Introduction
Bladder cancer is a common malignancy with high
rates of relapse, metastasis and mortality (1). An estimated 72,570 novel cases of
bladder cancer were diagnosed, and ~15,210 patients succumbed to
bladder cancer in the United States in 2013 (1). In China, bladder cancer represents
3.2% of all malignant tumors (2).
Histologically, >95% of bladder tumors are derived from the
urothelium. Bladder cancer can be classified as either
non-muscle-invasive bladder carcinoma or muscle-invasive bladder
carcinoma, and the majority of newly diagnosed patients present
with non-muscle-invasive bladder carcinoma. In ~2/3 of patients
with non-muscle-invasive bladder carcinoma, recurrence of the
cancer is observed after initial management, and 5–30% of these
cancers are transformed into muscle-invasive bladder carcinoma, for
which the five-year survival rate is ~50% (3).
Eukaryotic genomes encode numerous long non-coding
RNAs (lncRNAs), which are defined as endogenous cellular RNAs with
>200 nucleotides that lack open reading frames of significant
length (4). Within four years, the
number of identified lncRNAs has increased to >8,000 (5). Although the function of the majority
of lncRNAs is currently elusive, a rapidly increasing number of
studies have provided accumulating evidence for their involvement
in numerous biological processes, supporting the hypothesis that
dysregulation of lncRNAs may be associated with cancer development,
as well as invasion and metastasis of malignant cells (5–7).
In human urothelial carcinoma of the bladder, a gene
microarray analysis of the lncRNA expression profile detected 1,122
differentially expressed (≥2-fold) lncRNAs; of these, 734 lncRNAs
were upregulated and 388 were downregulated (8). Urothelial cancer associated-1
(9), linc-upregulated in bladder
cancer 1 (10) and
metastasis-associated lung adenocarcinoma transcript-1 (11,12)
have been shown to be upregulated in invasive bladder cancer
tissues. Conversely, maternally expressed gene-3 (13) and growth arrest-specific 5 (GAS5)
(14) were found to be
downregulated in bladder cancer tissues as compared with adjacent
normal tissues. lncRNA-GAS5 has recently been identified to be
involved in the tumorigenesis of various types of cancer, including
breast cancer (15), lung cancer
(16), hepatocellular carcinoma
(17), and renal cancer (4). Overexpression of GAS5 has been shown
to reduce the rate of cell cycle progression, whereas
downregulation of GAS5 inhibited apoptosis and accelerated cell
cycle progression. Furthermore, overexpression of GAS5 has been
reported to induce growth arrest and apoptosis, independent of
other stimuli (14). To date, the
underlying mechanisms of the effects of GAS5 on the regulation of
cancer cell apoptosis have remained to be fully elucidated.
Although the effects of GAS5-induced tumor apoptosis
have been studied in certain cancer types, the role of GAS5 in the
apoptosis of bladder transitional cell lines and the possible
involvement of chemokine (C-C) ligand 1 (CCL1) expression have
largely remained elusive. Therefore, the aim of the present study
was to investigate the involvement of GAS5 on the apoptosis of a
bladder transitional cell line (BLX).
Materials and methods
Cell culture
The BLX human bladder cancer cells were obtained
from the Cell Resource Center, Shanghai Institutes for Biological
Sciences (Shanghai, China), and maintained in RPMI-1640 (Invitrogen
Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (Invitrogen Life Technologies, Carlsbad, CA, USA) at
37°C in a humidified incubator (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 5% CO2 and 95% air. The
medium was replaced every day.
Transfection
The small interfering (si)RNAs specific for human
GAS5 and CCL1, and scramble siRNA were obtained from Dharmacon
(Lafayette, CO, USA). The siRNA sequences were as follows: GAS5
forward, 5′-CAC UCU GAG UGG GAC AAG CUC UUCA-3′ and reverse, 5′-UGA
AGA GCU UGU CCC ACU CAG AGUG-3′; CCL1 forward, 5′-GCU AUC AGU CCA
CUG UGC UUG UGGU-3′ and reverse, 5′-ACC ACA AGC ACA GUG GAC UGA
UAGC-3′, and scramble forward. 5′-CAC GAG UGG GUA ACA CUC GUC
UUCA-3′ and reverse, 5′-UGA AGA CGA GUG UUA CCC ACU CGUG-3′. The
BLX cells were plated in 6-well plates (40, 000 cells/well).
Following 24 h incubation, the cells were transfected with
GAS-siRNA and CCL1-siRNA, respectively, using Lipofectamine 2000
(Invitrogen Life Technologies), according to the manufacturer's
instructions. The final siRNA concentration was 100 nM. Untreated
cells and scramble siRNA-transfected cells were used as a negative
control (NC) and Mock group, respectively.
To overexpress GAS5 and CCL1, the pcDNA-GAS5 and
pcDNA-CCL1 plasmids were constructed by introducing a
KpnI-XhoI fragment containing GAS5 and CCL1 cDNA into
the same sites in a pcDNA3.1 plasmid. The GAS5 and CCL1 genes were
amplified from cDNA prepared from BLX cells by polymerase chain
reaction (PCR) using the following forward and reverse primers:
GAS5 forward, 5′-CAC UCU GAG UGG GAC AAG CUC UUCA-3′ and reverse,
5′-UGA AGA GCU UGU CCC ACU CAG AGUG-3′; and CCL1 forward, 5′-GCU
AUC AGU CCA CUG UGC UUG UGGU-3′ and reverse, 5′-ACC ACA AGC ACA GUG
GAC UGA UAGC-3′. The empty pcDNA3.1 vector was used as a control.
The BLX cells were transfected with pcDNA-GAS5 and pcDNA-CCL1
plasmids (2 µg/1.5×1,000,000 cells) respectively, using
Lipofectamine 2000 (Invitrogen Life Technologies), according to the
manufacturer's instructions.
