Introduction
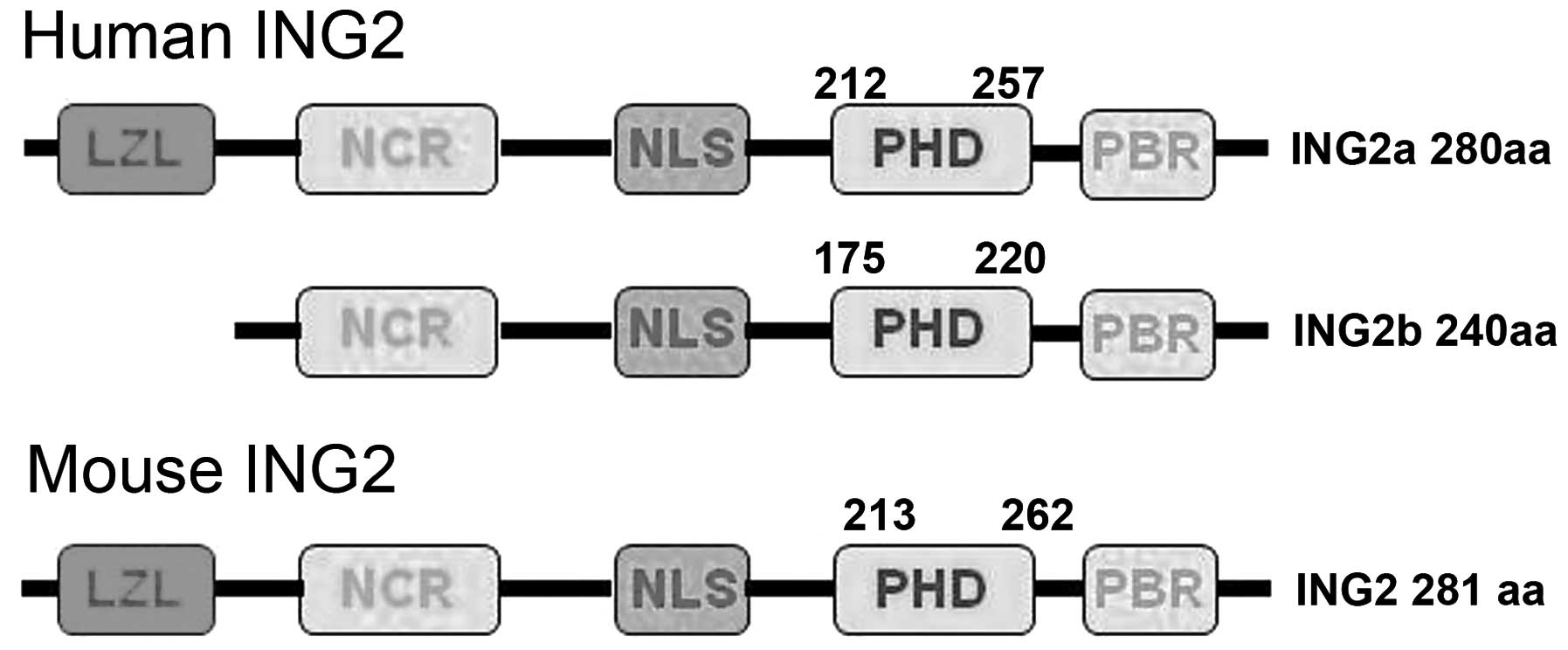
The gene encoding inhibitor of growth protein 2
(ING2) is made up of three exons (1a, 1b and 2) and encodes two
transcript isoforms (ING2a and ING2b) by alternative splicing. The
ING2 C-terminal domain contains a plant homeodomain (PHD) with a
zinc-binding motif, which has a high affinity for the histone 3
tri-methylated on lysine 4 and a nuclear localization sequence
(NLS) (1). Initially, ING2a was
shown to regulate cell proliferation, cell-cycle arrest, senescence
and apoptosis due to enhancing p53 transcription and acetylation on
lysine 382 by interaction with p300 acetyltransferase, which
promotes p53 target genes (eg. p21 and Bcl-2 associated X protein)
(2–4). In response to exogenous stresses, the
p38 kinase pathway is activated to regulate phosphorylation of
phosphatidylinositol 5-phosphate, resulting in an induction of ING2
accumulation to the chromatin and subsequent modulation of p53
acetylation and ING2 targets (3–7).
ING2 protein recruits and stabilizes the mSin3a/higher histone
deacetylase 1 (HDAC)1–2/Sin3a associated protein complex, and
regulates target gene expression (1), which has been enhanced by
phosphorylation-dependent ING2a sumoylation by small ubiquitin-like
modifier 1 on lysine 195 (8),
while ubiquitin ligase SMAD specific E3 ubiquitin protein ligase 1
targets the tumor suppressor ING2 for ubiquitination and
degradation (9). In addition,
ING2a binds to Ski-like oncogene (SnoN), a SMAD interacting
transcriptional modulator, through its PHD domain to finally
suppress cell proliferation, suggesting that ING2 collaborates with
SnoN to mediate transforming growth factor β-induced SMAD-dependent
transcription and cellular responses (10).
ING2a mRNA is highly expressed in skeletal muscle,
lung, thymus, pancreas and testis (11,12).
Although human ING2a and 2b have a molecular weight of 33 and 28
kDa, respectively, ING2b protein has never been experimentally
detected (1,12,13).
