Introduction
Diabetes mellitus (DM) is the most severe metabolic
disease in the developed world, affecting a large number of people.
A major symptom of DM is hyperglycemia, which leads to severe
complications (1). Among patients
with DM, ~15% exhibit impaired skin wound healing (2), and high blood sugar is linked to skin
ulceration by altering angiogenesis (3), but the underlying mechanism remains
unclear.
Skin wound repair requires the coordination of
several cell types, including keratinocytes, fibroblasts,
endothelial cells, macrophages and platelets. Fibroblast cell
proliferation and migration, collagen deposition and remodeling,
wound contraction and angiogenesis are important steps during wound
repair (4,5). Extracellular matrix (ECM) forms the
largest component of the dermal skin layer, therefore, the repair
of damaged ECM is a key step for wound healing (6). Fibroblasts constitute one of the
important cell layers that participate in the production and
remodeling the ECM and fibroblast proliferation and migration are
important for the formation of granulation tissue and further skin
repair (7,8). The impaired wound healing during DM
is attributed to altered protein and lipid metabolism and the
associated abnormal formation of granulation tissue (9). Higher glucose levels in the blood
result in abnormal attachment of aldose sugars to a protein or
lipid, which affects normal glycosylation modifications (9). The aberrantly glycosylated products
[advanced glycation end products (AGEs)] then accumulate in cells.
AGEs attached to ECM proteins may lead to a reduction in their
turnover rate (9). Nitric oxide
(NO) is an important mediator of cell proliferation, maturation and
differentiation and serves a key role in wound healing (10). Fibroblasts isolated from diabetic
ulcers are usually large and widely spread during in vitro
culture compared with normal fibroblasts in age-matched controls.
They often exhibit abnormal endoplasmic reticulum, increased
numbers of vesicular bodies and lost microtubular structure.
Therefore, DM affects protein turnover, autonomous trafficking and
normal protein secretion in diabetic ulcer fibroblasts (11,12).
Fibroblasts from diabetic ulcers have defects in cell
proliferation, which may result in decreased ECM protein production
and further delayed wound healing (13). High glucose-induced fibroblast
migration was previously identified to be a result of reduced JNK
activity (14). However, few
molecular studies have investigating the underlying mechanisms of
DM-mediated fibroblast cell damage.
In the present study, RNA sequencing (RNA-Seq) was
used to analyze the alterations of large numbers of transcripts
following HG stimulation of human fibroblast cells, and the genes
and pathways associated with HG stress were identified.
Additionally, the inflammatory response pathway and Wnt signaling
were further analyzed for their role in the protection of
fibroblasts from HG damage. The results of current study may be
important for understanding the mechanisms of DM-mediated skin
ulceration and may provide a theoretical basis for repair of skin
damage in patients with DM in the future.
Materials and methods
Human foreskin fibroblast cell
culture
Human fibroblast cells were isolated and
subsequently cultured for analysis of the effects of HG treatment.
All the procedures followed for purification and culture of human
fibroblasts were described by Xuan et al (14). Human foreskin samples were
collected from 3 patients in Department of Dermatology, the First
Affiliated Hospital, Wenzhou Medical University (Wenzhou, China).
This study was approved by the ethics committee of Wenzhou Medical
University (Wenzhou, China) and written informed consent was
obtained from all the patients involved. The fat was removed from
the tissue and was cut into 3 by 2 mm strips and were incubated
overnight at 4°C in 0.05% Dispase I (Sigma-Aldrich, St. Louis, MO,
USA). The epidermis was removed and the dermis was placed in
25-cm2 flasks pre-treated with FBS, and placed
horizontally for 1 h and then vertically for 3 h in a culture
chamber with 5% CO2 at 37°C. The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA) containing 5.5 mM glucose with 10%
fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences) and
1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific Inc.,
Watham, MA, USA) the medium was changed every 3 days. When cell
confluence reached 70–80% the cells were digested and passaged with
0.25% trypsin (Gibco; Thermo Fisher Scientific Inc.) Cells were
cultured for 3 days in 5.5 mM glucose medium and transferred to the
media containing either 5.5 mM glucose (LG) or 30 mM glucose (HG).
Cells at passage 3–6 were used for the LG and HG treatment. Cells
were harvested after 1 h of LG and HG treatment.
Cell proliferation assay
Cell proliferation was assayed using a Cell Counting
Kit-8 (CCK-8) kit (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). The fibroblast cell culture and measurement of cell
densities under different treatments were followed as previously
described (14). In summary, 50 ml
of the cell resuspension solution (1×103 cells/well)
were transferred into 96-well plates following digestion with
trypsin, and five parallel wells were used for each treatment.
Subsequent to attachment to the culture plate, the cells were
subjected to the different glucose treatments for 72 h in a 5%
CO2 incubator at 37°C. Then, 5 ml of CCK-8 was added to
each well, and the cells were cultured for another 3 h. Cell
density was determined by quantifying the absorbance at 450 nm
using a Varioskan Flash Multimode Reader (Thermo Fisher Scientific,
Inc.) using the following formula: Cell dens
ity=(Acell+CCK8+medium-ACCK8+medium/Acell+CCK8-ACCK8+medium)
× 100.
RNA deep sequencing
Total RNA was extracted from human foreskin
fibroblasts for RNA-Seq experiments following treatment with a low
(5.5 mM) or high (30 mM) concentration of glucose. RNA-Seq
experiments and data analysis were performed by the NovelBio
Bio-Pharm Technology Co., Ltd. (Shanghai, China). The RNA-Seq data
is available upon request.
