Introduction
Colorectal cancer (CRC) is a common malignancy
worldwide and is a major cause of cancer-associated mortality
(1). Despite the marked
improvements in surgical and chemoradiothera-peutic techniques that
are currently in use, the prognosis of patients with advanced CRC
remains poor, and the morbidity remains high (2). In China, the 5-year survival rate of
stage IV CRC patients is 8%, and very few survive for 10 years
(3). A notable number of patients
with CRC who undergo surgery to remove the cancer develop local
recurrence or distant metastasis, resulting in lower survival rates
(4). Early diagnosis and surgery
are effective ways to cure CRC (5). Rapid integration of novel endoscopic
and molecular techniques into clinical practice has established
colorectal medicine at the forefront of translational research
(6,7), therefore, investigating the molecular
mechanisms of CRC and identifying biomarkers for early diagnosis
and of prognostic value may aid the selection of suitable
therapeutic strategies and enable regular surveillance.
Forkhead box (FOX)M1 is a member of the FOX family
of transcription factors (8,9).
FOXM1 is key in cell cycle progression, where endogenous FOXM1
expression regulates the transition from the G1 to the S
phase, as well as the progression to mitosis (10,11).
Abnormal upregulation of FOXM1 is observed in a variety of human
cancers, including liver, breast, prostate, brain, cervix and lung
cancer (12–15). These findings associate FOXM1 with
the tumorigenesis and progression of various types of malignancy.
However, the role of FOXM1 in the metastatic process of CRC remains
to be elucidated. In the present study, the mRNA and protein
expression of FOXM1 were investigated in tissue samples from
patients with primary CRC using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and immunohistochemistry, and
the current study also aimed to elucidate the effect of FOXM1 on
cell invasion and metastasis in CRC cell lines.
Materials and methods
Tumor tissues
A cohort of 96 patients (57 men and 39 women) with
CRC diagnosed between March 2012 and January 2014 were selected.
Patient consent and approval from the Ethics Committee of
Affiliated Hospital of Weifang Medical University (Weifang, China)
were obtained prior to use of these clinical materials for
research. Tumor and paired normal samples (10 cm from colorectal
tumor) were surgically obtained from these patients. No local or
systemic treatment was administered in these patients before
surgery. In each selected case, pathological diagnosis was
performed in the Department of Pathology, Affiliated Hospital of
Weifang Medical University.
Cell culture
The human colorectal cancer cell lines (HCT116,
SW480, SW620, LoVo, CaCo-2 and HT-29) were purchased from the
American Type Culture Collection (Manassas, VA, USA). SW620 and
SW480 cells were cultured in Leibovitz's L-15 media (Invitrogen;
Thermo Fisher Scientific, Inc. Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA);
Caco-2 and HT-29 cells were maintained in RPMI-1640 (Invitrogen;
Thermo Fisher Scientific, Inc.) with 10% FBS, and LoVo cells were
cultured in Ham's F-12 Kaighn's medium (Invitrogen; Thermo Fisher
Scientific, Inc.) with 10% FBS.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissues were cut
into 3-µm sections. The tissues were obtained from the
Department of Pathology of the Affiliated Hospital of Weifang
Medical University, where the formalin fixation process was carried
out. The immunohistochemistry steps were performed using
streptavidin-biotin peroxidase complex immunostain kit (Wuhan
Boster Biological Technology, Ltd., Wuhan, China) according to the
manufacturer's protocols. The sections were incubated in a moist
chamber with primary rabbit anti-human FOXM1 monoclonal antibody
(cat. no., sc-501; dilution, 1:100; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) for 30 min at room temperature, followed by a
horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G
secondary antibody (cat. no. SA00001-2; (dilution, 1:2,000;
Proteintech Group, Inc., Chicago, IL, USA) for 30 min at room
temperature. Rabbit serum (HyClone, Logan, UT, USA) served as a
negative control. The images were collected and analyzed using a
Leica DC200 image system (Leica Microsystems, Inc., Buffalo Grove,
IL, USA).
Immunohistochemical analysis
evaluation
Five random microscopic fields (magnification, ×200)
were examined per slide, and 100 cells were evaluated per field.