Saito et al (12) have
reported that ING2−/− mice develop soft-tissue sarcomas with an
incidence of 46%, among which histiocytic sarcomas were most
frequent (28%). The ING2−/− male mice appeared infertile because of
deficient spermatogenesis, small testes, seminiferous tubule
degeneration, abnormal spermatozoal motility and morphology from
the age of eight weeks. A high frequency of loss of heterozygosity
(LOH) of the ING2 chromosomal region 4q32-35.1 has been found in
basal cell carcinomas (14), head
and neck squa mous cell carcinomas (15,16),
and hepatocellular carcinomas (HCC) (17). LOH of ING2 is associated with
advanced tumor stages in head and neck squamous cell carcinomas
(15). ING2 expression is
downregulated in various cancer types, including melanoma and HCC
(17), non small cell lung
carcinomas (18) and melanoma
(19). The expression levels of
ING2 protein were negatively correlated with tumor size,
histopathological classification and serum alpha-fetoprotein in
HCC, but may be employed as an independent prognostic factor to
indicate favorable prognosis (17). By contrast, Kumamoto et al
(20) demonstrated that ING2 mRNA
overexpression was associated with colon carcinogenesis by
upregulating the expression of matrix metalloproteinase 13 via the
formation of an ING2-HDAC1-mSin3A complex.
Identification of normal tissues or cell types which
express ING2 may contribute towards clarifying the physiological
function of ING2, while the elucidation of its expression pattern
as well as its heterogeneity among tumor cases or between tumor and
normal tissues may lead to its identification as a target for gene
therapy, which may be evaluated using animal models of conditional
ING2 knockout (1). A previous
study employed intermittent microwave irradiation for
immunohistochemical analysis of ING2, during which >2.4 billion
vibrations per minute enhanced the probability of specific
antibody-antigen recognition (21). The present study adopted this
technique to perform immunohistochemical expression profiling of
ING2 protein in normal mouse and human tissues as well as in human
cancer tissues.
Materials and methods
Specimens
Three male and three female C57BL/6 mice (eight
weeks old; 25 g) were sacrificed under sodium pentobarbital
anesthesia and brain, heart, liver, spleen, lung, kidney, breast,
stomach and intestinal tissue samples were excised. All tissues
were fixed in 10% neutral formalin and embedded in paraffin. An
array of normal human tissues (cerebrum, cerebellum, brain stem,
aorta, tongue, thyroid, esophagus, stomach, intestine, liver,
pancreas, lung, trachea, appendix, smooth, muscle, skeletal muscle,
heart, testis, bladder and prostate) and cancer tissues (62
hepatocellular carcinomas, 62 renal clear cell carcinomas, 62
pancreatic carcinomas, 45 esophageal squamous cell carcinomas and
31 cervical squamous cell carcinomas) were purchased from Shanghai
Outdo Biotech Co., Ltd. (Shanghai, China). Normal human cervical,
endometrial, ovary and breast tissues, as well as breast (n=144),
gastric (n=196), colorectal (n=96), ovarian (n=208), endometrial
(n=96) and lung carcinoma (n=192) tissues were obtained at The
First Affiliated Hospital of Liaoning Medical University (Jinzhou,
China). The normal mouse and human tissues, and the cancer tissues
were also subjected to a tissue microarray using a tissue
microarray apparatus (KIN-1; Azumaya, Tokyo, Japan). No patients
with cancer had undergone chemotherapy, radiotherapy, or adjuvant
treatment prior to surgery. The patients or their relatives
provided written consent for the use of tumor tissues for clinical
research, and the research protocol was approved by the Ethical and
Animal Experimentation Committees of Liaoning Medical University
(Jinzhou, China).
Immunohistochemistry
Consecutive sections were de-waxed with xylene,
debenzolization with ethanol in water and subjected to antigen
retrieval by irradiation in target retrieval solution (DAKO,
Glostrup, Denmark) in a microwave oven for 15 min (500 W; Oriental
Rotor Ltd Co., Tokyo, Japan). Sections were then blocked in 5%
bovine serum albumin (Shanghai Chemical Technology Co., Ltd.,
Shanghai, China) for 5 min to prevent non-specific antibody
binding. The sections were incubated with rabbit anti-ING2 antibody
(cat no. 11560-1-AP; Proteintech, Chicago, IL USA; 1:50 dilution)
for 15 min, followed by incubation with anti-rabbit secondary
antibody conjugated to horseradish peroxidase (DAKO) for 15 min.
All of the incubations were cross-linked in a microwave oven at
37°C in order to allow intermittent irradiation as previously
described (21). After each
treatment, the slides were washed with Tris-buffered saline
containing Tween 20 (3×1 min). Bound antibodies were then stained
using 3,3′-diaminobenzidine (Sangon Biotech Co., Ltd., Shanghai,
China). After counterstaining with Mayer's hematoxylin, the
sections were de-hydrated, cleared and mounted. Normal mouse
immunoglobulin G (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) was used instead of the primary antibody as a negative
control.
Immunostaining evaluation
As indicated in Figs.
1Figure 2–3, ING2 protein was localized to the
cytoplasm or/and nuclei. Initially, the high-expression field was
selected at low magnification and one hundred cells were randomly
counted from five different representative fields at a high
magnification by two independent examiners (Miss S Zhao and
Professor HC Zheng). In the case of conflicting data, the two
examiners continued until a final agreement was reached. The
percentages of counted cells were scored as follows: 0–10%,
negative (−); and 11–100%, positive (+). The density of
immunostaining was assessed using ImagePro Plus software, version
16.0 (Media Cybernetics, Rockville, MD, USA). The percentage of
ING2-positive cells was graded semi-quantitatively according to a
four-tier scoring system: (−), negative; (+), weakly positive; (++)
moderately positive; and (+++) strongly positive.
