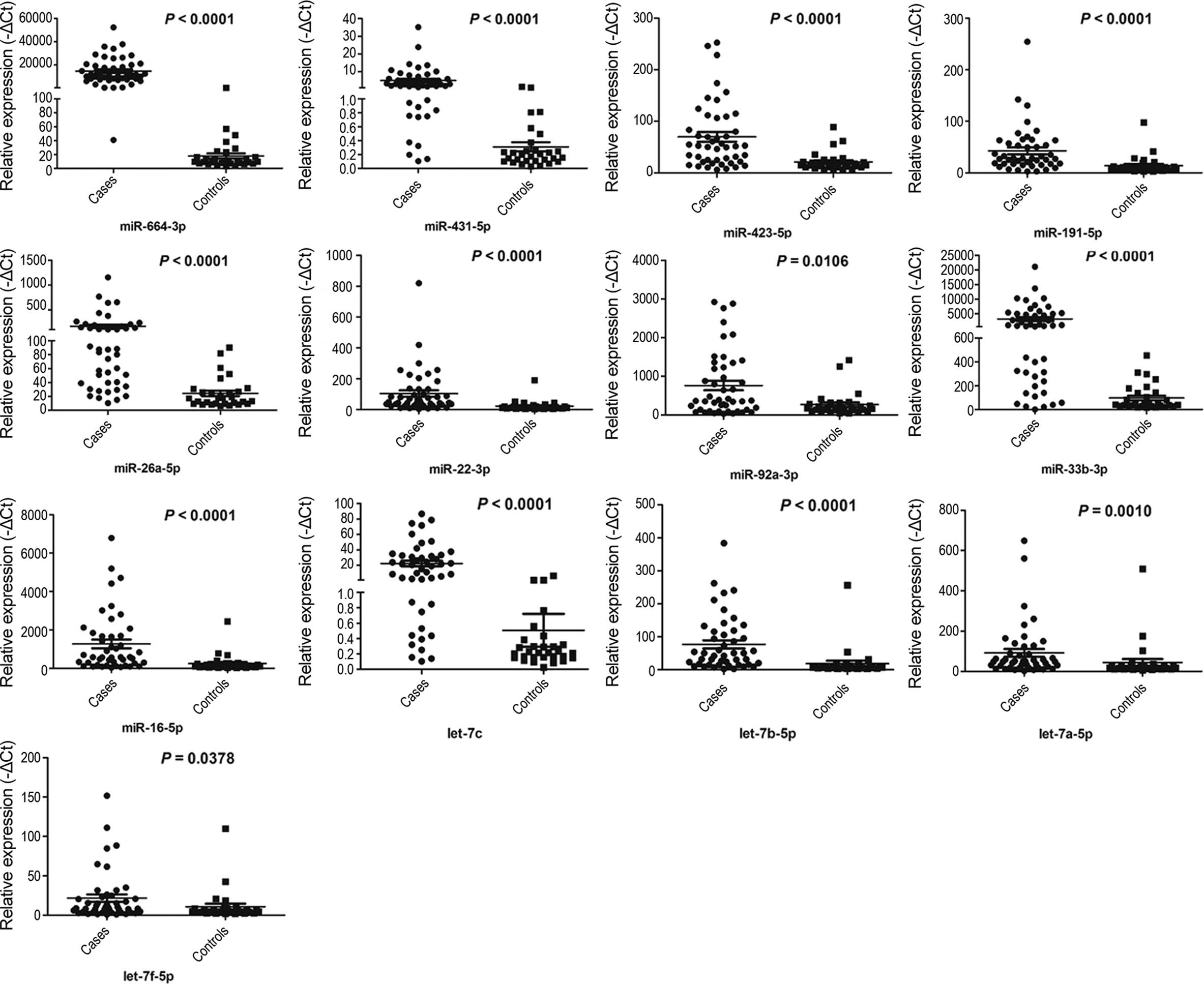
Within the validation set, the miRNAs of individual
serum samples from 46 UTUC patients and 30 controls were measured,
and only miRNAs with a Ct value <35 in either the UTUC or
control groups were selected for further analysis. Furthermore,
miRNAs with a detection rate of <80% and P>0.05 were excluded
from further analysis. These criteria were used to generate a list
of 13 miRNAs (miR-664a-3p, miR-423-5p, miR-431-5p, miR-191-5p,
miR-92a-3p, miR-22-3p, miR-26a-5p, miR-33b-3p, miR-16-5p,
let-7a-5p, let-7b-5p, let-7f-5p and let-7c) that showed different
miRNA patterns between the UTUC patients and controls (Fig. 3).