The expression of FOXM1 was classified into five groups according
to the percentage of positively staining cells: 0=absent; 1=1–25%;
2=26–50%; 3=51–75%; and 4=≥76%. The staining intensity was
categorized as follows: 0=negative; 1=weak; 2=moderate; and
3=strong. The proportion and intensity scores were subsequently
multiplied to obtain a total score. Where the product of
multiplication between staining intensity and the percentage of
positive cells was ≤4, it was defined as low expression; by
contrast, where the overall score was >4, it was defined as high
expression.
Small interfering RNA (siRNA)
transfection
Three FOXM1 siRNAs were designed and synthesized by
Qiagen GmbH (Hilden, Germany) to generate effective FOXM1-siRNA
oligonucleotides for gene knockdown studies. An siRNA with the
sequence CUCUUCUCCCUCAGAUAU AdTdT was determined to exert the
greatest effect on inhibition of FOXM1 expression. The LoVo CRC
cells were transfected with FOXM1-siRNA oligonucleotides or control
siRNA oligonucleotides (50 nmol/l; Santa Cruz Biotechnology, Inc.)
with the use of Lipofectamine® 2000 Transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols.
RT-qPCR assay
Total RNA was isolated from the cell lines using
TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
concentration and quality of the extracted total RNA was determined
by measuring the optical density at 260 and 280 nm, and then
calculating the OD260:OD280 ratio. Complementary DNA (cDNA) was
synthesized from 2 µg of total RNA using a Reverse
Transcription System kit (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Briefly, the samples were
pre-incubated at 70°C for 10 min, cooled on ice and then added to a
reaction mixture consisting of 10 mmol/l deoxynucleotide
triphosphate, 25 mmol/l MgCl2, 15 units avian myoblastosis virus
reverse transcriptase, 10X reverse transcription buffer, 0.5 units
RNasin® and 0.5 µg oligo-(dT)15 primer
(all provided with the Reverse Transcription System kit). A final
volume of 20 l reaction mixture was incubated successively at 44°C
for 15 min, 99°C for 5 min and 4°C for 5 min. The cDNA was
maintained at 20°C prior to use. Then, it was reverse-transcribed
from 5 µg of total RNA. RT-qPCR was performed using a SYBR
Master Mix (Takara Bio Inc., Otsu, Japan) on an ABI Prism 7900HT
Sequence Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primer sequences of FOXM1 were as follows:
Sense, 5′-GGAGGAAATGCCACACTTAGCG-3′; and reverse,
5′-TAGGACTTCTTGGGTCTTGGGGTG-3′ (designed by Guangzhou Ruibo Trading
Co., Ltd., Guangzhou, China). The products of PCR were separated by
1.5% agarose gel electrophoresis, and the expression levels of the
mRNA were detected, with GAPDH serving as the control. The primer
sequences for GAPDH were as follows: Sense,
5′-GCAGTGGCAAAGTGGAGATT-3′; and reverse,
5′-TGAAGTCGCAGGAGACAACC-3′. The cycling conditions used were as
follows: 94°C for 2 min for 2 cycles, followed by 40 cycles of 94°C
for 15 sec, 58°C for 25 sec, and 72°C for 30 sec. qPCR was
conducted on an Applied Biosystems 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Each
experiment was performed three times, each time in duplicate. The
relative expression levels of Trop2 mRNA were calculated using the
2−ΔΔCt method (16).
Western blot analysis
Whole cell lysates were prepared from the colon
cells as described previously (17). Standard western blot analysis of
the lysates was conducted with the primary rabbit anti-human FOXM1
monoclonal antibody and a second anti-IgG antibody (GE Healthcare
Life Sciences, Chalfont, UK). The membranes were then stripped and
blotted, and an anti-β-actin antibody (Sigma-Aldrich) served as a
loading control. Blots were visualized using an enhanced
chemiluminescence kit (Santa Cruz Biotechnology., Inc.) according
to the manufacturer's protocol, and exposed to film (X-OMAT BT;
Kodak, Rochester, NY, USA).
Cell survival assay
The effects of FOXM1 on CRC cell survival were
determined using the
3-(4,5-dimethylthi-azol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Four groups of cells were seeded into 96-well plates (Nunc
A/S Plastfabrikation, Roskilde, Denmark; 5×103
cells/well) and cultured for 120 h. Following treatment, cells were
incubated with MTT (20 µl/well; Sigma-Aldrich) at 37°C for 4
h, and then 200 µl dimethyl sulfoxide was added to each
well. The absorbance of the cells was measured at a wavelength of
570 nm using a microplate reader (Bio-Tek Instruments, Inc.,
Winooski, VT, USA). The percentage of residual cell viability was
determined by: [(optical density (OD) of experimental group − OD of
blank group)/(OD of negative group − OD of blank group)] ×100%. The
assays were performed three times.