Analysis of the pathway and gene ontology
(GO) category
Differentially expressed genes were identified by
analyzing for association with biological process gene ontology
(GO) terms (15). Fisher's exact
test was used to classify the GO category, and the false discovery
rate (FDR) was calculated to correct the P-value (16). Enrichment of GO members among
differentially expressed gene sets was identified using the
one-tailed Fisher's exact test for 2×2 contingency tables (17), which measures the significance of
the function that as the enrichment increases, the corresponding
function is more specific, which aids the identification of GOs
with a more concrete function description in the experiment.
Pathway analysis was used to determine the significant pathways of
the differential genes according to Kyoto Encyclopedia of Genes and
Genomes (KEGG) (18), BioCarta
(http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways and
Reactome (19). Fisher's exact
test was followed by Benjamini-Hochberg multiple testing correction
to select the significant pathway and the threshold of significance
was defined by P-value and FDR (20).
Total RNA extraction, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from stimulated or
unstimulated fibroblasts treated with high-concentration glucose
(30 mM), Bay11-7082 (0.5 μM; Sigma-Aldrich) or inhibitor of
Wnt response (IWR) (0.5 μM; Sigma-Aldrich). The cell
monolayer was rinsed with ice-cold phosphate-buffered saline once.
Each sample was treated with RQ1-DNAse (Promega Corporation,
Madison, WI, USA). The cells were then lysed directly in a culture
dish by adding 1 ml TRIzol (Thermo Fisher Scientific, Inc.) per
each 3.5 cm diameter dish, scraped with a cell scraper and then 0.2
ml chloroform was added per 1 ml TRIzol. RNA (2 μg) was
reverse transcribed at 42°C for 60 min, 70°C for 5 min and
following stop the reaction at 8°C using a GoScript Reverse
Transcription System (Promega Corporation) following the
manufacturer's protocol. A SYBR Green Master Mix (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used to perform the qPCR
on an Illumina Eco 3.0 (Illumina, Inc., San Diego, CA, USA). A
typical reaction consisted of an initial denaturation at 95°C for 3
min, followed by 40 cycles of denaturation for 30 sec at 95°C,
annealing for 30 sec at 58°C, and extension at 72°C for 30 sec,
followed by a final extension at 72°C for 5 min. The transcription
levels were normalized against those of GAPDH using the
2−ΔΔCq method (21).
The gene-specific primer sequences used for RT-qPCR are described
in Table I. Each experiment was
repeated at least 3 times. A unreversed transcribed RNA was used as
a PCR template control.
 | Table IReverse transcription-quantitative
polymerase chain reaction primer sequences. |
Table I
Reverse transcription-quantitative
polymerase chain reaction primer sequences.
| Primer | Sequence |
|---|
| Caspase 3 | F:
TGATGATGACATGGCGTGTC |
| R:
GTTGCCACCTTTCGGTTAAC |
| PAI1 | F:
GAGACTGAAGTCGACCTCAG |
| R:
CTGTCCATGATGATCTCCTC |
| GAPDH | F:
GACCTGCCGTCTAGAAAAAC |
| R:
CTGTAGCCAAATTCGTTGTC |
| CCL13 | F:
CGTCCCATCTACTTGCTGCT |
| R:
TCAAGTCTTCAGGGTGTGAGC |
| IL8 | F:
GGTGCAGTTTTGCCAAGGAG |
| R:
TTCCTTGGGGTCCAGACAGA |
| FZD8 | F:
CTGGTGGAGATCCAGTGCTC |
| R:
TTGTAGTCCATGCACAGCGT |
| EGR2 | F:
TCGCAAGTACCCCAACAGAC |
| R:
CTCATCACTCCGGGCAAACT |
Western blot analysis
For extraction of total protein, the cells were
lysed in an ice-cold lysis solution containing 7 M urea, 2
Mthiourea, 2% CHAPS detergent, 40 mM Trizma base, 40 mM
dithiothreitol, 1% protease inhibitor, the lysates were centrifuged
for 15 min at 15,000 × g. All reagents were sourced from
Sigma-Aldrich. The supernatant from each tube was moved to a new
tube. The total proteins were separated on a 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis gel (Sigma-Aldrich) at
100 V for 2 h following extraction. Then transferred onto
Immobilon-P Transfer Membranes (Merck Millipore, Tokyo, Japan). The
membranes were incubated in Tris-buffered saline containing 5%
skimmed milk and 0.05% Tween-20 (EMD Milipore, Billerica, MA, USA)
for 1–2 h and reacted with the corresponding primary antibodies at
4°C overnight. The following primary antibodies were purchased from
Abcam, all at dilution of 1:2,000 (Cambridge, MA, USA): p-IKBα
(mouse monoclonal; cat no. 39A1431, reactivity - mouse, rat, cow,
human), IKBα (rabbit polyclonal; cat no. ab7217; reactivity -
mouse, rat, human) and GAPDH (mouse monoclonal; cat no. mAbcam
9484; reactivity - mouse, rat, rabbit, chicken, cow, dog, human,
pig). The membranes were incubated for 1 h with an anti-mouse or
polyclonal anti-rabbit horseradish peroxidase-linked secondary
antibody (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA).
Statistical analysis
Statistical calculations were performed with Prism 5
software package (GraphPad Software, Inc., La Jolla, CA, USA).
Significant differences are expressed as the mean ± standard error.