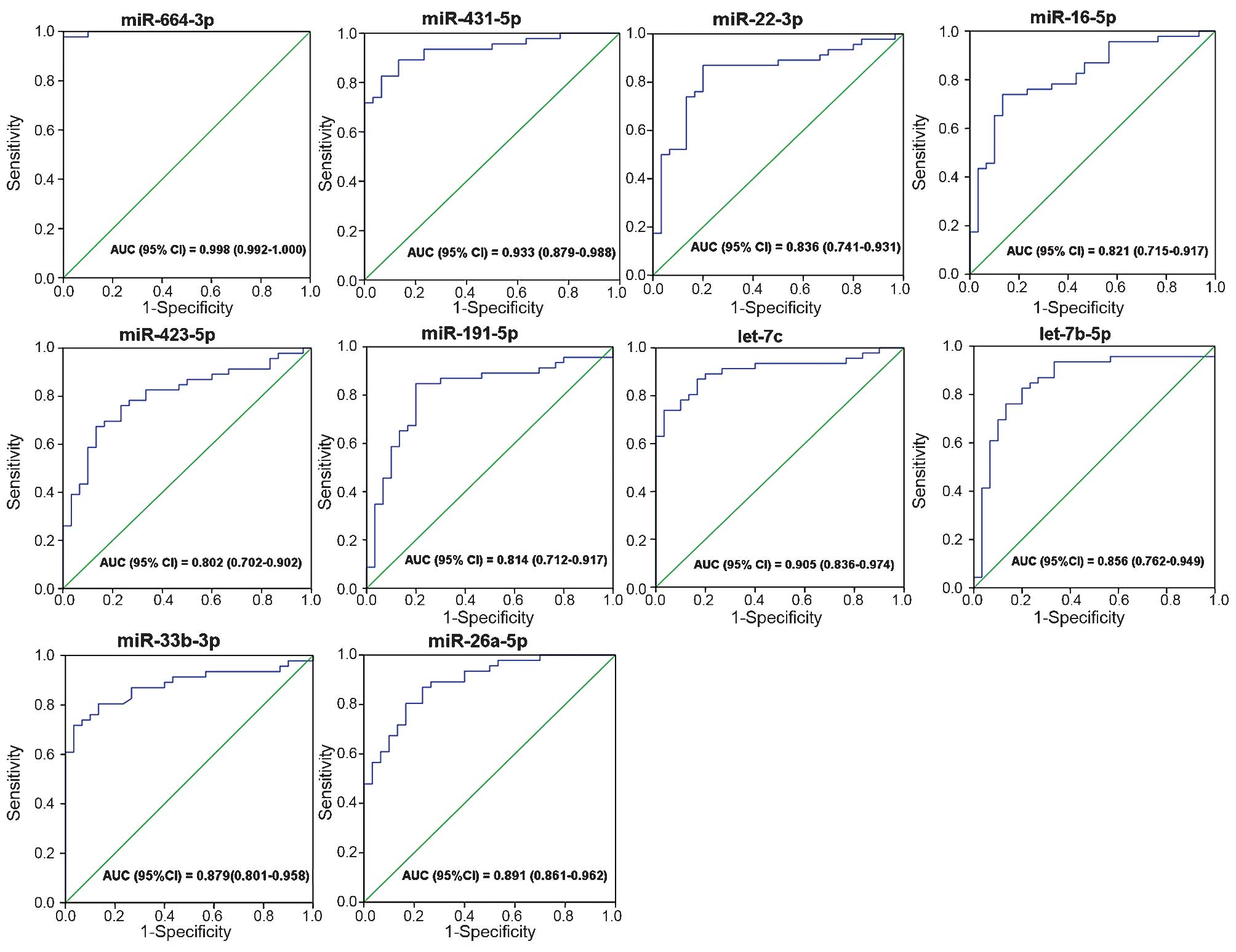
ROC curve analysis
The ROC curves that were constructed to compare the
relative concentrations of the 13 miRNAs between the UTUC patients
and controls yielded the following AUCs: miR-664a-3p, 0.998, 95% CI
(0.992–1.000); miR-423-5p, 0.802, 95% CI (0.702–0.902); miR-431-5p,
0.933, 95% CI (0.879–0.988); miR-191-5p, 0.814, 95% CI
(0.712–0.917); miR-92a-3p, 0.675, 95% CI (0.552–0.797); miR-22-3p,
0.836, 95% CI (0.741–0.931); miR-26a-5p, 0.891, 95% CI
(0.861–0.962); miR-33b-3p, 0.879, 95% CI (0.801–0.958); miR-16-5p,
0.821, 95% CI (0.715–0.917); let-7a-5p, 0.725, 95% CI
(0.605–0.845); let-7b-5p, 0.856, 95% CI (0.762–0.949); let-7f-5p,
0.642, 95% CI (0.516–0.768); and let-7c, 0.905, 95% CI
(0.836–0.974). These results suggested the potential of miRNAs for
discriminating patients with UTUC from control subjects (Fig. 4).
Association with clinical
characteristics
To demonstrate the increase in serum miRNA
expression during UTUC development, UTUC cases were further
stratified according to tumor stage and grade. The patients were
divided into two groups (muscle invasive and non muscle invasive)