Results
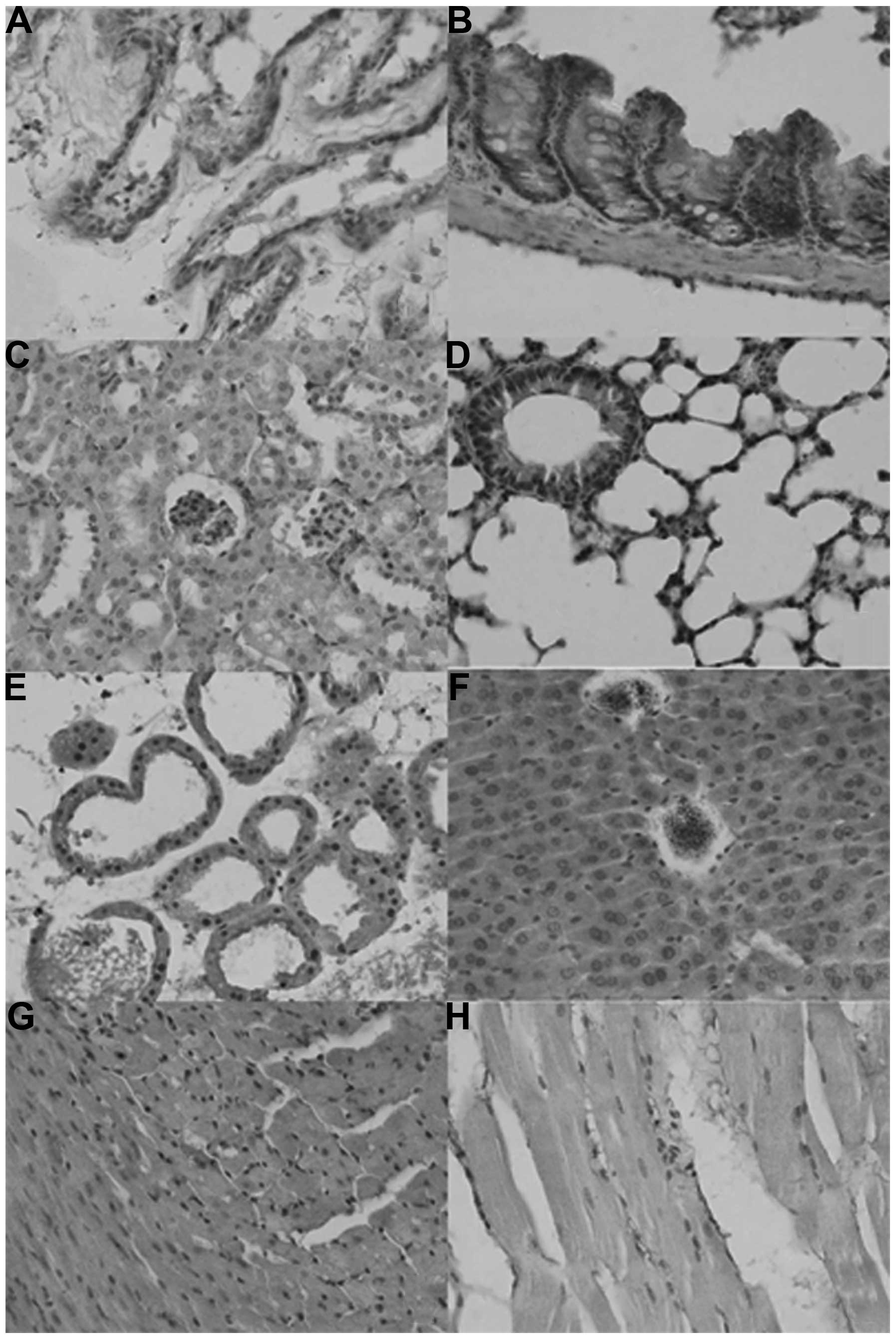
As shown in Fig. 1,
the sub-cellular location of ING2 was in the nuclei, cytoplasm or
nucleocytoplasm in the mouse tissues in either sporadic or
localized patterns, although expression levels differed among
tissues and cell populations. ING2 reactivity was detectable in the
nuclei as well as the cytoplasm of the glandular epithelium of the
breast, liver, intestine, bronchium and alveolum, the squamous
epithelium of the skin, glomeruli, and in myocardial cells, while
it was located in the cytoplasm of renal tubules and striated
muscle cells. ING2 protein was scattered in the brain and spleen
(Table I).
 | Table IImmunohistochemical detection of
inhibitor of growth protein 2 expression in normal mouse
tissues. |
Table I
Immunohistochemical detection of
inhibitor of growth protein 2 expression in normal mouse
tissues.