Assessment of cell viability using Cell
Counting kit (CCK)8
Bladder cancer cells were plated
(5.0×103/well) and treated in 96-well plates (three
wells per group) with various stimuli for 24, 48 or 72 h.
Subsequently, 10 µl CCK8 (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) was added to the cells, and the optical
density value of the cells was measured at 450 nm using an ELX 808
Ultra microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA), according to the manufacturer's instructions.
Quantification of apoptosis by flow
cytometry
Apoptosis was assessed using annexin V, a protein
that binds to phosphatidylserine (PS) residues, which are exposed
on the cell surface of apoptotic cells. The Annexin V-fluorescein
isothiocyanate (FITC) apoptosis detection kit was purchased from
Sigma-Aldrich (St. Louis, MO, USA). The BLX cells were plated
(5.0×105/well, 1 ml) and treated in six-well plates
(three wells per group) with various stimuli for 48 h. After
treatment, the cells were washed twice with phosphate-buffered
saline (pH 7.4), and re-suspended in staining buffer containing 10
µl propidium iodide (PI) and 5 µl annexin V-FITC
Double-staining was performed at room temperature for 15 min in the
dark prior to flow cytometric analysis. The cells were immediately
analyzed using a FACScan flow cytometer (Becton Dickinson, Franklin
Lakes, NJ, USA) and CellQuest pro software version 5.2 (BD
Biosciences, San Jose, CA, USA). Quantitative analysis of apoptotic
cells was performed using the terminal deoxynucleotidyl
transferase-mediated deoxyuridine triphosphate nick end labeling
method, which examines DNA-strand breaks during apoptosis, using
the BD ApoAlert™ DNA Fragmentation Assay kit (cat. no. 630107; BD
Biosciences). Briefly, the cells were trypsinized, fixed with 4%
paraformaldehyde and permeabilized with 0.1% Triton X-100 in 0.1%
sodium citrate. After being washed, the cells were incubated with
the reaction mixture for 60 min at 37°C. The stained cells were
then analyzed using a flow cytometer.
Reverse transcription-quantitative
(RT-q)PCR
The bladder cancer cells were plated
(5.0×105/well) in six-well plates (three wells per
group) and after 24 h, cells were transfected with GAS5-siRNAs for
48 h. Total RNA was extracted from the bladder cancer cells using
TRIzol® (Invitrogen Life Technologies), according to the
manufacturer's instructions. Synthesis of cDNA was performed by RT
with 2 µg total RNA using Moloney Murine Leukemia Virus
Reverse Transcriptase (Promega Corp. Madison, WI, USA) with oligo
dT (15) primers (Fermentas,
Thermo Fisher Scientific), according to the manufacturer's
instructions. qPCR was performed using a SYBR Green PCR kit (Toyobo
Co., Ltd., Osaka, Japan) on an ABI 7300 Real Time PCR system
(Applied Biosystems, Life Technologies, Thermo Fisher Scientific),
according to the manufacturer's instructions of the kit. The PCR
cycling conditions were as follows: 2 min polymerase activation at
95°C, followed by 40 cycles of denaturation at 95°C for 15 sec,
annealing at 55°C for 60 sec, and extension at 72°C for 20 sec. The
sequences of the primers were as follows: GAS5 forward, 5′-TTG CGA
TTC TGT TTT GTGCT-3′ and reverse, 5′-GTG GGG TCC TCA GTG GG-3′; and
β-actin, forward, 5′-ACA GGG GAG GTG ATA GCATT-3′ and reverse,
5′-GAC CAA AAG CCT TCA TAC ATC TC-3′. β-actin was used as an
internal control to normalize the data and to determine the
relative expression levels of the target genes. Following
completion of the reaction, the amplification curve and melting
curve were analyzed. Gene expression values were determined using
the 2−ΔΔCT method (17).
Western blot analysis
For the whole cell extracts, the bladder cancer
cells were washed with ice-cold phosphate-buffered saline. The cell
pellets were homogenized in extraction buffer, containing 8 M urea,
10% glycerol, 10 mM Tris-HCl (pH 6.8), 1% SDS, 5 mM DTT, 0.5 mM
phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, 10
µg/ml pepstatin and 10 µg/ml leupeptin]. Protein was
extracted using NP-40 lysis buffer (cat. no. FNN0021; Thermo Fisher
Scientific). Protein concentration was determined using a Pierce
BCA protein assay (Pierce Biotechnology, Inc., Rockford, IL, USA).
Subsequently, the samples were boiled for 5–10 min and centrifuged
at 12,000 × g for 10 min in order to obtain the supernatant.
Aliquots of 50 µg protein were separated by 10% SDS-PAGE and
transferred onto polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA). Following saturation with 5% (w/v) non-fat dry
milk in Tris-buffered saline with 0.1% (w/v) Tween 20 (TBST), the
membranes were incubated with monoclonal rat anti-CCL1 (cat. no.
sc-74092; Santa Cruz Biotechnology, Inc., Dallas, TX, USA.) at
dilutions of 1: 1,000 at 4°C overnight. After three washes with
TBST, the membranes were incubated at 37°C for 1 h with secondary
antibodies conjugated to IRDye® 800 CW Infrared Dye
(LI-COR Biosciences, Lincoln, NE, USA), including polyclonal donkey
anti-goat IgG (cat. no. A-11056; Thermo Fisher Scientific) and
polyclonal donkey anti-mouse IgG (cat. no. A-21202; Thermo Fisher
Scientific) at dilutions of 1:10,000. The membranes were then
washed three times with TBST. The blots were visualized using the
Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE,
USA). Signals were densitometrically assessed (Odyssey Application
Software version 3.0; LI-COR Biosciences) and normalized to the
β-actin signals, in order to correct for unequal loading, using a
mouse monoclonal anti-β-actin antibody (cat. no. AP0060; 1:1,000;
Bioworld Technology, Inc., St. Louis Park, MN, USA).