The comparison between two groups was analyzed by t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
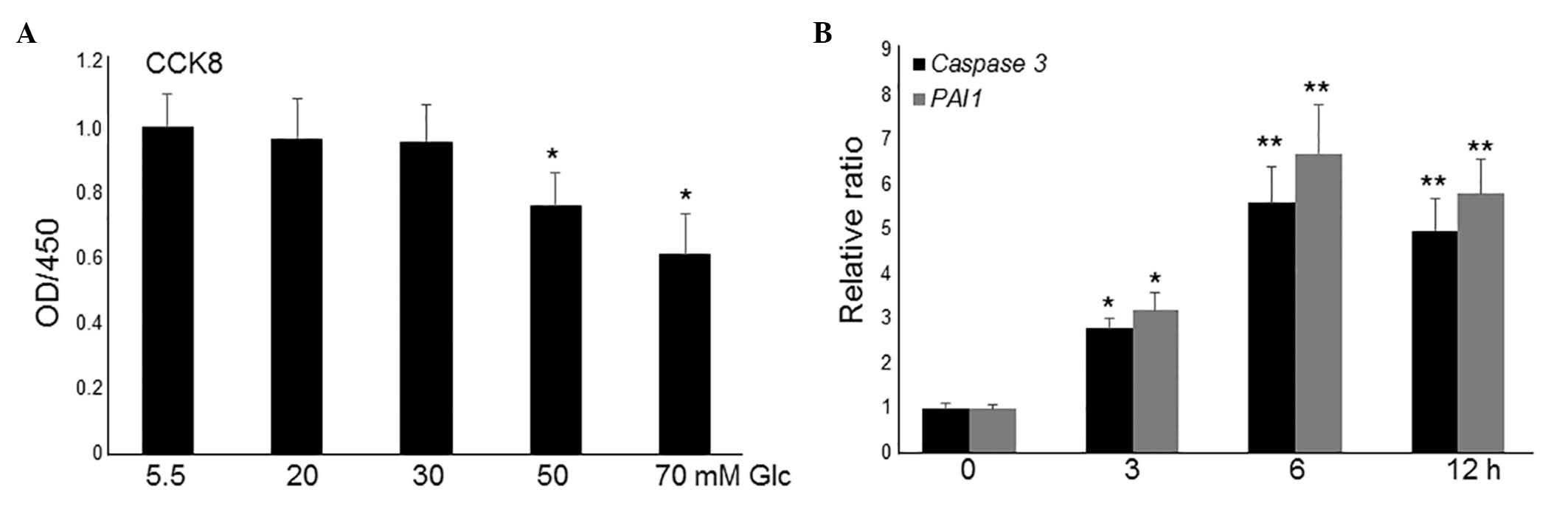
HG induces apoptosis and inflammatory
responses in fibroblasts
To simulate diabetes, HG was utilized to study its
effects on the fibroblasts (22).
To analyze the effects of different concentrations of HG on
fibroblasts, cell proliferation was monitored in different
concentrations of glucose-containing media with 10% fetal bovine
serum. HG treatment up to 30 mM did not markedly alter cell
proliferation, whilst culture in 50 and 70 mM glucose significantly
inhibited cell proliferation compared with culture in low-glucose
(LG; 5.5 mM; P<0.01; Fig. 1A).
However, this concentration of glucose is much higher than the
levels recorded in patients' blood; therefore, 20 and 30 mM were
used to further analyze gene expressions. Compared to 30 mM, 20 mM
of glucose did not significantly affect the expression levels of
caspase 3 and PAI1 (data not shown). Therefore, 30 Mm of glucose
was selected for transcriptome analysis. As presented in Fig. 1A, 30 mM glucose treatment did not
affect cell proliferation activity, thus, the expression levels of
two apoptosis and inflammation marker genes, caspase 3 and
plasminogen activator inhibitor 1 (PAI1), were further monitored at
this concentration. RT-qPCR results indicated that the application
of 30 mM glucose led to an increase in the expression levels of
caspase 3 and PAI1 following 3-h treatment, which reached a peak at
6 h (Fig. 1B). These data indicate
that HG culture damages fibroblast cells.
Identification of HG-regulating
transcriptome in fibroblasts
To identify HG-regulated genes and pathways, RNA-Seq
experiments were performed using human fibroblast cells cultured in
LG (5.5 mM) and HG (30 mM). Since Caspase 3 and PAI1 expression
levels were highest at 6 h subsequent to HG stress, the fibroblast
cells stimulated for 6 h with LG and HG were collected for RNA-Seq
analysis. The RNA-Seq results demonstrated that 814 genes were
differentially expressed (>1.5-fold change; P<0.05) in the
HG-treated fibroblasts compared with LG-treated cells. Among them,
351 genes were downregulated, and 463 genes were upregulated
(Fig. 2A), determined from
statistical outcomes by analysis for association with biological
process GO terms. To verify the RNA-Seq data, HG-mediated
expression levels of the following four genes were assessed by
RT-qPCR: Interleukin 8 (IL8), chemokine (C-C motif) ligand 13
(CCL13), frizzled class receptor 8 (FZD8) and early growth receptor
2 (EGR2). The inflammatory response genes (IL8 and CCL13) were
upregulated, while the Wnt signaling gene (FZD8) and putative SUMO
E3 ligase (EGR2) were repressed by HG stimulation, and the RT-qPCR
results were similar to RNA-Seq data (Fig. 2B). GO analysis indicated that 31 GO
terms were enriched (P<0.01; Table
II). These genes were associated with multiple biological
processes, including cellular triglyceride homeostasis, positive
regulation of cholesterol efflux, the canonical Wnt signaling
pathway and transcription (Table
II). Further pathway analysis was performed with 814 genes that
were altered by HG stress. The results demonstrated that these
genes were divided into 20 different pathways, including NF-κB,
tumor necrosis factor (TNF), Wnt, ECM/receptor interaction and
hedgehog signaling pathways. Among them, certain inflammatory
response pathways involving NF-κB and TNF were upregulated by HG
stress, while Wnt signaling genes were downregulated (Table III). Together, these data
indicate that HG regulates a large number of genes involved in
various biological processes.