according to pathological stage, and three groups according to
pathological grade. No statistically significant differences were
observed among the miRNAs; miR-664a-3p, miR-431-5p, miR-423-5p,
miR-191-5p, miR-92a-3p, miR-16-5p and let-7b-5p were all
upregulated in the muscle invasive group. Similarly, no statis
tically significant difference was identified when members of the
UTUC group were stratified according to tumor grade (data not
shown).
Discussion
The present study evaluated the use of circulating
miRNA levels as diagnostic and prognostic factors for UTUC in order
to improve currently used diagnostic tools. The results showed that
UTUC was associated with a significant increase in the serum levels
of 13 circulating miRNAs, namely miR-664a-3p, miR-423-5p,
miR-431-5p, miR-191-5p, miR-92a-3p, miR-22-3p, miR-26a-5p,
miR-33b-3p, miR-16-5p, let-7a-5p, let-7b-5p, let-7f-5p and let-7c.
Further ROC curve analysis showed that ten of the 13 miRNAs
(miR-664a-3p, miR-431-5p, miR-423-5p, miR-191-5p, miR-33b-3p,
miR-26a-5p, miR-22-3p, miR-16-5p, let-7b-5p and let-7c) were
potentially suitable for distinguishing UTUC individuals from the
controls (AUC >0.8).