Bioinformatics analysis
The total RNA from the CCL1-none (DMSO) and CCL1
transfer (CCL1) groups were individually hybridized with gene chips
of the mouse lncRNA microarray V2.0 (8x 60K; Arraystar). Briefly,
RNA was purified from 1 µg total RNA following removal of
rRNA. The RNA was then transcribed into fluorescent cRNA with the
entire length of the transcripts, without 3′ bias, using random
primers. The labeled cRNAs were hybridized to the gene microarray.
Finally, an Agilent Scanner (G2505B; Agilent Technologies, Inc.,
Santa Clara, CA, USA) was used to scan the arrays and the array
results were analyzed using Agilent Feature Extraction software
(version 10.7.3.1). The GeneSpring GX v11.5.1 software package
(Agilent Technologies, Inc.) was used to analyze the subsequent
data processing. Microarray hybridization was performed by Kangchen
Biology Engineering Co., Ltd. (Shanghai, China). Differentially
expressed lncRNAs of statistical significance were identified
through Volcano Plot filtering. The threshold used to identify
upregulated or downregulated RNAs was a fold-change >2.0
(P<0.05). The resulting differentially expressed genes were
subjected to hierarchical clustering (Cluster 3.0) and TreeView
analysis (Stanford University, Stanford, CA, USA).
Statistical analysis
Values are expressed as the mean ± standard error of
the mean for each group. All statistical analyses were performed
using GraphPad Prism version 4.0 (GraphPad Software, Inc., La
Jolla, CA, USA). Inter-group differences were analyzed by one-way
analysis of variance, followed by Tukey's multiple comparison test
as a post-hoc test to compare the group means if overall P<0.05.
P<0.05 was considered to indicate a statistically significant
difference.
Results
GAS5 inhibits bladder cancer cell
proliferation in vitro
To evaluate the potential role of GAS5 in regulating
bladder cancer cell proliferation, BLX cells transfected with
scramble-siRNA or GAS5-siRNAs were subjected to a CCK8 cell
viability assay. The viability of the BLX cells transfected with
GAS5-siRNA was significantly higher as compared with that in the
untransfected group or the scramble-siRNA group, and GAS5 knockdown
was able to increase the cell count by ~20% in the
GAS5-siRNA-transfected groups (Fig.
1A). Furthermore, the expression levels of GAS5 were decreased
in the GAS5-siRNA-transfected BLX cells, demonstrating the
knockdown efficiency of the siRNAs (Fig. 1B). The present study further
investigated whether GAS5 knockdown was able to induce cell
apoptosis and influence cell cycle progression. Following knockdown
of GAS5 expression, the cell cycle distribution was analyzed by
flow cytometry. As compared with the untransfected group, GAS5
knockdown resulted in an increased percentage of cells in S and G2
phase, and a decreased percentage of cells in G1 phase (Fig. 1C). This result inversely
demonstrated the tumor-suppressive function of GAS5, as cancer
cells may be prevented from proliferating by G1 cell cycle arrest.
All of these results indicated that downregulation of GAS5
expression in bladder cancer contributed to bladder cancer cell
proliferation.
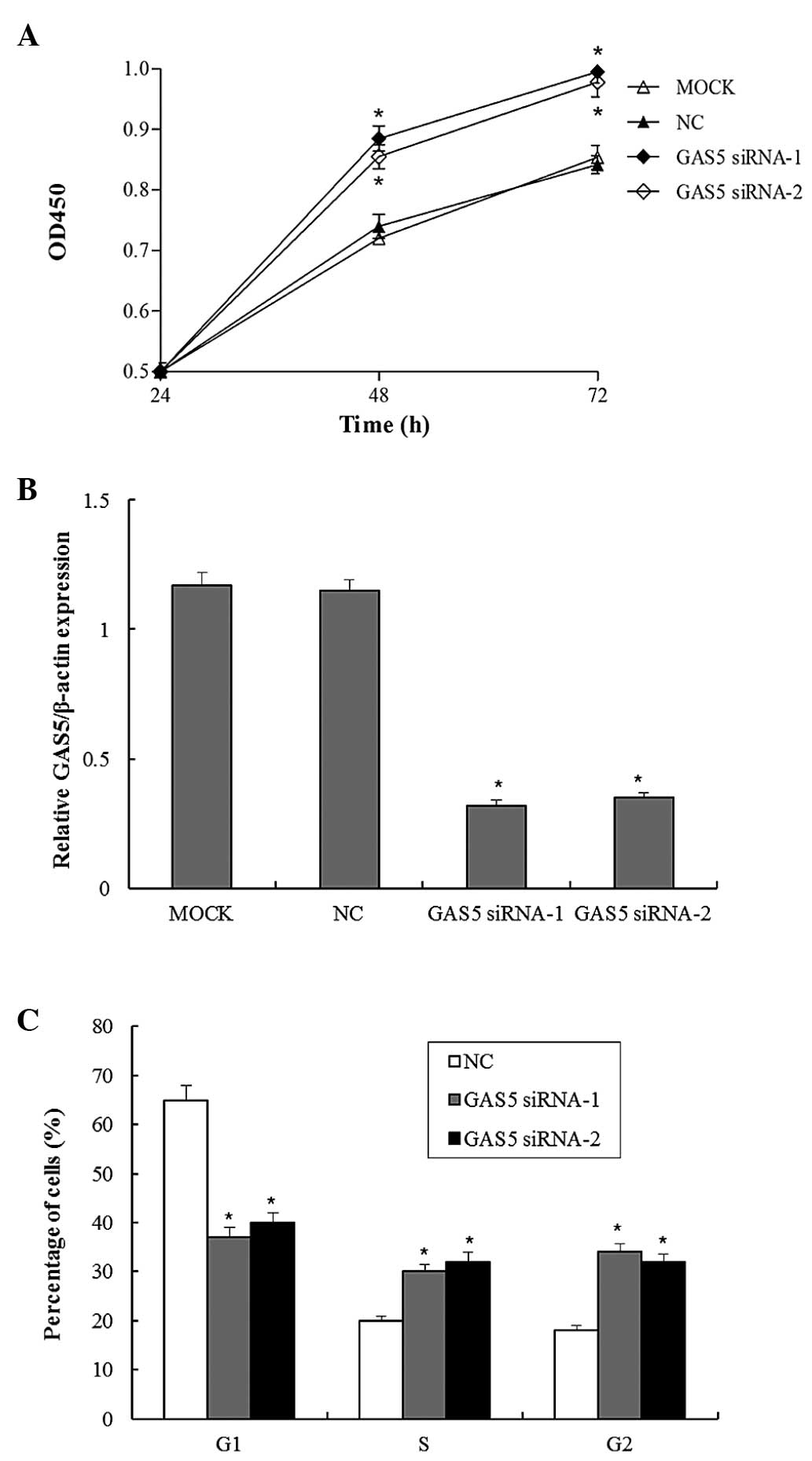 | Figure 1GAS5 inhibits bladder cancer cell
proliferation. (A) Bladder cancer cells were transfected with
GAS5-siRNA for 24, 48 or 72 h, and cell viability was examined
using the Cell Counting kit-8 assay. (B) Bladder cancer cells were
transfected with GAS5-siRNA, and the GAS5 expression levels were
detected after 48 h of incubation by reverse
transcription-quantitative polymerase chain reaction. (C) Bladder
cancer cells were transfected with GAS5-siRNA, and following 48 h
of incubation, the relative number of cells in each cell cycle
phase was detected by propidium iodide staining and
fluorescence-activated cell sorting analysis. Values are expressed
as the mean ± standard error of the mean (n=3/group).