Cell migration assay
Motility in vitro was assessed using
Transwell® chambers (Corning Incorporated, Corning, NY,
USA). Four groups of cells (5×105) were seeded on the
upper wells with Ham's F-12K Kaighn's medium. Medium (500
µl) with 20% FBS was plated in the bottom wells as
chemoattractants. Following a 48-h incubation, cells were fixed
with methanol and stained with 1% crystal violet (Sigma-Aldrich)
for 30 min at 37°C. Cells on the upper filter of the membranes were
wiped, while cells that had penetrated to the lower filter were
stained with hematoxylin (Sigma-Aldrich). Cells in 15 randomized
fields were counted and images were captured using an inverted
microscope (magnification, ×50; Olympus BX51; Olympus Corp., Tokyo,
Japan).
Statistical analysis
Data analyses were performed using SPSS version 15.0
(SPSS, Inc., Chicago, IL, USA). Patient characteristics were
expressed as the mean ± standard deviation for continuous
variables, and as the count and percentage for discrete variables.
The data were analyzed using the Pearson's chi-square test and
Fisher's exact test, and P<0.05 was considered to indicate a
statistically significant difference.
Results
To investigate the clinical significance of FOXM1
expression in human CRC progression and metastasis, FOXM1
expression was investigated using immunohistochemistry, western
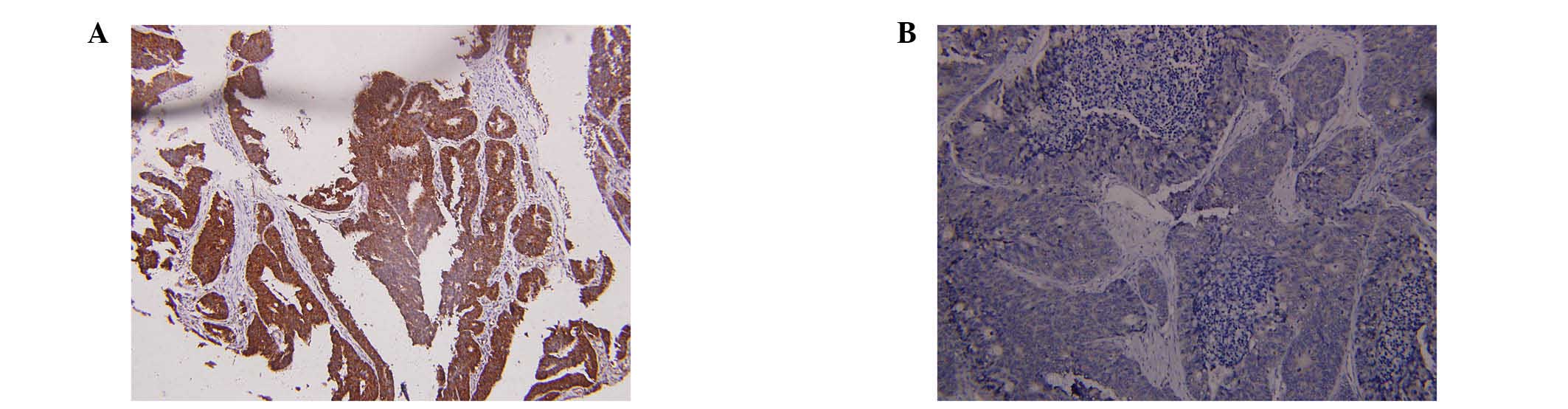
blotting and RT-qPCR in 96 CRC tissue samples. As presented in
Fig. 1A and B, the expression of
FOXM1 was higher in 82 of 96 randomly selected human CRC tissues