Circulating miRNAs are present in body fluids and
dysregulated miRNAs may be used as disease markers due to their
small size, stability and pivotal involvement in the regulation of
cell function (22). Tumor derived
miRNAs in serum or plasma are easily accessible and the detection
technology is widely used in clinical settings. Therefore,
circulating miRNAs may emerge as potentially useful blood based
tools for monitoring human cancers, particularly at an early stage
(23). In the present study, the
deep sequencing platform Illumina HiSeq™ 2000 was used to
preliminarily characterize an index for estimating the abundance of
circulating miRNAs from 12 UTUC patients and 12 controls as an
initial screening stage, allowing for the determination of the
differential expression of 21 miRNAs between UTUC patients and
controls. Deep sequencing approaches for miRNA analysis are
becoming routine, and the number of studies targeting miRNAs as
potential diagnostic biomarkers for diseases is increasing
(24). The major benefits of
next-generation sequencing for miRNA profiling include detection of
novel as well as known miRNAs and precise identification of miRNA
sequences (25). Deep sequencing
technology is an effective high throughput tool for initial serum
miRNA screening with high accuracy and sensitivity. However, the
sequencing results should be validated by RT-qPCR using a large
number of individual serum samples, because the sequencing results
were obtained from pooled serum samples and may include inaccurate
information caused by individual variations. After the initial
stage of sequencing circulating miRNA based cancer biomarkers, the
present study used RT-qPCR to assess the relative miRNA expression.
Subsequently, a list of 13 miRNAs with differential expression
between the UTUC cases (n=46) and controls (n=30) was generated.
The present study retrospectively assessed the diagnostic capacity
of the 13 miRNAs and identified 10 miRNAs that exhibited
significant differences (AUC >80%) between the UTUC cases and
the controls, therefore being suitable for distinguishing between
the two groups. Circulating miRNAs identified by deep sequencing
technology were recently considered as fingerprints for the
diagnosis of other types of solid tumor, including gastric cancer
(15), papillary thyroid carcinoma
(16), hepatocarcinoma (17) and malignant astrocytomas (18). Furthermore, miRNAs isolated from
urine or blood in bladder cancer patients were also reported to
show potential as non invasive biomarkers for diagnosis and
prognosis (26–28).
Among the 10 serum miRNAs identified in the present
study, several have already been reported to function in detecting
disease, creating prognosis or monitoring treatment response. The
expression of circulating miRNA precursor let-7b was reported to be
diverse among various cancer types. Let-7b was found to be
upregulated in breast tumors (29)
and downregulated in ovarian cancer (30). The same trend was found for the
expression of circulating let-7c, which was increased in esophageal
cancer (31) but decreased in
nasopharyngeal carcinoma patients with poor prognosis (32). Circulating let-7b and let-7c
expression is cancer specific. Let-7b is highly expressed in
platelets (33), which may obscure
the diagnostic value of measuring let-7b expression levels in
serum. Circulating miR-22 was first reported in esophageal squamous
cell carcinoma (ESCC) and serves as a non invasive biomarker for
ESCC diagnosis (34). High
expression of miR-22 can explain the lack of response in
pemetrexed-treated non small cell lung cancer (NSCLC) patients.
However, in primary plasma cell leukemia, miR-22 is associated with
a positive clinical outcome (35).
One study suggested that circulating miR-431 can contribute to the
detection of colorectal cancer (36). The present study found that serum
let-7b, miR-22, and miR-431 were suitable for distinguishing UTUC
cases from the controls with AUCs of 85.6, 83.6 and 93.3%,
respectively. As mentioned above, circulating let-7b, miR-22 and
miR-431 have been linked to the pathogenesis of other cancer types,
indicating the value of these three miRNAs in detecting cancer,
including UTUC.
Circulating miR-16 and miR-191 have been reported as
reference genes in colorectal adenocarcinoma (37). However, miR-16 and miR-191 are
associated with the pathogenesis of other cancer types, including
NSCLC (38) and lymphoma (39). Of note, several serum miRNAs
identified in the present study are also correlated with urinary
cancers. For instance, serum miR-16 and miR-26a allow for the
discrimination of prostate cancer and benign prostate hyperplasia
patients with PSA >2.5 ng/ml (AUCs of 83.7 and 91.8%,
respectively) (40). Serum miR-191
expression was found to be 5.3 fold higher in renal cell carcinoma
patients than that in controls (41). Plasma miR-33b was significantly
lower in bladder cancer cases (28) than that in the controls, whereas
the serum expression of miR-33b was enhanced in the UTUC cases in
the present study. This disparity should be cautiously interpreted
according to the possibly different cellular and molecular
mechanisms underlying UTUC and lower tracturothelial carcinoma,
even if the pathogenetic similarity of UTUC and bladder cancer has
been previously reported (4,42).