*P<0.05, compared with the control. OD, optical
density; NC, negative control; Mock, scramble siRNA-transfected
group; GAS5, growth arrest specific 5; GAS5 siRNA-1 and GAS5
siRNA-2, transfected with GAS5-small interfering RNA. |
GAS5 negatively regulates CCL1 expression
in vitro
The present study investigated the possible
mechanisms underlying the regulation of bladder cancer cell
proliferation by lncRNAs. A hierarchical cluster analysis of
differentially expressed lncRNAs in bladder cancer cells was
performed, which identified that CCL1 is associated with lncRNA
expression (Fig. 2A).
Overexpression of CCL1 was associated with lncRNA-GAS5, the
expression of which was highest among the 30 lncRNAs assessed in
vitro. This indicated that the overexpression of CCL1
significantly upregulated the expression of GAS5. CCL1 was
previously shown to promote M2 macrophage and T-helper cell type 2
polarization in tumors that subvert the immune system by
establishing a microenvironment of immune cells and cytokines that
suppress specific anti-tumor responses (18). To further validate the interaction
between GAS5 and CCL1, gene knockdown and overexpression of GAS5
were implemented. GAS5 knockdown markedly increased the mRNA and
protein expression levels of CCL1. Conversely, overexpression of
GAS5 decreased the mRNA and protein expression levels of CCL1 in
bladder cancer cells (Fig. 2B and
C).
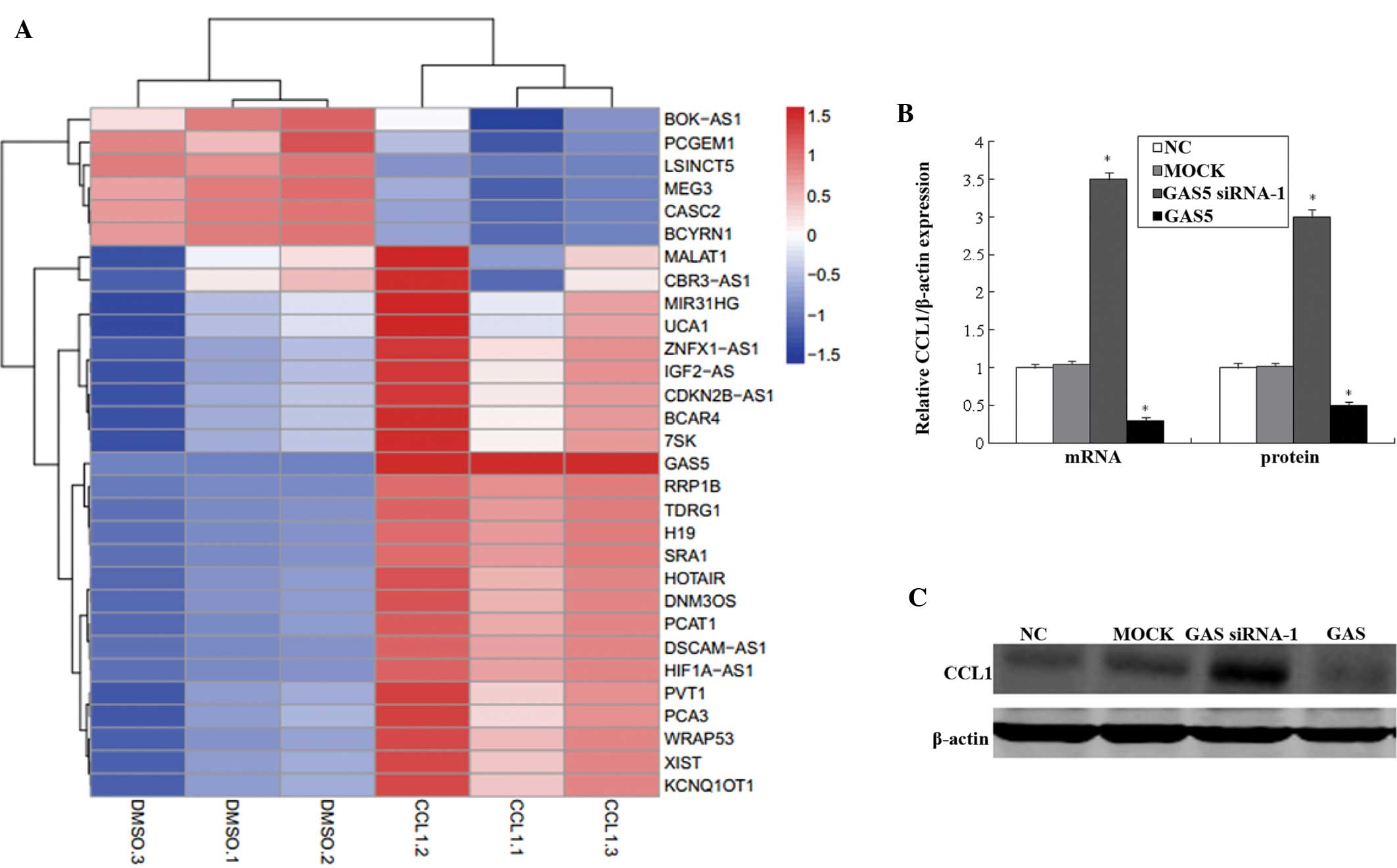 | Figure 2GAS5 regulates the expression of CCL1.