| Tissue source | Tissue type | Localization |
|---|
| Brain | | Scattered |
| Heart | Myocardial
fibroblasts | Nuclei and
cytoplasm |
| Lung | Bronchial glandular
epithelial cells | Nuclei and
cytoplasm |
| Alveolar glandular
epithelial cells | Nuclei and
cytoplasm |
| Kidney | Renal tubules | Cytoplasm |
| Glomeruli | Nuclei and
cytoplasm |
| Intestine | Glandular
epithelium | Nuclei and
cytoplasm |
| Spleen
endothelium | | Scattered |
| Skin | Squamous
epithelium | Nuclei and
cytoplasm |
| Striated muscle | Striated muscle
cells | Cytoplasm |
| Liver | Glandular
epithelium | Nuclei and
cytoplasm |
| Breast | Glandular
epithelium | Nuclei and
cytoplasm |
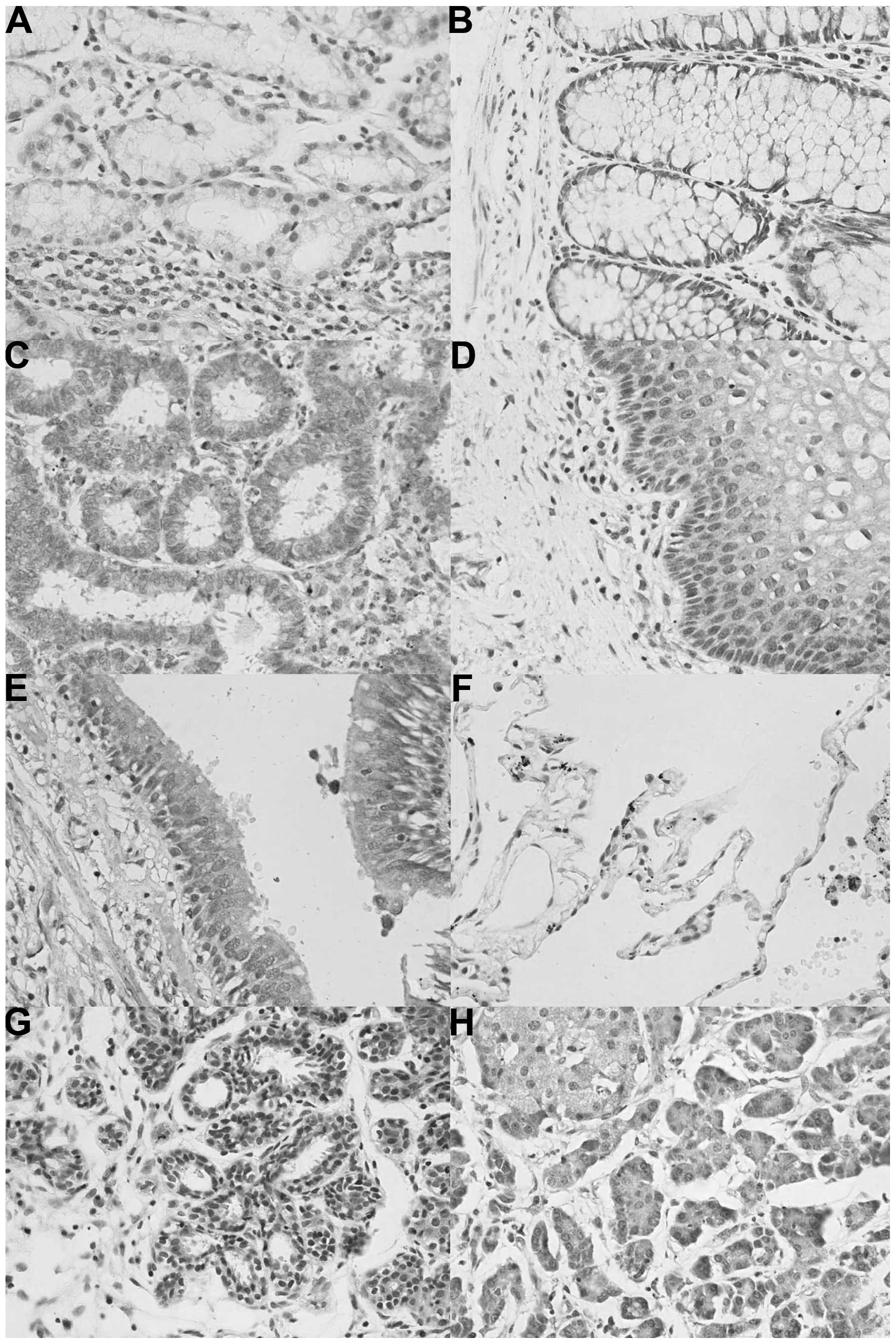
In human tissues, ING2 protein was primarily
distributed in the cytoplasm; however, it was located in the
cytoplasm and nuclei of tissues from the stomach, intestine,
cervix, trachea, breast and pancreas. In the stomach, the nuclear
location of ING2 was more prominent than that in the cytoplasm
(Fig. 2). According to the density
of the immunostaining, ING2 was highly expressed in the tongue,
stomach, skin, pancreas, cervix and breast, whereas it was weakly
expressed in the brain stem, thymus, thyroid, lung, striated
muscle, testis, bladder and ovary (Table II).
 | Table IIImmunohistochemical detection of ING2
expression and localization in normal human tissues. |
Table II
Immunohistochemical detection of ING2
expression and localization in normal human tissues.