These miRNAs may serve as a 'fingerprint' to identify the presence
of urinary cancers. Thus, the diagnostic capacity of these four
circulating miRNAs (miR-16, miR-26a, miR-33b and miR-191) in the
precise detection of UTUC may be impaired by their involvement in
other urinary cancers. Further studies should be performed to
confirm the functions of these four circulating miRNAs in urinary
cancers.
In the present study, UTUC was found to be
associated with serum miR-423 and miR-664. Circulating miR-423 may
be useful as a stable blood based biomarker for numerous diseases,
including heart failure (43) and
acute graft vs host disease (44).
However, few studies have demonstrated the implication of serum miR
423 in cancers. Previous studies have shown that miR-423 is
increased in head and neck squamous cell carcinomas (45), and is highly expressed in
infiltrating ductal carcinomas in women, who later developed
metastasis (46). These findings
suggested that miR-423 has a function in cancer development. To the
best of our knowledge, no previous study has revealed the
association between circulating miR-664 and disease prediction.
However, the upregulation of miR-664 contributed to enhanced
tumorigenesis in a hepatocellular carcinoma cell line (47), suggesting its potential oncogenic
behavior. The lack of results showing the association between
cancer and circulating miR-664 and miR-423 levels indicates marked
cancer specificity of these two miRNAs in UTUC diagnosis. Of note,
the high diagnostic capacity (AUC=99.8%) of serum miR-664 in
detecting UTUC may be applied in future studies.
Other molecular markers have also been investigated
in samples from UTUC patients using immunohistochemistry or in
situ hybridization. E cadherin and hypoxia inducible factor 1α
are prognostic factors in UTUC, and the expression of human
telomerase RNA is a useful indicator for the diagnosis and
prognosis of UTUC (48–50). However, these molecular markers are
more associated with the prognosis rather than the diagnosis of
UTUC, and depend on conventional detection methods. Compared with
circulating miRNA detection, the conventional diagnostic methods
for UTUC, including ureteros copy, urine cytology, excretory
urography and ultrasonography, cause discomfort among patients and
are time consuming and invasive. Such methods also lack sensitivity
and specificity, which may delay diagnosis and treatment. By
contrast, the circulating miRNAs identified in the present study
could allow for early diagnosis with high sensitivity and
specificity.
However, the results of the present study need to be
interpreted with caution, as the diagnosis as UTUC has several
limitations. First, as UTUC is an uncommon disease, the sample size
was relatively small, which may have limited the diagnostic
capacity of serum miRNA. Second, the miRNA expression levels were
higher in the case group than those in the control group, even if
the initial sequencing stage showed that the levels of several
miRNAs were downregulated in the case group. The discrepancy
between the results of the two stages may be attributed to the
relatively low abundance of reduced miRNAs, which was beyond the
threshold of RT-qPCR detection. Third, the results of the present
study were based on a retrospective study. Thus, a future
prospective cohort is required to validate the results of the
present study. Finally, other factors, including race and
methodology, possibly affected the final results.
In conclusion, a 10 serum miRNA-based expression
profile that was able to accurately discern UTUC individuals from
the controls was identified. Although the manner by which UTUC
specific miRNAs enter the circulation is unknown, the identified
serum miRNAs are useful biomarkers for UTUC detection. Such serum
based assays may result in improved screening compared with stool
based or endoscopic screening methods. Circular miRNA based
screening for UTUC must be tested on large cohorts to confirm and
increase their sensitivity and specificity.