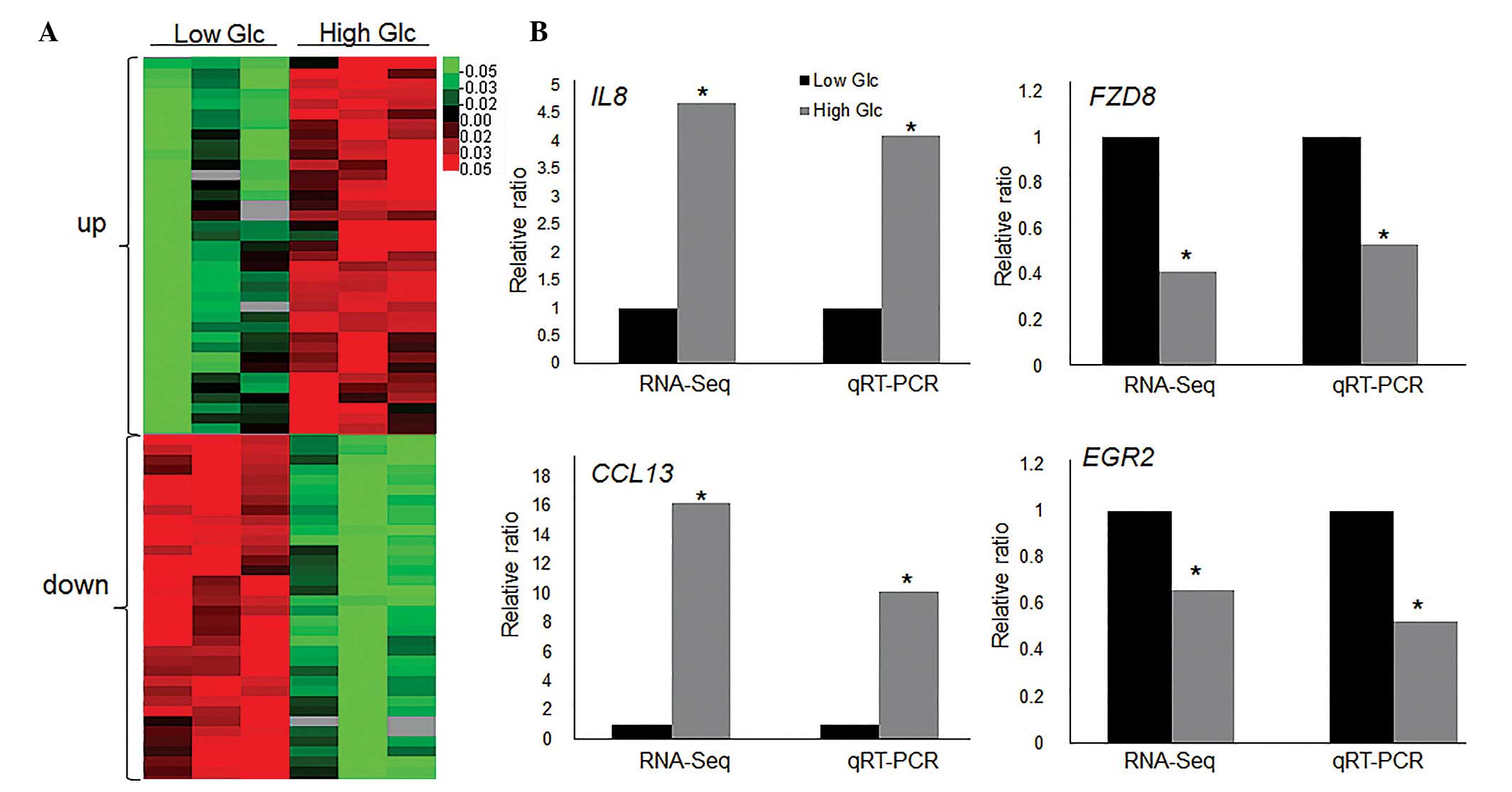 | Figure 2High Glc regulated transcriptome
profile. (A) Heat map represents the differentially expressed genes
following high Glc (30 mM) treatment for 6 h in human fibroblast
cells. Low Glc treatment was 5.5 mM. Gene expression is presented
as a pseudocolor scale with red denoting higher gene expression
levels and green denoting lower levels. Significant differences
between low and high glucose treated groups were compared
(P<0.05). (B) RT-qPCR was performed to verify the expression
levels of IL8, CCL13, FZD8 and EGR2 and the data was compared with
RNA-Seq results. Significant differences of IL8, FZD8, CCL13 and
EGR2 expression levels between low and high glucose treated groups
in both RNA-Seq and qRT-PCR analyses (*P<0.05) GAPDH
was used as an internal control. Glc, glucose; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; IL8,
interleukin 8; FZD8, frizzled class receptor 8; CCL13, chemokine
(C-C motif) ligand 13; EGR2, early growth response 2; RNA-Seq, RNA
sequencing. |
 | Table IIGO classification. |
Table II
GO classification.