| Tissue type | ING2 expression
|
|---|
| Nucleus | Cytoplasm |
|---|
| Cerebrum | − | ++ |
| Cerebellum | − | ++ |
| Brain stem | − | + |
| Thymus | − | + |
| Skeletal muscle | − | ++ |
| Aorta | − | ++ |
| Tongue | +++ | +++ |
| Thyroid | − | + |
| Esophagus | ++ | ++ |
| Stomach | +++ | ++ |
| Intestine | ++ | ++ |
| Liver | − | ++ |
| Pancreas | + | +++ |
| Lung | − | + |
| Trachea | − | ++ |
| Skin | ++ | +++ |
| Appendix | + | ++ |
| Muscle | − | + |
| Striated
muscle | − | + |
| Smooth muscle | − | ++ |
| Testis | − | + |
| Bladder | − | + |
| Prostate | − | ++ |
| Cervix | ++ | +++ |
| Endometrium | +++ | ++ |
| Ovary | − | + |
| Breast | +++ | ++ |
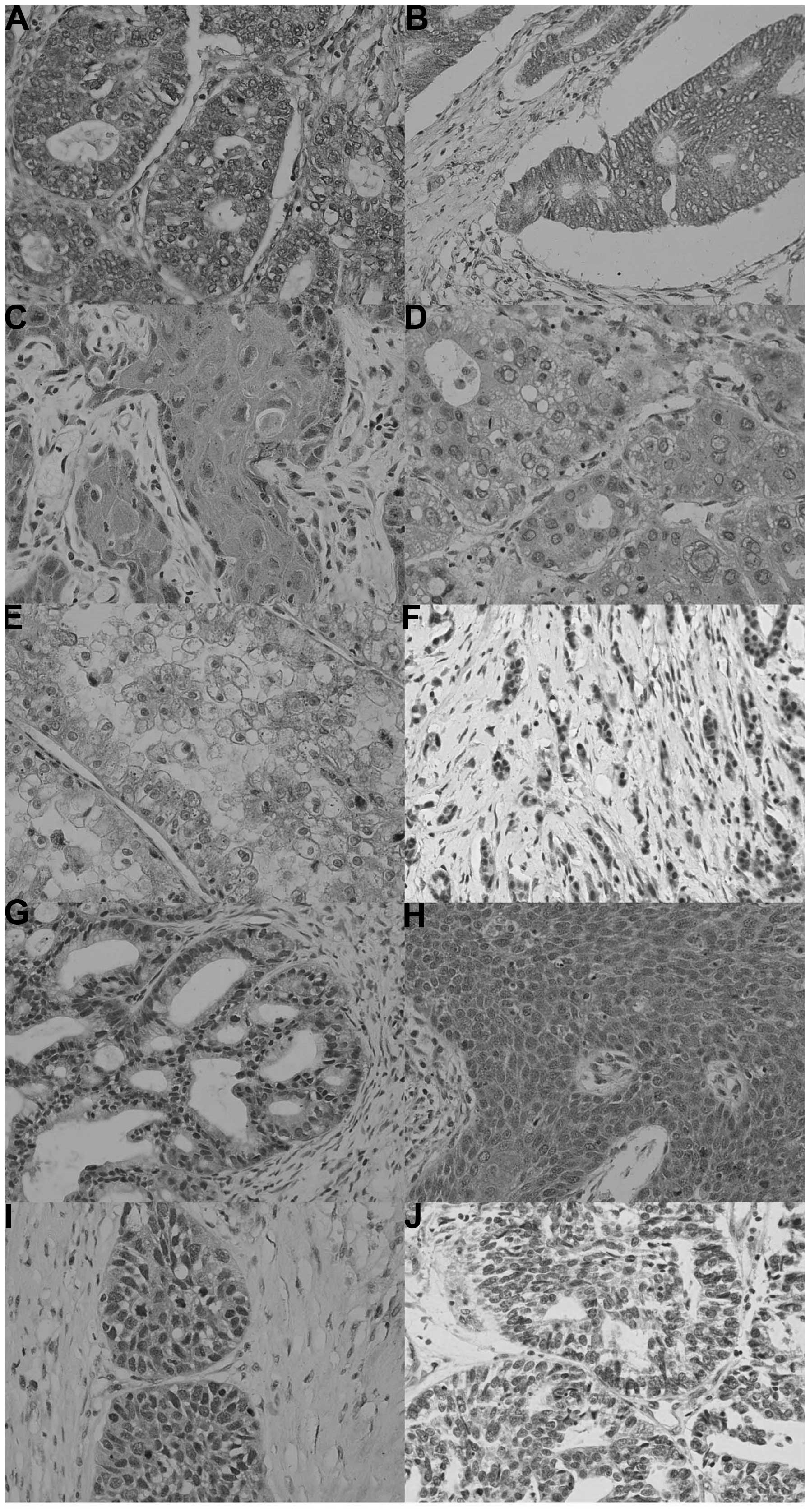
In total, 617 out of 1,194 cancer tissues assessed
in the present study were ING2-positive (51.7%), with a homogeneity
in their expression pattern (Fig.