Acknowledgments
This work was supported by the Program for
Development of Innovative Research Team of the First Affiliated
Hospital of Nanjing Medical University, the Provincial Initiative
Program for Excellency Disciplines of Jiangsu Province, the
National Natural Science Foundation of China (grant nos. 81272832
and 81201997), the Natural Science Foundation of Jiangsu Province
(grant no. BK2011848), the Six Major Talent Peak Project of Jiangsu
Province (grant no. 2011-WS-121), the Priority Academic Program
Development of Jiangsu Higher Education Institutions (PAPD) and
Jiangsu Provincial Special Program of Medical Science (grant no.
BL2012027). The funders had no role in study design, data
collection and analysis, decision to publish, or preparation of the
manuscript.
References
|
1
|
Hall MC, Womack S, Sagalowsky AI, Carmody
T, Erickstad MD and Roehrborn CG: Prognostic factors, recurrence
and survival in transitional cell carcinoma of the upper urinary
tract: A 30-year experience in 252 patients. Urology. 52:594–601.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Munoz JJ and Ellison LM: Upper tract
urothelial neoplasms: Incidence and survival during the last 2
decades. J Urol. 164:1523–1525. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rouprêt M, Zigeuner R, Palou J, Boehle A,
Kaasinen E, Sylvester R, Babjuk M and Oosterlinck W: European
guidelines for the diagnosis and management of upper urinary tract
urothelial cell carcinomas: 2011 update. Eur Urol. 59:584–594.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krabbe LM, Lotan Y, Bagrodia A, Gayed BA,
Darwish OM, Youssef RF, Bolenz C, Sagalowsky AI, Raj GV, Shariat
SF, et al: Prospective comparison of molecular signatures in
urothelial cancer of the bladder and the upper urinary tract is
there evidence for discordant biology? J Urol. 191:926–931. 2014.
View Article : Google Scholar
|
|
5
|
Hautmann RE, de Petriconi RC, Pfeiffer C
and Volkmer BG: Radical cystectomy for urothelial carcinoma of the
bladder without neoadjuvant or adjuvant therapy: Long term results
in 1100 patients. Eur Urol. 61:1039–1047. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Margulis V, Shariat SF, Matin SF, Kamat
AM, Zigeuner R, Kikuchi E, Lotan Y, Weizer A and Raman JD; The
Upper Tract Urothelial Carcinoma Collaboration: Outcomes of radical
nephroureterectomy: A series from the upper tract urothelial
carcinoma collaboration. Cancer. 115:1224–1233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Visone R and Croce CM: MiRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregu lation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar
|
|
12
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang X, Liang M, Dittmar R and Wang L:
Extracellular microRNAs in urologic malignancies: Chances and
challenges. Int J Mol Sci. 14:14785–14799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Izquierdo L, Ingelmo Torres M, Mallofré C,
Lozano JJ, Verhasselt Crinquette M, Leroy X, Colin P, Comperat E,
Roupret M, Alcaraz A and Mengual L: Prognostic value of microRNA
expression pattern in upper tract urothelial carcinoma. BJU Int.