| GO ID | GO term | Enrichment | (−log2P) |
Up/downregulated |
|---|
| 0071371 | Cellular response
to gonadotropin stimulus | 19.12899 | 10.23295 | Up |
| 0034421 | Post-translational
protein acetylation | 63.76331 | 9.872556 | Up |
| 0071320 | Cellular response
to cAMP | 9.564497 | 9.733907 | Up |
| 0001508 | Regulation of
action potential | 15.94083 | 9.562348 | Up |
| 0050890 | Cognition | 15.10184 | 9.362226 | Up |
| 0034088 | Maintenance of
mitotic sister chromatid cohesion | 47.82249 | 9.297728 | Up |
| 0006642 | Triglyceride
mobilization | 38.25799 | 8.822425 | Up |
| 0035356 | Cellular
triglyceride homeostasis | 38.25799 | 8.822425 | Up |
| 0071356 | Cellular response
to tumor necrosis factor | 7.501566 | 8.538068 | Up |
| 0042471 | Ear
morphogenesis | 31.88166 | 8.417503 | Up |
| 0042158 | Lipoprotein
biosynthetic process | 31.88166 | 8.417503 | Up |
| 0045444 | Fat cell
differentiation | 6.596205 | 7.912633 | Up |
| 0048752 | Semicircular canal
morphogenesis | 23.91124 | 7.753206 | Up |
| 0030728 | Ovulation | 23.91124 | 7.753206 | Up |
| 0006955 | Immune
response | 2.610657 | 7.576953 | Up |
| 0006351 | Transcription,
DNA-templated | 1.628693 | 7.526714 | Up |
| 0042472 | Inner ear
morphogenesis | 5.710147 | 7.220235 | Up |
| 0010875 | Positive regulation
of cholesterol efflux | 17.38999 | 6.989887 | Up |
| 0002237 | Response to
molecule of bacterial origin | 17.38999 | 6.989887 | Up |
| 0006334 | Nucleosome
assembly | 7.598543 | 21.94307 | Down |
| 0035115 | Embryonic forelimb
morphogenesis | 12.5653 | 13.38364 | Down |
| 0007275 | Multicellular
organism development | 2.149419 | 11.51722 | Down |
| 0032688 | Negative regulation
of interferon-beta production | 53.61194 | 9.387513 | Down |
| 0060675 | Ureteric bud
morphogenesis | 53.61194 | 9.387513 | Down |
| 2001181 | Positive regulation
of interleukin-10 secretion | 53.61194 | 9.387513 | Down |
| 0048706 | Embryonic skeletal
system development | 8.247991 | 8.969668 | Down |
| 0009952 | Anterior/posterior
pattern specification | 4.684538 | 8.623385 | Down |
| 0060070 | Canonical Wnt
signaling pathway | 5.154994 | 8.014638 | Down |
| 0010042 | Response to
manganese ion | 26.80597 | 7.938039 | Down |
| 0009954 | Proximal/distal
pattern formation | 10.05224 | 7.809469 | Down |
| 0046628 | Positive regulation
of insulin receptor signaling pathway | 22.97655 | 7.587427 | Down |
 | Table IIIPathway classification. |
Table III
Pathway classification.
| Pathway ID | Pathway term | Enrichment | (−log2P) |
Up/downregulated |
|---|
| 05034 | Alcoholism | 8.39875 | 32.3993 | Up |
| 05322 | Systemic lupus
erythematosus | 9.129076 | 29.151 | Up |
| 05217 | Basal cell
carcinoma | 7.635227 | 10.23381 | Up |
| 04512 | ECM-receptor
interaction | 4.772017 | 7.471521 | Up |
| 04916 | Melanogenesis | 4.157797 | 6.698221 | Up |