3; Table III). ING2
expression was detected in the cytoplasm of all cancer types,
whereas it was present in the nuclei of certain cancer tissues,
including lung, breast and endometrial cancers. Among them, ING2
was more frequently expressed in breast cancer (67.4%; 97/144) and
gynecological cancers, including ovarian cancer (61.5%; 128/208)
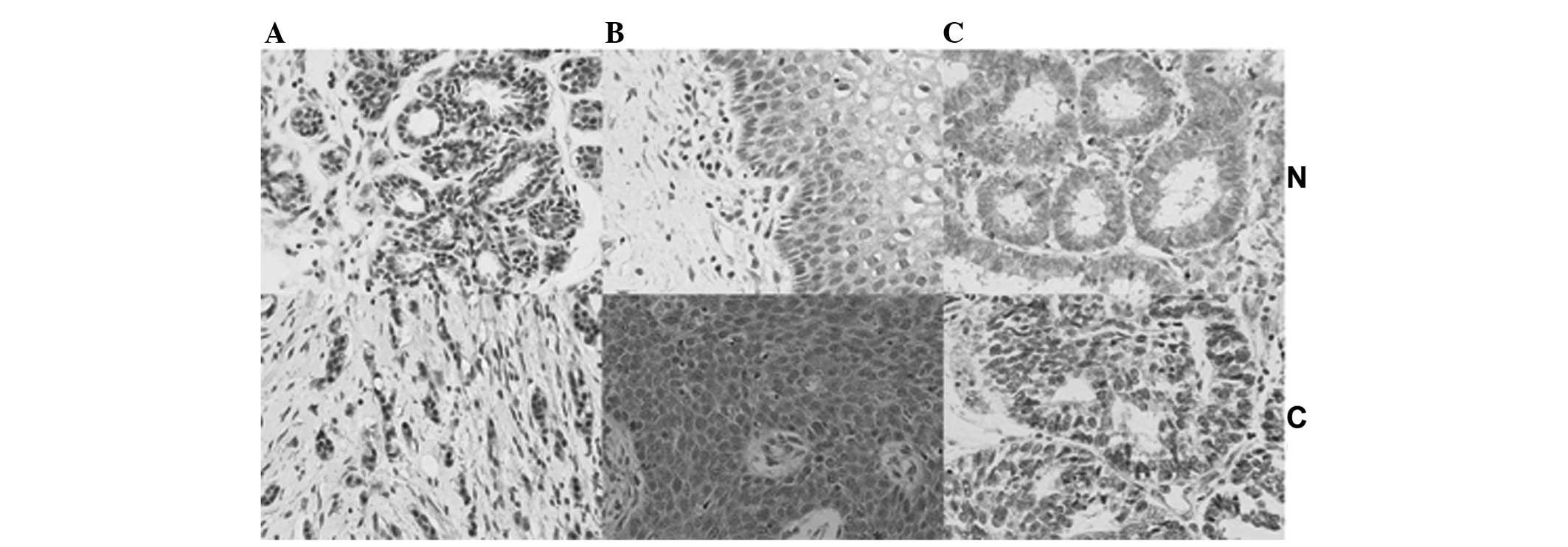
and endometrial cancer (57.3%; 55/96). Compared with their
respective normal tissues, ING2 expression in breast cancer tissues
was decreased, while it was upregulated in the nuclei as well as in
the cytoplasm of cervical cancer tissues, whereas ING2 was
increased in the nuclei and declined in the cytoplasm of
endometrial cancer tissues (Fig.
4). By contrast ING2-positive cases were less frequent in renal
clear cell carcinoma (17.7%; 11/62).
 | Table IIIImmunohistochemical detection of ING2
expression in human cancer tissues. |
Table III
Immunohistochemical detection of ING2
expression in human cancer tissues.
| Cancer type | n | Positive cases
(n) | PR (%) | ING2 expression
|
|---|
| Nucleus | Cytoplasm |
|---|
| Hepatocellular
carcinoma | 62 | 25 | 40.3 | − | + |
| Renal cell
carcinoma | 62 | 11 | 17.7 | − | + |
| Pancreatic
cancer | 62 | 20 | 32.3 | − | + |
| Esophageal
carcinoma | 45 | 19 | 42.2 | + | + |
| Cervical
carcinoma | 31 | 15 | 48.4 | + | + |
| Breast cancer | 144 | 97 | 67.4 | + | + |
| Gastric
carcinoma | 196 | 85 | 43.4 | + | + |
| Colorectal
carcinoma | 96 | 56 | 58.3 | + | + |
| Ovarian cancer | 208 | 128 | 61.5 | + | + |
| Endometrial
carcinoma | 96 | 55 | 57.3 | + | + |
| Lung carcinoma | 192 | 106 | 55.2 | + | + |
Discussion
ING2 has been identified and characterized as a
Type-II tumor suppressor gene as its involvement in cancer appears
to be linked to its level of expression rather than its mutational
status (1,18). ING2 protein contains NLS and PHD
finger motifs in its C-terminus (1). In the present study, the expression
levels and cellular localization of ING2 protein were determined in
a tissue array comprising normal mouse and human tissues, as well
as human cancer tissues. Positive expression of ING2 was observed
in the cytoplasm of normal mouse and human tissues as well as human
cancer tissues; however, ING2 was present in the cytoplasm as well
as in the nuclei in certain tissue types. It has been reported that
LOH of the ING2 chromosomal region or the ING2 gene may result in
the downregulation of its expression (14–17).
Cenzig et al (22) found
that the mutation or deletion of ING5 NLS is responsible for the
nucleocytoplasmic translocation. Zhang et al (17) demonstrated that immunostaining for
ING2 was mostly located to the cytoplasm, while weak nuclear
staining was also observed in HCC tissues and normal hepatocytes,
in line with a study by Gozani et al (5), who reported that phosphatidylinositol
5-phosphate 4-kinase type II β expression decreased endogenous
nuclear ING2 in HT1080 fibrosarcoma cells. It has been reported
that ING1 phosphorylation by 14-3-3 family members (23) or Src (24) caused its cytoplasmic
re-localization leading to apoptotic induction. Future studies
should investigate the purpose of ING2 restoration in the cytoplasm
as well as its function.