compared with the adjacent normal mucosa tissues. High expression
of FOXM1 was observed in significantly more of the CRC tissue
samples [85.42% (82/96) of the CRC tissues] compared with 18.75%
(18/96) of adjacent normal mucosa tissues (P<0.001). The FOXM1
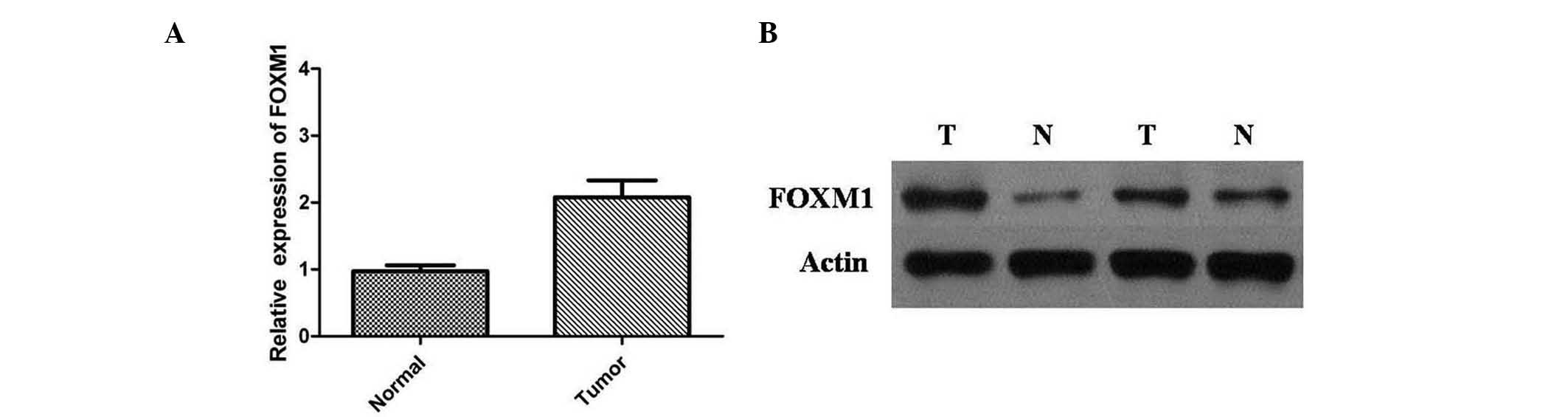
mRNA expression levels were normalized to GAPDH mRNA and detected
by RT-qPCR (Fig. 2A). The results
indicated that the relative expression level of FOXM1 mRNA in CRC
tissues was significantly higher compared with adjacent normal
mucosa tissues (P<0.01). These results were supported by western
blot analysis, and protein expression levels of FOXM1 were
significantly higher in CRCs than in adjacent normal mucosa tissues
(P<0.01) (Fig. 2B). These data
suggest that, as FOXM1 is overexpressed in CRC tissue samples, it
may be important in CRC progression.
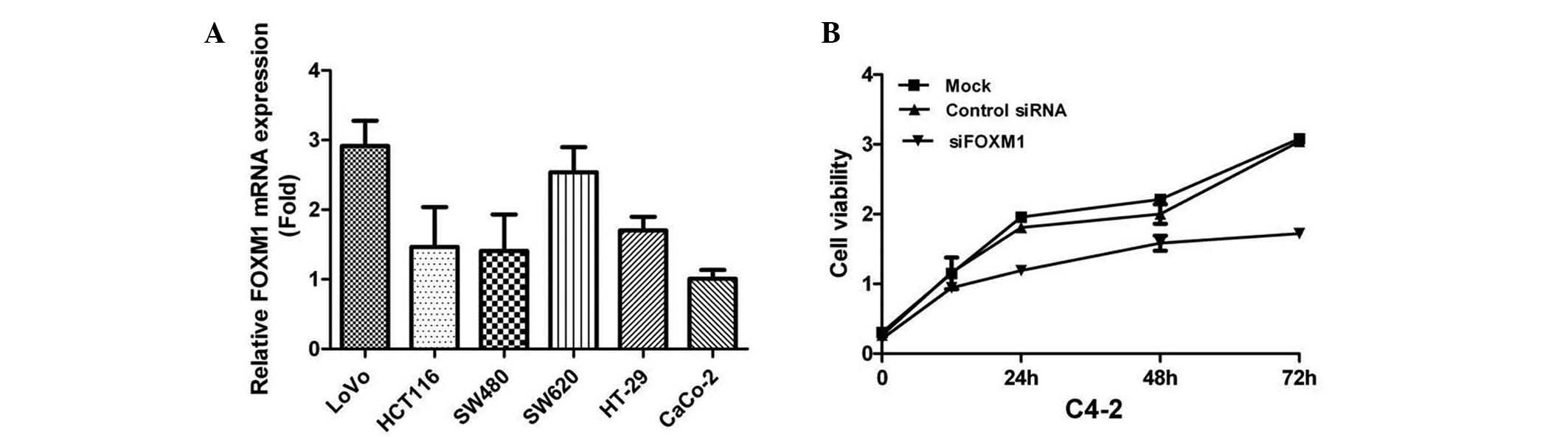
RT-qPCR analysis was used to determine the mRNA
expression levels of FOXM1 in CRC cell lines. The results indicated
that FOXM1 was highly expressed in LoVo cells, so this cell line,
among others, was used in the present study (Fig. 3A). To investigate the effects of
FOXM1 silencing on cell proliferation, FOXM1 was downregulated in
LoVo cells using siRNA against FOXM1 transcripts. The cells were
assessed for cell proliferation using the MTT assay, and the
results indicated that FOXM1-siRNA-transfected LoVo cells
demonstrated a significantly lower proliferation rate compared with
the control groups (P<0.05) (Fig.