| 04390 | Hippo signaling
pathway | 3.27224 | 6.165099 | Up |
| 05203 | Viral
carcinogenesis | 2.840157 | 5.914846 | Up |
| 05202 | Transcriptional
misregulation in cancer | 2.799583 | 5.229064 | Up |
| 04910 | Insulin signaling
pathway | 2.957306 | 4.892077 | Up |
| 04340 | Hedgehog signaling
pathway | 4.49933 | 4.869559 | Up |
| 04977 | Vitamin digestion
and absorption | 6.998958 | 4.700239 | Up |
| 04064 | NF-κB signaling
pathway | 6.15293 | 8.938564 | Up |
| 04668 | TNF signaling
pathway | 5.090152 | 7.82519 | Up |
| 04060 | Cytokine-cytokine
receptor interaction | 3.803208 | 9.593708 | Down |
| 04310 | Wnt signaling
pathway | 2.74469 | 4.521127 | Down |
| 05202 | Transcriptional
misregulation in cancer | 3.732778 | 6.955703 | Down |
| 05166 | HTLV-I
infection | 2.94693 | 6.11439 | Down |
| 04621 | NOD-like receptor
signaling pathway | 5.418548 | 5.528479 | Down |
| 05132 | Salmonella
infection | 3.952353 | 4.424635 | Down |
| 05161 | Hepatitis B | 3.068037 | 4.341831 | Down |
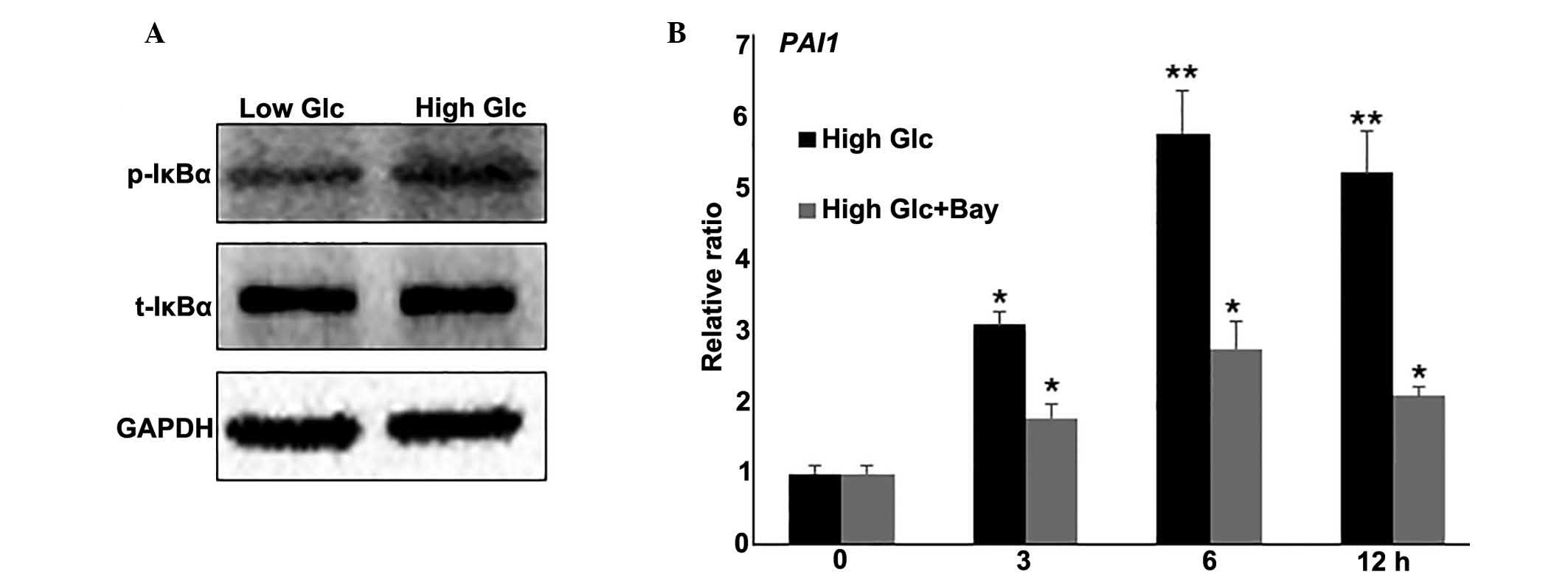
Regulatory role of the inflammatory
response in HG-mediated fibroblast cell damage
The NF-κB pathway was identified to be involved in
HG-regulated biological processes, the effect of the inflammatory
response in HG-mediated fibroblast cell damage was further
examined. To further evaluate the effects of HG on NF-κB signaling,
the activity of IκBα, the most characterized and studied NF-κB
regulator, was examined. IκBα is phosphorylated by IκB kinases
(IKK), resulting in the translocation of NF-κB to the nucleus and
transcription of target genes (23). Western blot analysis indicated that
HG induced IκBα phosphorylation (p-IκBα), but did not change total
IκBα (t-IκBα) levels (Fig. 3A). To
further analyze the effect of the inflammatory response on
HG-mediated gene expression, a combination of HG stress and
Bay11-7082 (0.5 μM), a representative NF-κB pathway
inhibitor, was used to treat fibroblasts. RNA was extracted and
RT-qPCR was performed to monitor PAI1 gene expression. The results
indicated that HG induced an increase in PAI1 mRNA levels at 3
(P<0.05), 6 and 12 h (P<0.01) compared with the levels
observed at 0 h, and this induction was blocked by inhibiting NF-κB
with Bay11-7082 (Fig. 3B). Taken
together, these results suggest that the inflammatory response is
inversely correlated with HG-regulated gene expression.
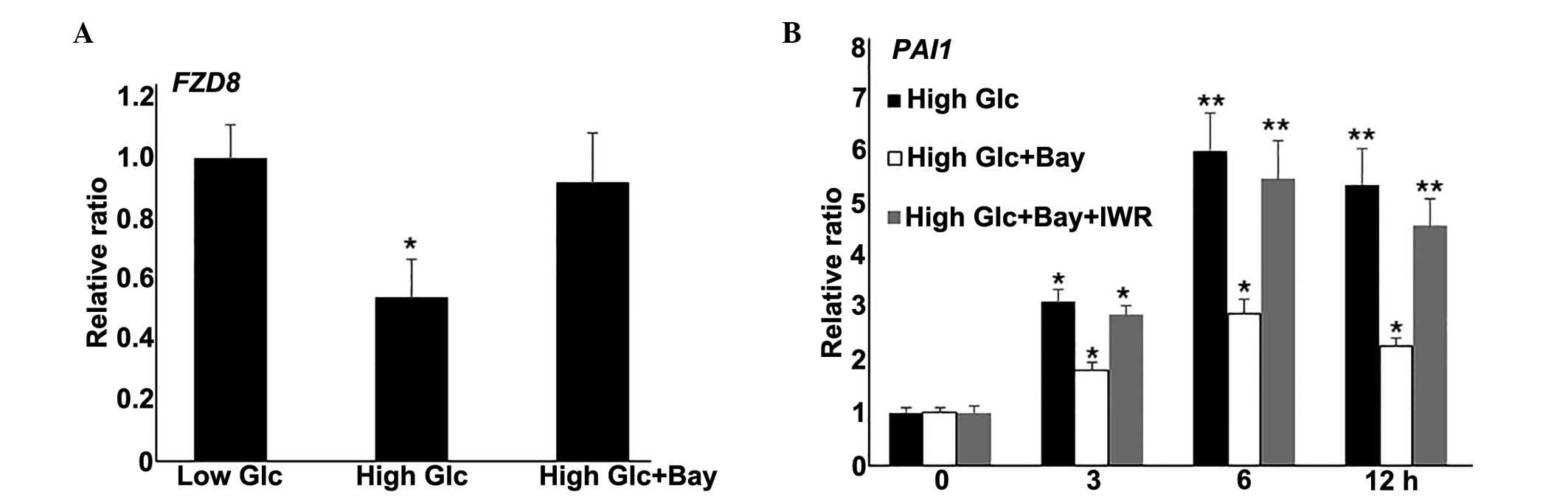
Wnt signaling is downstream of the NF-κB
pathway
Wnt signaling is known to regulate diverse aspects
of numerous biological processes (24). The RNA-Seq data from the present
study demonstrated repressed expression of a number of Wnt
signaling genes following HG stimulation in fibroblasts. NF-κB
pathway inhibition partially rescued HG-mediated fibroblast damage.