Amino acid sequence alignment has revealed a high
similarity between human ING2a and mouse ING2 (96% identity)
(Fig. 5) (1); this may explain for the findings of
the present study, which revealed no substantial differences in the
patterns of ING2 expression between mouse and human samples, with
the exception of sub-cellular location in certain cell types. In
human tissues, high expression of ING2 protein was detected in the
tongue, stomach, skin, pancreas, cervix and breast, while its
expression was low in the brain stem, thymus, thyroid, lung,
striated muscle, testis, bladder and ovary, suggesting the
functional involvement of ING2 in the functions of specific cell
types or in cells in a specific functional state. In order to
further investigate the roles of ING2, our group has inhibited ING2
using a cell-specific promoter in order to establish an animal
model of ING2-negative cancer. Previous studies have highlighted
the anti-proliferative and apoptotic functions of ING2 (4,8).
ING2 overexpression in the stomach, tongue, skin, pancreas, cervix
and breast may indicate its association with regenerative processes
regardless of the identity of the cells being glandular or squamous
epithelium, which is supported by the finding of relatively low
expression in organs with a decreased capacity for cell repair and
renewal, including the brain stem, thymus, thyroid, skeletal muscle
and testis. Of note, ING2 has been involved in muscle
differentiation by regulating myogenin transcription (25), providing an explanation for ING2
overexpression in muscle cells.
ING2 is a candidate tumor suppressor gene, whose
expression is frequently lost in tumors. The present study focused
on common epithelial cell-derived tumors and demonstrated that
breast cancer and gynecological cancer types, including ovarian and
endometrial carcinoma, showed a high incidence of ING2 expression,
indicating that ING2 protein may be closely linked to estrogen
production. By contrast, the positive rate for ING2 expression in
renal clear cell carcinoma was <20%. This finding may indicate
that ING2 may be used as a therapeutic target in renal clear cell
carcinoma. ING2 was reported to interact with proliferating cell
nuclear antigen and regulates its quantity of the chromatin
fraction, thereby directly maintaining DNA replication and
integrity (26,27). The ING2 C-terminus recruits histone
methyltransferase activity and trimethylates histone H3 at lysine 9
(28). The leucine zipper-like
motif of ING2 is critical for the proper functioning of ING2 in DNA
repair, apoptosis and p53-dependent chromatin remodeling by acting
as a scaffold protein to mediate the interaction between p53 and
p300 (3–7). The profiling of ING2 expression
performed in the present study provided useful hints regarding the
disruption of DNA replication, proliferation and apoptosis in
various epithelial cancer types.
In conclusion, the present study not only elucidated
the abundant expression of ING2 in normal mouse and human tissues
as well as human cancer tissues, but also demonstrated the
differential expression and/or sub-cellular location of ING2 among
various tissues, cell types and single cells, suggesting
differential functional involvement. According to the results of
the present study, it is hypothesized that ING2 may be involved in
the repair and regeneration of organs or tissues and have an
important role in gynecological carcinogenesis.
Acknowledgments
The present study was supported by the Natural
Science Foundation of Liaoning Province (no. 2013022070) and the
National Natural Scientific Foundation of China (no. 81172371).
References
|
1
|
Guérillon C, Larrieu D and Pedeux R: ING1
and ING2: Multifaceted tumor suppressor genes. Cell Mol Life Sci.
70:3753–3772. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pedeux R, Sengupta S, Shen JC, Demidov ON,
Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfeld SH, McMenamin
M, et al: ING2 regulates the onset of replicative senescence by
induction of p300-dependent p53 acetylation. Mol Cell Biol.
25:6639–6648. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bua DJ, Martin GM, Binda O and Gozani O:
Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability
at discrete chromatin targets in response to DNA damage. Sci Rep.
3:21372013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Larrieu D, Ythier D, Brambilla C and
Pedeux R: ING2 controls the G1 to S-phase transition by regulating
p21 expression. Cell Cycle. 9:3984–3990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gozani O, Karuman P, Jones DR, Ivanov D,
Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al:
The PHD finger of the chromatin-associated protein ING2 functions
as a nuclear phosphoinositide receptor. Cell. 114:99–111. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang W, Zhang H, Davrazou F, Kutateladze
TG, Shi X, Gozani O and Prestwich GD: Stabilized
phosphatidylinositol-5-phosphate analogues as ligands for the
nuclear protein ING2: Chemistry, biology and molecular modeling. J
Am Chem Soc. 129:6498–6506. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jones DR, Bultsma Y, Keune WJ, Halstead
JR, Elouarrat D, Mohammed S, Heck AJ, D'Santos CS and Divecha N:
Nuclear PtdIns5P as a transducer of stress signaling: An in vivo
role for PIP4Kbeta. Mol Cell. 23:685–695. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ythier D, Larrieu D, Binet R, Binda O,
Brambilla C, Gazzeri S and Pedeux R: Sumoylation of ING2 regulates
the transcription mediated by Sin3A. Oncogene. 29:5946–5956. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nie J, Liu L, Wu M, Xing G, He S, Yin Y,
Tian C, He F and Zhang L: HECT ubiquitin ligase smurf1 targets the
tumor suppressor ING2 for ubiquitination and degradation. FEBS
Lett. 584:3005–3012. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sarker KP, Kataoka H, Chan A, Netherton
SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K and Bonni S: ING2
as a novel mediator of transforming growth factor-beta-dependent
responses in epithelial cells. J Biol Chem. 283:13269–13279. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walzak AA, Veldhoen N, Feng X, Riabowol K
and Helbing CC: Expression profiles of mRNA transcript variants
encoding the human inhibitor of growth tumor suppressor gene family
in normal and neoplastic tissues. Exp Cell Res. 314:273–285. 2008.