113:813–821. 2014. View Article : Google Scholar
|
|
15
|
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang
C, Huang D, Chen X, Zhang H, Zhuang R, et al: A five-microRNA
signature identified from genome-wide serum microRNA expression
profiling serves as a fingerprint for gastric cancer diagnosis. Eur
J Cancer. 47:784–791. 2011. View Article : Google Scholar
|
|
16
|
Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F,
Zhang Y, Huang K, Li Y, Song E, et al: Circulating microRNA
profiles as potential biomarkers for diagnosis of papillary thyroid
carcinoma. J Clin Endocrinol Metab. 97:2084–2092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY,
Zhang JF, Shen HB, Zhang CY and Zen K: Serum microRNA profiles
serve as novel biomarkers for HBV infection and diagnosis of HBV
positive hepatocarcinoma. Cancer Res. 70:9798–9807. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang C, Wang C, Chen X, Chen S, Zhang Y,
Zhi F, Wang J, Li L, Zhou X, Li N, et al: Identification of seven
serum microRNAs from a genome-wide serum microRNA expression
profile as potential noninvasive biomarkers for malignant
astrocytomas. Int J Cancer. 132:116–127. 2013. View Article : Google Scholar
|
|
19
|
Sauter G, Algaba F, Amin M, et al: Tumours
of the urinary system: Non-invasive urothelial neoplasia. Eble JN,
Sauter G, Epstein JI and Sesterhenn IA: The WHO Classification of
Tumours of the Urinary System and Male Genital Organs. IARC Press;
Lyon: pp. 110–114. 2004
|
|
20
|
Sobin DH and Witteking CH: TNM
Classification of Malignant Tumours. 6th edition. Wiley-Liss; New
York, NY: pp. 199–202. 2002
|
|
21
|
Sanders I, Holdenrieder S,
Walgenbach-Brunagel G, Walgenbach-Brünagel G, von Ruecker A,
Kristiansen G, Müller SC and Ellinger J: Evaluation of reference
genes for the analysis of serum miRNA in patients with prostate
cancer, bladder cancer and renal cell carcinoma. Int J Urol.
19:1017–1025. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cortez MA, Bueso Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zen K and Zhang CY: Circulating microRNAs:
A novel class of biomarkers to diagnose and monitor human cancers.
Med Res Rev. 32:326–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brosnan JA and Iacobuzio Donahue CA: A new
branch on the tree: Next-generation sequencing in the study of
cancer evolution. Semin Cell Dev Biol. 23:237–242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: Approaches and considerations. Nat Rev Genet.
13:358–369. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mlcochova H, Hezova R, Stanik M and Slaby
O: Urine microRNAs as potential noninvasive biomarkers in urologic
cancers. Urol Oncol. 32:41 e41–e49. 2014. View Article : Google Scholar
|
|
27
|
Mengual L, Lozano JJ, Ingelmo-Torres M,
Gazquez C, Ribal MJ and Alcaraz A: Using microRNA profiling in
urine samples to develop a non-invasive test for bladder cancer.
Int J Cancer. 133:2631–2641. 2013.PubMed/NCBI
|
|
28
|
Adam L, Wszolek MF, Liu CG, Jing W, Diao
L, Zien A, Zhang JD, Jackson D and Dinney CP: Plasma microRNA
profiles for bladder cancer detection. Urol Oncol. 31:1701–1708.
2013. View Article : Google Scholar
|
|
29
|
Cookson VJ, Bentley MA, Hogan BV, Horgan
K, Hayward BE, Hazelwood LD and Hughes TA: Circulating microRNA
profiles reflect the presence of breast tumours but not the
profiles of microRNAs within the tumours. Cell Oncol (Dordr).
35:301–308. 2012. View Article : Google Scholar
|
|
30
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
Kim T and Lee KW: Detection of microRNA as novel biomarkers of
epithelial ovarian cancer from the serum of ovarian cancer
patients. Int J Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tanaka K, Miyata H, Yamasaki M, Sugimura
K, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M and
Doki Y: Circulating miR-200c levels significantly predict response
to chemotherapy and prognosis of patients undergoing neoadjuvant
chemotherapy for esophageal cancer. Ann Surg Oncol. 20(Suppl 3):
S607–S615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang HY, Yan LX, Shao Q, Fu S, Zhang ZC,
Ye W, Zeng YX and Shao JY: Profiling plasma microRNA in
nasopharyngeal carcinoma with deep sequencing. Clin Chem.
60:773–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kannan M, Mohan KV, Kulkarni S and Atreya
C: Membrane array-based differential profiling of platelets during
storage for 52 miRNAs associated with apoptosis. Transfusion.