Therefore, the relationship between NF-κB and Wnt signaling were
investigated further. In the fibroblast cells stimulated with
Bay11-7082 (0.5 μM) and HG, expression levels of the Wnt
signaling gene, FZD8, were similar to the levels in the LG cells
(P>0.05; Fig. 4A). To further
evaluate the role of Wnt signaling in HG-mediated gene expression,
IWR, a typical Wnt signaling inhibitor, was also used to treated
fibroblasts alongside Bay11-7082 and HG culture, then PAI1 gene
expression was measured. Notably, IWR application reduced the
effect of Bay11-7082 on HG-induced PAI1 expression levels (Fig. 4B). Together, these data demonstrate
that NF-κB inhibition blocked the gene expression changes induced
by HG. Additionally, Wnt signaling inhibition reversed
Bay11-7082-mediated PAI1 repression under HG conditions.
Discussion
Fibroblasts are important for synthesizing ECM and
collagen, the structural framework (stroma) for skin tissues, and
serve a key role in wound healing (25). Skin wound healing requires the
involvement of several cell types, including keratinocytes,
fibroblasts, endothelial cells, macrophages and platelets (26). Therefore, understanding the
underlying mechanisms by which fibroblast cells protect themselves
from DM is important for the treatment of skin ulcers (4). One of the strategies that all living
organisms utilize to adapt to environmental changes is the rapid
reprogramming of transcriptional regulations via cell signaling
mechanisms (27–29). Therefore, analysis of
transcriptomic changes under certain stress is a method to clarify
the regulation of these mechanisms. In the present study, RNA-Seq
was utilized to analyze transcriptomes, providing an efficient
experimental basis to extract information regarding gene
expression, somatic mutations and novel gene fusions (30) using HG-cultured human primary
fibroblast cells. The results demonstrated a large population of
differentially expressed genes following HG stimulation. Among
them, 351 genes were downregulated and 463 were upregulated.
Further, analyses of the associated pathways using GO and KEGG
databases revealed various biological processes and pathways
(Tables II and III).
ECM synthesis is important for skin wound repair.
Various genes involved in ECM/receptor interactions were identified
as undergoing changes in expression levels following HG stress
(data available upon request). Other pathways identified to be
altered by HG include NF-κB, TNF, Wnt, Hedgehog and Hippo
signaling. The Wnt signaling pathway and fibroblast growth factor
(FGF) regulate T-box family transcription factors, control cell
fate within the zebrafish tailbud and are involved in axis
elongation (31); FGF positively
regulates Hedgehog signaling during embryonic tracheal cell
migration (32); Hippo signaling
and EGFR pathways control growth and activate tumorigenesis when
dysregulated. Epidermal growth factor receptor (EGFR) activates
Yorkie, a key Hippo pathway transcription factor that has been
indicated to influence cell proliferation in Drosophila (33); and bFGF previously inhibited
TNF-mediated activation of NF-κB by blocking phosphorylation and
degradation of IκBα, leading to the repression of leukocyte
adhesion in tumor vessels (34).
HG has been demonstrated to affect FGF and downstream JNK activity,
resulting in delay to human fibroblast cell migration (14). Therefore, those pathways that are
regulated by HG may be partially connected to FGF signaling, which
is known to accelerate DM-induced skin wound repair. Further
biochemical and molecular studies are required to specify how these
pathways are connected.
In the current study, inhibition of the NF-κB
pathway through treatment with Bay11-7082 repressed the HG-induced
PAI1 levels, suggesting that HG stimulation may activate
inflammatory response pathways and cause damage to cells. Notably,
Bay11-7082 application reversed the repression of FZD8 expression
levels resulting from HG stress.
In addition, treatment with the Wnt signaling
inhibitor, IWR, together with Bay11-7082 diminished the effects of
Bay11-7082 on PAI1 repression under HG conditions, indicating that
Wnt signaling functions downstream of the NF-κB pathway to regulate
HG-mediated gene expression. HG stress negatively and positively
regulated the NF-κB and Wnt signaling pathways, respectively, and
this suggests that Wnt activation is important for the protection
of fibroblasts from DM. The present study demonstrated HG-regulated
gene expression in fibroblasts, and a link between the NF-κB
pathway and Wnt signaling. In the future, these findings may be
notable for the treatment of DM-induced skin ulcers.
Acknowledgments
The present study was made possible by an initiative
grant from Wenzhou Medical University (Wenzhou, China).