(A) Hierarchical clustering of differentially expressed lncRNAs in
CCL1-none groups (DMSO.1, DMSO.2 and DMSO.3) and CCL1-transfer
groups (CCL1.1, CCL1.2 and CCL1.3). (B and C) Bladder cancer cells
were transfected with GAS5-siRNA or pcDNA-GAS5, and the expression
levels of CCL1 were examined following 48 h of incubation by
reverse transcription-quantitative polymerase chain reaction and
western blotting. Values are expressed as the mean ± standard error
of the mean (n=3/group). *P<0.05, compared with the
control. NC, negative control; Mock, scramble siRNA-transfected
group; CCL1, chemokine (C-C) ligand 1; LncRNA, long non-coding RNA;
GAS5, growth arrest-specific 5; siRNA, small interfering RNA;.
DMSO, treated with dimethyl sulfoxide; CCL1, transfected with
CCL1-siRNA group; GAS5 siRNA-1, transfected with GAS5-siRNA; GAS5,
transfected with pcDNA-GAS5 plasmid vector. |
GAS5 inhibits bladder cancer cell
proliferation by regulating CCL1
Knockdown of GAS5 expression increased BLX cell
proliferation, and an association was detected between GAS5 and
CCL1; therefore, it was hypothesized that the role of GAS5 in
regulating bladder cancer cell proliferation was mediated by
modulating CCL1 expression. As determined by CCK8 assay,
GAS5-siRNA-enhanced BLX cell proliferation was partially suppressed
by co-transfection with CCL1-siRNA (Fig. 3A). In addition, BLX cell
proliferation was partially suppressed by CCL1 knockout compared
with that in the control group. In addition, the present study
investigated whether GAS5 was able to induce cell death via an
apoptotic mechanism. Annexin V-PI double-staining was used for the
detection of PS externalization, which is a hallmark of early
apoptosis. In line with the results of the CCK-8 assay, the
proportion of early-phase apoptotic cells following transfection
with GAS5-siRNA was decreased, while it was markedly increased in
the group transfected with CCL1-siRNA. The apoptotic rate following
co-transfection was reduced as compared with that following