49:1443–1450. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lionetti M, Musto P, Di Martino MT, Fabris
S, Agnelli L, Todoerti K, Tuana G, Mosca L, Gallo Cantafio ME,
Grieco V, et al: Biological and clinical relevance of miRNA
expression signatures in primary plasma cell leukemia. Clin Cancer
Res. 19:3130–3142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kanaan Z, Roberts H, Eichenberger MR,
Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A and
Galandiuk S: A plasma microRNA panel for detection of colorectal
adenomas: A step toward more precise screening for colorectal
cancer. Ann Surg. 258:400–408. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng G, Wang H, Zhang X, Yang Y, Wang L,
Du L, Li W, Li J, Qu A, Liu Y and Wang C: Identification and
validation of reference genes for qPCR detection of serum microRNAs
in colorectal adenocarcinoma patients. PLoS One. 8:e830252013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Gu J, Roth JA, Hildebrandt MA,
Lippman SM, Ye Y, Minna JD and Wu X: Pathway-based serum microRNA
profiling and survival in patients with advanced stage non-small
cell lung cancer. Cancer Res. 73:4801–4809. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu C, Iqbal J, Teruya-Feldstein J, Shen
Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino
E, et al: MicroRNA expression profiling identifies molecular
signatures associated with anaplastic large cell lymphoma. Blood.
122:2083–2092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mahn R, Heukamp LC, Rogenhofer S, von
Ruecker A, Müller SC and Ellinger J: Circulating microRNAs (miRNA)
in serum of patients with prostate cancer. Urology. 77:1265e9–e16.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hauser S, Wulfken LM, Holdenrieder S,
Moritz R, Ohlmann CH, Jung V, Becker F, Herrmann E,
Walgenbach-Brünagel G, von Ruecker A, et al: Analysis of serum
microRNAs (miR-26a-2*, miR-191, miR-337-3p and miR-378)
as potential biomarkers in renal cell carcinoma. Cancer Epidemiol.
36:391–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Z, Furge KA, Yang XJ, Teh BT and
Hansel DE: Comparative gene expression profiling analysis of
urothelial carcinoma of the renal pelvis and bladder. BMC Med
Genomics. 3:582010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tijsen AJ, Creemers EE, Moerland PD, de
Windt LJ, van der Wal AC, Kok WE and Pinto YM: MiR423-5p as a
circulating biomarker for heart failure. Circ Res. 106:1035–1039.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xiao B, Wang Y, Li W, Baker M, Guo J,
Corbet K, Tsalik EL, Li QJ, Palmer SM, Woods CW, et al: Plasma
microRNA signature as a noninvasive biomarker for acute graft
versus host disease. Blood. 122:3365–3375. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Farazi TA, Horlings HM, Ten Hoeve JJ,
Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F,
van Kouwenhove M, et al: MicroRNA sequence and expression analysis
in breast tumors by deep sequencing. Cancer Res. 71:4443–4453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato
JM and Lu SC: MicroRNAs regulate methionine adenosyltransferase 1A
expression in hepatocellular carcinoma. J Clin Invest. 123:285–298.
2013. View Article : Google Scholar :
|
|
48
|
Fromont G, Rouprêt M, Amira N, Sibony M,
Vallancien G, Validire P and Cussenot O: Tissue microarray analysis
of the prognostic value of E-cadherin, Ki67, p53, p27, survivin and
MSH2 expression in upper urinary tract transitional cell carcinoma.
Eur Urol. 48:764–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nakanishi K, Kawai T, Hiroi S, Kumaki F,
Torikata C, Aurues T and Ikeda T: Expression of telomerase mRNA
component (hTR) in transitional cell carcinoma of the upper urinary
tract. Cancer. 86:2109–2116. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakanishi K, Hiroi S, Tominaga S, Aida S,
Kasamatsu H, Matsuyama S, Matsuyama T and Kawai T: Expression of
hypoxia inducible factor-1alpha protein predicts survival in
patients with transitional cell carcinoma of the upper urinary
tract. Clin Cancer Res. 11:2583–2590. 2005. View Article : Google Scholar : PubMed/NCBI
|