References
|
1
|
Brownlee M: Biochemistry and molecular
cell biology of diabetic complications. Nature. 414:813–820. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yach D, Stuckler D and Brownell KD:
Epidemiologic and economic consequences of the global epidemics of
obesity and diabetes. Nat Med. 12:62–66. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braiman-Wiksman L, Solomonik I, Spira R
and Tennenbaum T: Novel insights into wound healing sequence of
events. Toxicol Pathol. 35:767–779. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martin P: Wound healing - aiming for
perfect skin regeneration. Science. 276:75–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brem H and Tomic-Canic M: Cellular and
molecular basis of wound healing in diabetes. J Clin Invest.
117:1219–22. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wagner W and Wehrmann M: Differential
cytokine activity and morphology during wound healing in the
neonatal and adult rat skin. J Cell Mol Med. 11:1342–1351. 2007.
View Article : Google Scholar
|
|
8
|
Kanazawa S, Fujiwara T, Matsuzaki S,
Shingaki K, Taniguchi M, Miyata S, Tohyama M, Sakai Y, Yano K,
Hosokawa K and Kubo T: bFGF regulates PI3-kinase-Rac1-JNK pathway
and promotes fibroblast migration in wound healing. PLoS One.
5:e122282010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goldin A, Beckman JA, Schmidt AM and
Creager MA: Advanced glycation end products: Sparking the
development of diabetic vascular injury. Circulation. 114:597–605.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Obayashi K, Akamatsu H, Okano Y, Matsunaga
K and Masaki H: Exogenous nitric oxide enhances the synthesis of
type I collagen and heat shock protein 47 by normal human dermal
fibroblasts. J Dermatol Sci. 41:121–126. 2006. View Article : Google Scholar
|
|
11
|
Loots MA, Lamme EN, Mekkes JR, Bos JD and
Middelkoop E: Cultured fibroblasts from chronic diabetic wounds on
the lower extremity (non-insulin-dependent diabetes mellitus) show
disturbed proliferation. Arch Dermatol Res. 291:93–99. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rowe DW, Starman BJ, Fujimoto WY and
Williams RH: Abnormalities in proliferation and protein synthesis
in skin fibroblast cultures from patients with diabetes mellitus.
Diabetes. 26:284–290. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bachschmid MM, Xu S, Maitland-Toolan KA,
Ho YS, Cohen RA and Matsui R: Attenuated cardiovascular hypertrophy
and oxidant generation in response to angiotensin II infusion in
glutaredoxin-1 knockout mice. Free Radic Bio Med. 49:1221–1229.
2010. View Article : Google Scholar
|
|
14
|
Xuan YH, Huang BB, Tian HS, Chi LS, Duan
YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al: High-glucose
inhibits human fibroblast cell migration in wound healing via
repression of bFGF-regulating JNK phosphorylation. PLoS One.
9:e1081822014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006. View Article : Google Scholar :
|
|
16
|
Dupuy D, Bertin N, Hidalgo CA, Venkatesan
K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dunnick JK, Brix A, Cunny H, Vallant M and
Shockley KR: Characterization of polybrominated diphenyl ether
toxicity in Wistar Han rats and use of liver microarray data for
predicting disease susceptibilities. Toxicol Pathol. 40:93–106.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
19
|
Matthews L, Gopinath G, Gillespie M, Caudy
M, Croft D, de Bono B, Garapati P, Hemish J, Hermajakob H, Jassal
B, et al: Reactome knowlegdebase of human biological pathways and
processes. Nucleic Acids Res. 37:619–622. 2009. View Article : Google Scholar
|
|
20
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Lamers ML, Almeida ME, Vicente-Manzanares
M, Horwitz AF and Santos MF: High glucose-mediated oxidative stress
impairs cell migration. PLoS One. 6:e228652011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baeuerle PA: IkappaB-NF-kappaB structures:
At the interface of inflammation control. Cell. 95:729–731. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Malinauskas T and Jones EY: Extracellular
modulators of Wnt signalling. Curr Opin Struct Biol. 29:77–84.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krafts KP: Tissue repair: The hidden
drama. Organogenesis. 6:225–233. 2010. View Article : Google Scholar
|
|
26
|
Hinz B: Masters and servants of the force:
The role of matrix adhesions in myofibroblast force perception and
transmission. Eur J Cell Biol. 85:175–181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Greenhalgh DG: The role of apoptosis in
wound healing. Int J of Biochem Cell Biol. 30:1019–1030. 1998.
View Article : Google Scholar
|
|
28
|
Stashak TS, Farstvedt E and Othic A:
Update on wound dressings: Indications and best use. Clin Tech
Equine Prac. 3:148–163. 2004. View Article : Google Scholar
|
|
29
|
Versteeg HH, Heemskerk JW, Levi M and
Reitsma PH: New fundamentals in hemostasis. Physiological Reviews.
93:327–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meyerson M, Gabriel S and Getz G: Advances
in understanding cancer genomes through second-generation
sequencing. Nat Rev Genet. 11:685–696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stulberg MJ, Lin A, Zhao H and Holley SA:
Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud.
Dev Biol. 369:298–307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Butí E, Mesquita D and Araújo SJ: Hedgehog
is a positive regulator of FGF signalling during embryonic tracheal
cell migration. PLoS One. 9:e926822014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Reddy BV and Irvine KD: Regulation of
Hippo signaling by EGFR-MAPK signaling through Ajuba family
proteins. Dev Cell. 24:459–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Flati V, Pastore LI, Griffioen AW, Satijn
S, Toniato E, D'Alimonte I, Laglia E, Marchetti P, Gulino A and
Martinotti S: Endothelial cell anergy is mediated by bFGF through
the sustained activation of p38-MAPK and NF-kappaB inhibition. Int
J Immunopathol Pharmacol. 19:761–773. 2006.PubMed/NCBI
|