GAS5-siRNA transfection alone (Fig. 3B
and C). In the cells overexpressing GAS5, forced expression of
CCL1 was able to partially restore cell proliferation (Fig. 4A). Consistent with the results of
the CCK8 assay, the proportion of early-phase apoptotic cells was
increased in GAS5 overexpression, and decreased with CCL1
overexpression. The apoptotic rate of co-transfection was
decreased, compared with that transfected to overexpress GAS5 only
(Fig. 4B and C). These results
indicated that GAS5 may decrease bladder cancer progression, at
least in part, by regulating CCL1 expression.
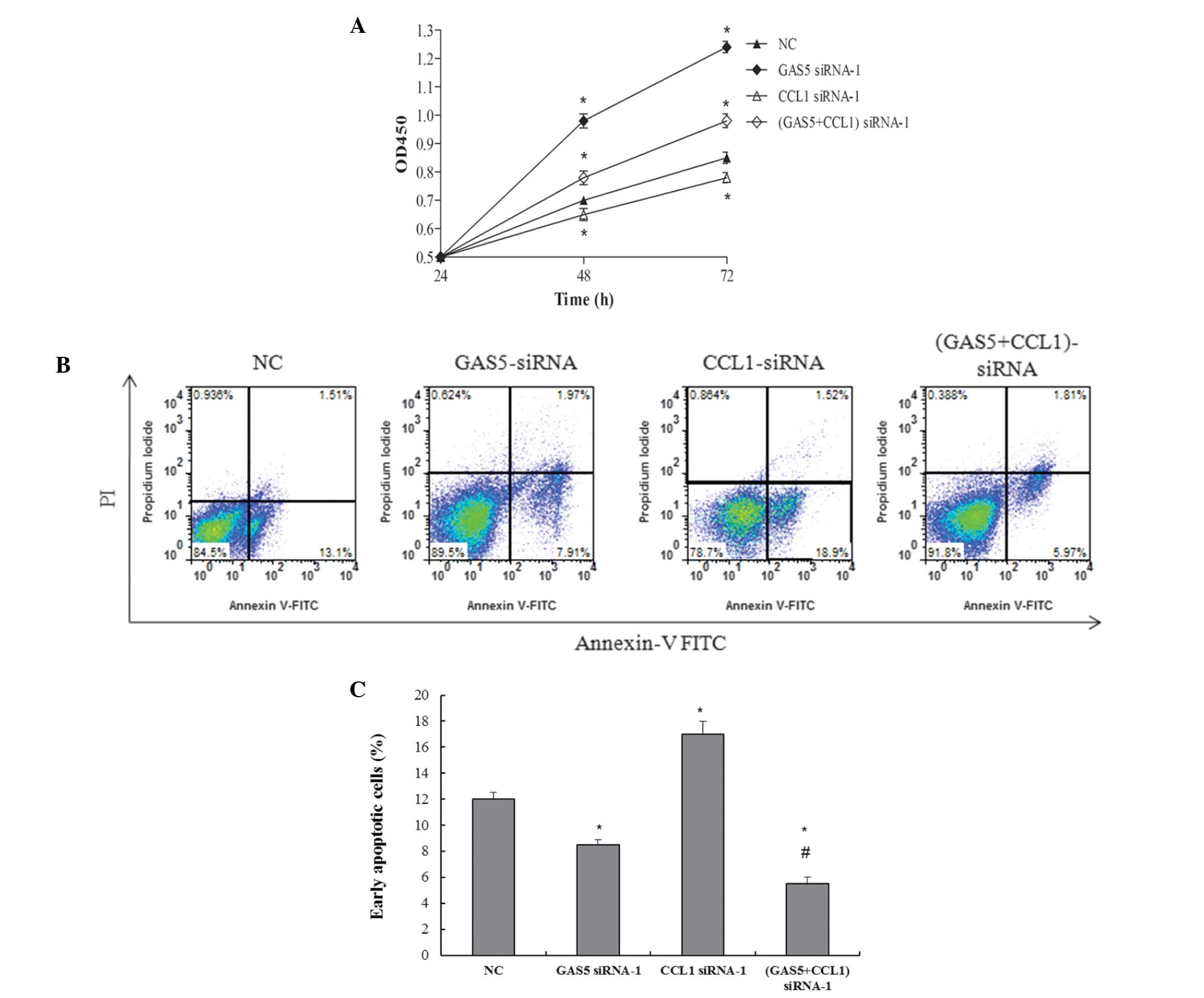 | Figure 3GAS5 inhibits bladder cancer cell
proliferation, partly via regulaton of CCL1. (A) Bladder cancer
cells were transfected with GAS5-siRNA and CCL1-siRNA, and cell
viability was examined using the Cell Counting kit-8 assay.
Apoptotic cells were detected using (B) flow cytometric analysis of
annexin V/propidium iodide double staining and (C) quantitative
analysis using a TUNEL assay. TUNEL-positive cells were examined
using flow cytometry. Values are expressed as the mean ± standard
error of the mean (n=3/group). *P<0.05, compared with
the NC group; #P<0.05, compared with the GAS5-siRNA
group. FITC, fluorescein isothiocyanate; OD, optical density; NC,
negative control; CCL1, chemokine (C-C) ligand 1; siRNA, small
interfering RNA; GAS5, growth arrest-specific 5; GAS5 siRNA-1,
transfected with GAS5-siRNA; CCL1 siRNA-1, transfected with
CCL1-siRNA; GAS5+CCL1 siRNA-1, co-trans-fected with GAS5-siRNA and
CCL1-siRNA. |
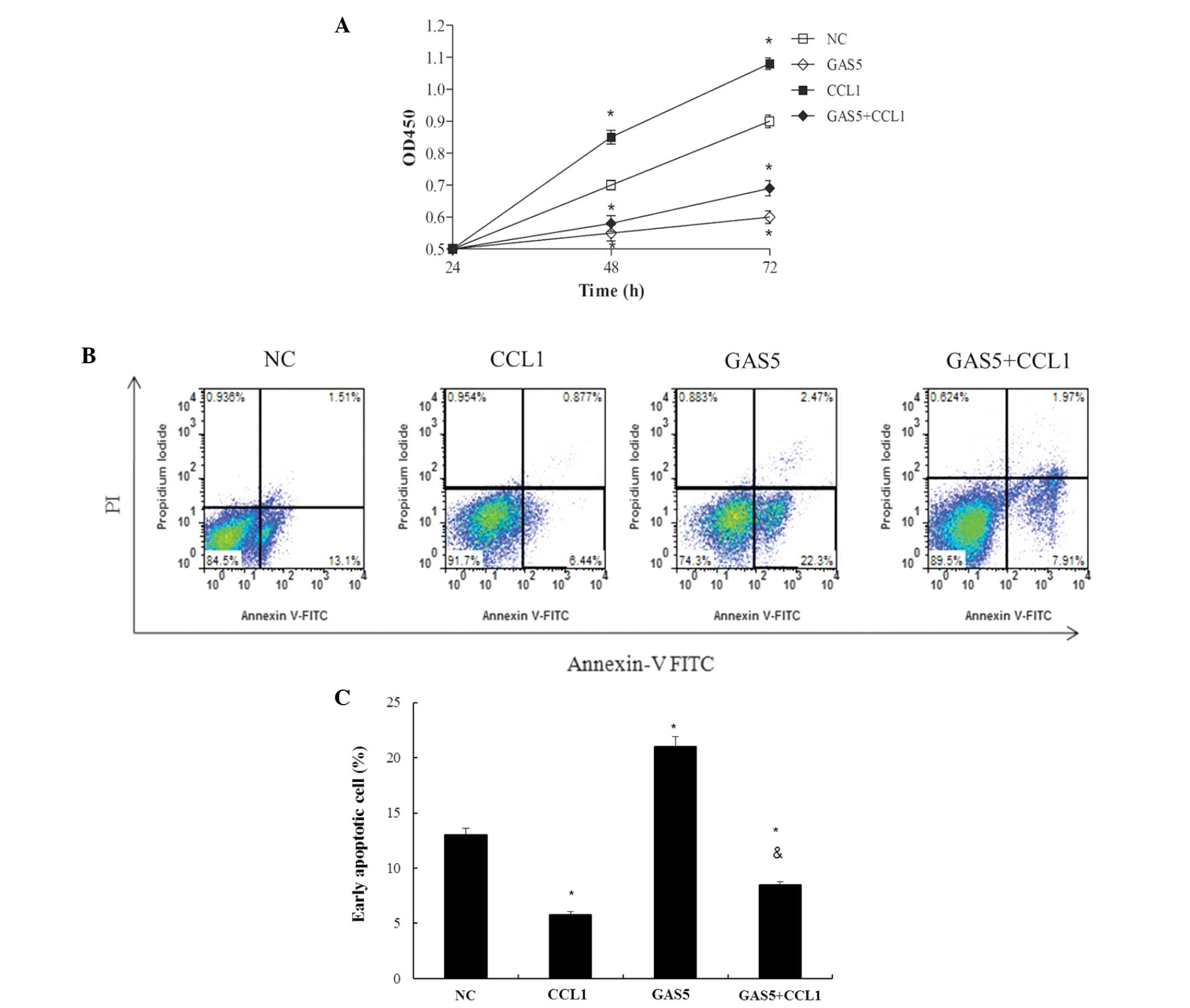 | Figure 4(A) Bladder cancer cells were
transfected with GAS5- and/or CCL1-overexpression vectors and the
number of cells/well was determined using the Cell Couting kit-8
assay (absorbance, 450 nm). (B) Apoptotic cells were detected by
flow cytometric analysis of annexin V/propidium iodide double
staining and (C) quantitative analysis using a TUNEL assay.