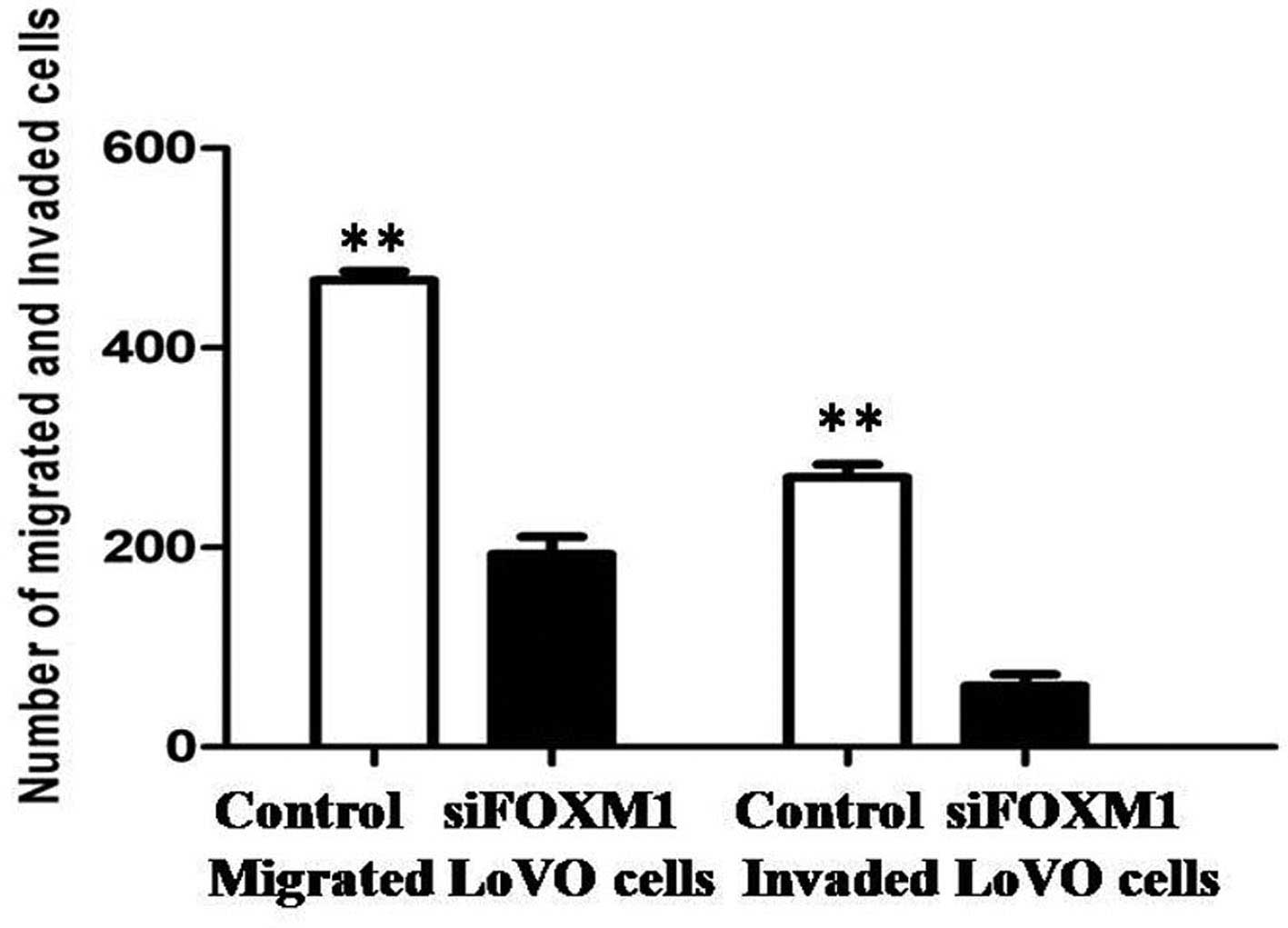
3B). The effects of FOXM1 silencing on tumor cell metastasis
were also investigated. Cellular invasiveness and migration were
analyzed by Transwell® assays, and the migratory and
invasive abilities were revealed to be attenuated in
FOXM1-siRNA-transfected LoVo cells (Fig. 4).