View Article : Google Scholar
|
|
12
|
Saito M, Kumamoto K, Robles AI, Horikawa
I, Furusato B, Okamura S, Goto A, Yamashita T, Nagashima M, Lee TL,
et al: Targeted disruption of Ing2 results in defective
spermatogenesis and development of soft-tissue sarcomas. PLoS One.
5:e155412010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagashima M, Shiseki M, Miura K, Hagiwara
K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K and Harris CC:
DNA damage-inducible gene p33ING2 negatively regulates cell
proliferation through acetylation of p53. Proc Natl Acad Sci USA.
98:9671–9676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sironi E, Cerri A, Tomasini D, Sirchia SM,
Porta G, Rossella F, Grati FR and Simoni G: Loss of heterozygosity
on chromosome 4q32–35 in sporadic basal cell carcinomas: Evidence
for the involvement of p33ING2/ING1L and SAP30 genes. J Cutan
Pathol. 31:318–322. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borkosky SS, Gunduz M, Nagatsuka H, Beder
LB, Gunduz E, Ali MA, Rodriguez AP, Cilek MZ, Tominaga S, Yamanaka
N, et al: Frequent deletion of ING2 locus at 4q35.1 associates with
advanced tumor stage in head and neck squamous cell carcinoma. J
Cancer Res Clin Oncol. 135:703–713. 2009. View Article : Google Scholar
|
|
16
|
Cetin E, Cengiz B, Gunduz E, Gunduz M,
Nagatsuka H, Bekir-Beder L, Fukushima K, Pehlivan D, N MO,
Nishizaki K, et al: Deletion mapping of chromosome 4q22–35 and
identification of four frequently deleted regions in head and neck
cancers. Neoplasma. 55:299–304. 2008.
|
|
17
|
Zhang HK, Pan K, Wang H, Weng DS, Song HF,
Zhou J, Huang W, Li JJ, Chen MS and Xia JC: Decreased expression of
ING2 gene and its clinicopathological significance in
hepato-cellular carcinoma. Cancer Lett. 261:183–192. 2008.
View Article : Google Scholar
|
|
18
|
Ythier D, Brambilla E, Binet R, Nissou D,
Vesin A, de Fraipont F, Moro-Sibilot D, Lantuejoul S, Brambilla C,
Gazzeri S and Pedeux R: Expression of candidate tumor suppressor
gene ING2 is lost in non-small cell lung carcinoma. Lung Cancer.
69:180–186. 2010. View Article : Google Scholar
|
|
19
|
Lu F, Dai DL, Martinka M, Ho V and Li G:
Nuclear ING2 expression is reduced in human cutaneous melanomas. Br
J Cancer. 95:80–86. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kumamoto K, Fujita K, Kurotani R, Saito M,
Unoki M, Hagiwara N, Shiga H, Bowman ED, Yanaihara N, Okamura S, et
al: ING2 is upregulated in colon cancer and increases invasion by
enhanced MMP13 expression. Int J Cancer. 125:1306–1315. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumada T, Tsuneyama K, Hatta H, Ishizawa S
and Takano Y: Improved 1-h rapid immunostaining method using
intermittent microwave irradiation: Practicability based on 5 years
application in Toyama medical and pharmaceutical university
hospital. Mod Pathol. 17:1141–1149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cengiz BI, Gunduz E, Gunduz M, Beder LB,
Tamamura R, Bagci C, Yamanaka N, Shimizu K and Nagatsuka H:
Tumor-specific mutation and downregulation of ING5 detected in oral
squamous cell carcinoma. Int J Cancer. 127:2088–2094. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong W, Russell M, Suzuki K and Riabowol
K: Subcellular targeting of p33ING1b by phosphorylation-dependent
14–3–3 binding regulates p21WAF1 expression. Mol Cell Biol.
26:2947–2954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu L, Thakur S, Leong-Quong RY, Suzuki K,
Pang A, Bjorge JD, Riabowol K and Fujita DJ: Src regulates the
activity of the ING1 tumor suppressor. PLoS One. 8:e609432013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eapen SA, Netherton SJ, Sarker KP, Deng L,
Chan A, Riabowol K and Bonni S: Identification of a novel function
for the chromatin remodeling protein ING2 in muscle
differentiation. PLoS One. 7:e406842012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Larrieu D, Ythier D, Binet R, Brambilla C,
Brambilla E, Sengupta S and Pedeux R: ING2 controls the progression
of DNA replication forks to maintain genome stability. EMBO Rep.
10:1168–1174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Goeman F, Otto K, Kyrylenko S, Schmidt O
and Baniahmad A: ING2 recruits histone methyltransferase activity
with methylation site specificity distinct from histone H3 lysines
4 and 9. Biochim Biophys Acta. 1783:1673–1680. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Wang J and Li G: Leucine
zipper-like domain is required for tumor suppressor ING2-mediated
nucleotide excision repair and apoptosis. FEBS Lett. 580:3787–3793.
2006. View Article : Google Scholar : PubMed/NCBI
|