TUNEL-positive cells were examined using flow cytometry.. Values
are expressed as the mean ± standard error of the mean (n=3/group).
*P<0.05, compared with the NC group;
#P<0.05, compared with the GAS5 group. OD, optical
density; FITC, fluorescein isothiocyanate; NC, negative control;
CCL1, chemokine (C-C) ligand 1; GAS5, growth arrest-specific 5;
GAS5, transfected with the pcDNA-GAS5 plasmid vector; CCL1,
transfected with the pcDNA-CCL1 plasmid vector; GAS5+CCL1,
co-transfected with the pcDNA-GAS5 and pcDNA-CCL1 plasmid
vectors. |
Discussion
Non-coding RNAs are the predominant transcripts in
the mammalian genome, exceeding the number of protein-coding genes.
It is thought that alterations in the expression of small
non-coding RNAs, particularly microRNAs, may contribute to the
pathogenesis of bladder cancer (19,20).
A novel class of non-coding RNAs, known as lncRNAs (>200
nucleotides), which are endogenous RNA transcripts with reduced or
absent protein-coding potential, have been reported to be
abundantly transcribed (1).
Previous studies have demonstrated the importance of lncRNAs in the
tumorigenesis and metastasis of malignant cells (5,6).
Downregulation of the lncRNA ncRuPAR has been shown to contribute
to tumor inhibition in colorectal cancer (21), and the lncRNA TARID directs
demethylation and activation of the tumor suppressor gene TCF21 via
GADD45A (22).
GAS5 is a recently identified lncRNA, which is
associated with cell cycle regulation (14). GAS5 has a crucial role in normal
growth arrest in T-cell lines as well as non-transformed
lymphocytes (23). In addition,
GAS5 knockdown is associated with renal cell carcinoma (RCC)
carcinogenesis and progression, whereas overexpression of GAS5 is
able to act as a tumor suppressor in RCC (4). Recent clinical studies demonstrated
that GAS5 is downregulated in the majority of patients with
hepatocellular carcinoma and bladder cancer (14,17).
However, there are few reports regarding the interaction between
GAS5 and bladder cancer. The present study performed a hierarchical
cluster analysis of the differentially expressed lncRNAs in the
bladder cancer cell and identified CCL1 as being associated with
GAS5 expression. Based on these findings, the present study aimed
to determine whether the expression of CCL1 was regulated by GAS5
in bladder cancer cell proliferation. Gain-of-function and
loss-of-function studies demonstrated that GAS5 regulated cell
cycle progression and inhibited bladder cancer cell proliferation,
partially via regulation of CCL1 expression.
The CCL1/chemokine (C-C motif) receptor (CCR)8 axis
is involved in leukemia as well as lymphoma. It has been
demonstrated in vitro that the CCL1/CCR8 autocrine loop
protects lymphoma and T-cell leukemia cells from apoptosis
(24). In the present study,
overexpression of CCL1 induced GAS5 upregulation, which may
enhanced the ability of the cells to regulate a stress response
in vitro. It has been indicated that inflammatory cytokines
and microbial products markedly induce the expression of CCL1
(25). In humans, CCL1 has been
detected in the lymph node lymphatic sinuses, but not in the
peripheral lymphatics. The CCL1 receptor CCR8 is highly expressed
in human malignant melanoma. Tumor cell migration to lymphatic
endothelial cells has been shown to be inhibited by suppression of
either CCR8 or CCL1 in vitro; furthermore, recombinant CCL1
promoted the migration of CCR8+ tumor cells (24). In a murine model, suppression of
CCR8 by a soluble antagonist or short hairpin RNA significantly
decreased lymph node metastasis. In addition, inhibition of CCR8
caused the arrest of tumor cells in the collecting lymphatic
vessels at the junction with the lymph node subcapsular sinus
(24). Of note, the
CCR8+ myeloid cell subset is increased in patients with
cancer. A previous study demonstrated that the expression of CCL1
was elevated in tumors, and the presence of CCR8+ myeloid cells was
increased in the peripheral blood and cancer tissues, thus
suggesting that the CCL1/CCR8 axis is involved in cancer-associated
inflammation and may have a role in immune evasion. GAS5 has been
reported to prevent glucocorticoid receptor localization to cognate
genes, regulating various immune response genes, including II-8,
CXCL10, CCL1 and CSF1 (26). In
the present study, hierarchical cluster analysis indicated that
overexpression of CCL1 was associated with lncRNA-GAS5. These
results suggested a correlation between GAS5 and CCL1. However, the
association between GAS5 and CCL1 requires further validation
through a luciferase reporter assay to confirm whether GAS5 can
regulate the CCL1 promoter.