Discussion
In developed countries, CRC is the third most
commonly diagnosed malignancy, and the second leading cause of
cancer-associated mortality (18).
A notable number of patients with CRC who undergo surgery to treat
the cancer develop local recurrence or distant metastasis, leading
to lower survival rates (19); the
same stage of the disease may present different clinical courses
and have a different prognosis due to intra- and intertumor
heterogeneity (20). Although the
regulation of CRC growth is well understood, the mechanism
underlying CRC proliferation, migration, invasion and metastasis
remains to be elucidated. In the present study, a high expression
of FOXM1 was observed in significantly more tumor tissue samples
[85.42% (82/96) of the CRC tissues vs. 18.75% (18/96) of adjacent
normal mucosa tissues] by immunohistochemical analysis. It was
demonstrated that the expression levels of FOXM1 were significantly
higher in CRC tissues than in adjacent normal mucosa tissues, at
the transcriptional and the translational level. The results of the
present study are consistent with previous studies that reported
the overexpression of FOXM1 in various human cancers.
FOXM1 is a proliferation-associated transcription
factor with important roles in cell proliferation, differentiation
and apoptosis (21). It is
expressed in actively dividing cells, and is critical for cell
cycle progression. It was first identified as a
proliferation-specific transcription factor expressed in various
tumor cell lines and embryonic tissues (22). Multiple signaling pathways are
associated with the FOXM1 signaling pathway, suggesting that FOXM1
is central to tumor aggressiveness (23). Previous studies have demonstrated
that FoxM1 is upregulated in a wide variety of malignant tumors
(23–26). The current study demonstrated that
FOXM1 is highly expressed in tissue samples of patients with CRC.
Thus, the biological function of FOXM1 was examined in greater
detail via in vitro analysis of the CRC cell lines. The mRNA
expression levels of FOXM1 in the CRC cell lines were investigated
by RT-qPCR analysis. LoVo cells demonstrated a relatively high
FOXM1 expression level, and these were used for further study.
siRNA was used to knockdown FOXM1 expression in LoVo cell lines. An
impaired proliferation capacity of LoVo cells was observed
following FOXM1 knockdown. Downregulation of FOXM1 was also
demonstrated to inhibit cell migration and invasion. Thus, the
present study suggested that FOXM1 is a potential therapeutic
target for the treatment of CRC.
In conclusion, the present study demonstrated the
clinical significance of overexpressed FOXM1 in patients with CRC.
Results from the current study indicate that FOXM1 may serve as a
promising therapeutic target for the inhibition of CRC progression,
and that targeting FOXM1 may provide a novel therapeutic strategy
for the treatment of CRC.
References
|
1
|
Perencevich M and Stoffel EM: A
multidisciplinary approach to the diagnosis and management of
multiple colorectal polyps. Gastroenterol Hepatol (NY). 7:420–423.
2011.
|
|
2
|
Shelton BK: Introduction to colorectal
cancer. Semin Oncol Nurs. 18(Suppl 2): S2–S12. 2002. View Article : Google Scholar
|
|
3
|
Shimada H, Tanaka K, Endou I and Ichikawa
Y: Treatment for colorectal liver metastases: A review. Langenbecks
Arch Surg. 394:973–983. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Maghrabi J, Emam E, Gomaa W, Saggaf M,
Buhmeida A, Al-Qahtani M and Al-Ahwal M: c-MET immunostaining in
colorectal carcinoma is associated with local disease recurrence.