In conclusion, the present study demonstrated that
GAS5 silencing promoted bladder cancer proliferation and suppressed
cell apoptosis. In addition, GAS5 knockdown resulted in an
increased percentage of cells in the S and G2 phases, and a
decreased percentage of cells in the G1 phase. Hierarchical cluster
analysis revealed that CCL1 overexpression resulted in an
upregulation of GAS5. Furthermore, knockdown of GAS5 increased the
mRNA and protein expression levels of CCL1 in the bladder cancer
cells, induced BLX cell proliferation and inhibited cancer cell
apoptosis. The overexpression of GAS5 suppressed the cell
proliferation and enhanced the apoptotic rate of the cancer cells.
In addition, BLX cell proliferation was partially suppressed by
CCL1 silencing, whereas CCL1 overexpression resulted in significant
cell proliferation. The results demonstrated that GAS5 suppressed
bladder cancer cell proliferation, at least partially, by
suppressing the expression of CCL1. These findings provide a basis
for the development of novel effective therapeutic strategies for
the treatment of bladder cancer.
References
|
1
|
Wang L, Fu D, Qiu Y, Xing X, Xu F, Han C,
Xu X, Wei Z, Zhang Z, Ge J, et al: Genome-wide screening and
identification of long noncoding RNAs and their interaction with
protein coding RNAs in bladder urothelial cell carcinoma. Cancer
Lett. 349:77–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L, Parkin DM, Li LD, Chen YD and Bray
F: Estimation and projection of the national profile of cancer
mortality in China: 1991–2005. Br J Cancer. 90:2157–2166.
2004.PubMed/NCBI
|
|
3
|
Tong ZT, Wei JH, Zhang JX, Liang CZ, Liao
B, Lu J, Fan S, Chen ZH, Zhang F, Ma HH, et al: AIB1 predicts
bladder cancer outcome and promotes bladder cancer cell
proliferation through AKT and E2F1. Br J Cancer. 108:1470–1479.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladde, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo H, Zhao X, Wan X, Huang S and Wu D:
Gene microarray analysis of the lncRNA expression profile in human
urothelial carcinoma of the bladder. Int J Clin Exp Med.
7:1244–1254. 2014.PubMed/NCBI
|
|
9
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He W, Cai Q, Sun F, Zhong G, Wang P, Liu
H, Luo J, Yu H, Huang J and Lin T: linc-UBC1 physically associates
with polycomb repressive complex 2 (PRC2) and acts as a negative
prognostic factor for lymph node metastasis and survival in bladder
cancer. Biochim Biophys Acta. 1832:1528–1537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar
|
|
12
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
noncoding RNA MALAT1. FEBS Lett. S0014–S5793. 2013.
|
|
13
|
Ying L, Huang Y, Chen H, Wang Y, Xia L,
Chen Y, Liu Y and Qiu F: Downregulated MEG3 activates autophagy and
increases cell proliferation in bladder cancer. Mol Biosyst.
9:407–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng
Y, Tao L and Qiu J: Downregulation of GAS5 promotes bladder cancer
cell proliferation, partly by regulating CDK6. PLoS One.
8:e739912013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pickard MR and Williams GT: Regulation of
apoptosis by long non-coding RNA GAS5 in breast cancer cells:
Implications for chemotherapy. Breast Cancer Res Treat.
145:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F
and Song Y: A critical role for the long non-coding RNA GAS5 in
proliferation and apoptosis in non-small-cell lung cancer. Mol
Carcinog. 54(Suppl 1): E1–E12. 2015. View
Article : Google Scholar
|
|
17
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
18
|
Eruslanov E, Stoffs T, Kim WJ, Daurkin I,
Gilbert SM, Su LM, Vieweg J, Daaka Y and Kusmartsev S: Expansion of
CCR8(+) inflammatory myeloid cells in cancer patients with
urothelial and renal carcinomas. Clin Cancer Res. 19:1670–1680.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Drayton RM, Peter S, Myers K, Miah S,
Dudziec E, Bryant HE and Catto JW: MicroRNA-99a and 100 mediated
upregulation of FOXA1 in bladder cancer. Oncotarget. 5:6375–6386.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Drayton RM, Dudziec E, Peter S, Bertz S,
Hartmann A, Bryant HE and Catto JW: Reduced expression of miRNA-27a
modulates cisplatin resistance in bladder cancer by targeting the
cystine/glutamate exchanger SLC7A11. Clin Cancer Res. 20:1990–2000.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan B, Gu W, Yang Z, Gu Z, Yue X, Gu Q and
Liu L: Downregulation of a long noncoding RNA-ncRuPAR contributes
to tumor inhibition in colorectal cancer. Tumour Biol.
35:11329–11335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arab K, Park YJ, Lindroth AM, Schäfer A,
Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et
al: Long noncoding RNA TARID directs demethylation and activation
of the tumor suppressor TCF21 via GADD45A. Mol Cell. 55:604–614.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Das S, Sarrou E, Podgrabinska S, Cassella
M, Mungamuri SK, Feirt N, Gordon R, Nagi CS, Wang Y, Entenberg D,
et al: Tumor cell entry into the lymph node is controlled by CCL1
chemokine expressed by lymph node lymphatic sinuses. J Exp Med.
210:1509–1528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gombert M, Dieu-Nosjean MC, Winterberg F,
Bünemann E, Kubitza RC, Da Cunha L, Haahtela A, Lehtimäki S, Müller
A, Rieker J, et al: CCL1-CCR8 interactions: An axis mediating the
recruitment of T cells and Langerhans-type dendritic cells to sites
of atopic skin inflammation. J Immunol. 174:5082–5091. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu AD, Wang ZC and Morris KV: Long
noncoding RNAs: a potent source of regulation in immunity and
disease. Immunol Cell Biol. 93:277–283. 2015. View Article : Google Scholar : PubMed/NCBI
|


