BMC Cancer. 15:6762015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jagadish N, Parashar D, Gupta N, Agarwal
S, Purohit S, Kumar V, Sharma A, Fatima R, Topno AP, Shaha C and
Suri A: A-kinase anchor protein 4 (AKAP4) a promising therapeutic
target of colorectal cancer. J Exp Clin Cancer Res. 34:1422015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
7
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology. 138:2073–2087.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Elzagallaai AA, Garcia-Bournissen F,
Finkelstein Y, Bend JR, Rieder MJ and Koren G: Severe bullous
hypersensitivity reactions after exposure to carbamazepine in a
Han-Chinese child with a positive HLA-B*1502 and
negative in vitro toxicity assays: Evidence for different
pathophysiological mechanisms. J Popul Ther Clin Pharmacol.
18:e1–e9. 2011.
|
|
9
|
Raychaudhuri P and Park HJ: FoxM1: A
master regulator of tumor metastasis. Cancer Res. 71:4329–4333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalin TV, Ustiyan V and Kalinichenko VV:
Multiple faces of FoxM1 transcription factor: Lessons from
transgenic mouse models. Cell Cycle. 10:396–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang IC, Chen YJ, Hughes D, Petrovic V,
Major ML, Park HJ, Tan Y, Ackerson T and Costa RH: Forkhead box M1
regulates the transcriptional network of genes essential for
mitotic progression and genes encoding the SCF (Skp2-Cks1)
ubiquitin ligase. Mol Cell Biol. 25:10875–10894. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun H, Teng M, Liu J, Jin D, Wu J, Yan D,
Fan J, Qin X, Tang H and Peng Z: FOXM1 expression predicts the
prognosis in hepatocellular carcinoma patients after orthotopic
liver transplantation combined with the Milan criteria. Cancer
Lett. 306:214–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Deshmukh H, Payton JE, Dunham C,
Scheithauer BW, Tihan T, Prayson RA, Guha A, Bridge JA, Ferner RE,
et al: Array-based comparative genomic hybridization identifies
CDK4 and FOXM1 alterations as independent predictors of survival in
malignant peripheral nerve sheath tumor. Clin Cancer Res.
17:1924–1934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Balli D, Zhang Y, Snyder J, Kalinichenko
VV and Kalin TV: Endothelial cell specific deletion of
transcription factor FoxM1 increases urethane-induced lung
carcinogenesis. Cancer Res. 71:40–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calvisi DF, Pinna F, Ladu S, Pellegrino R,
Simile MM, Frau M, De Miglio MR, Tomasi ML, Sanna V, Muroni MR, et
al: Forkhead box M1B is a determinant of rat susceptibility to
hepatocarcinogenesis and sustains ERK activity in human HCC. Gut.
58:679–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Chen GQ, Tang CF, Shi XK, Lin CY, Fatima
S, Pan XH, Yang DJ, Zhang G, Lu AP, Lin SH and Bian ZX:
Halofuginone inhibits colorectal cancer growth through suppression
of Akt/mTORC1 signaling and glucose metabolism. Oncotarget.
6:24148–24162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rasool S, Kadla SA, Rasool V and Ganai BA:
A comparative overview of general risk factors associated with the
incidence of colorectal cancer. Tumour Biol. 34:2469–2476. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Van Cutsem E, Nordlinger B, Adam R, Köhne
CH, Pozzo C, Poston G, Ychou M and Rougier P; European Colorectal
Metastases Treatment Group: Towards a pan-European consensus on the
treatment of patients with colorectal liver metastases. Eur J
Cancer. 42:2212–2221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poston GJ, Figueras J, Giuliante F, Nuzzo
G, Sobrero AF, Gigot JF, Nordlinger B, Adam R, Gruenberger T, Choti
MA, et al: Urgent need for a new staging system in advanced
colorectal cancer. J Clin Oncol. 26:4828–4833. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–95. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai Y, Balli D, Ustiyan V, Fulford L,
Hiller A, Misetic V, Zhang Y, Paluch AM, Waltz SE, Kasper S and
Kalin TV: Foxm1 expression in prostate epithelial cells is
essential for prostate carcinogenesis. J Biol Chem.
288:22527–22541. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fan Q, Cai Q and Xu Y: FOXM1 is a
downstream target of LPA and YAP oncogenic signaling pathways in
high grade serous ovarian cancer. Oncotarget. 6:27688–27699. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu G, Zhou A, Xue J, Huang C, Zhang X,
Kang SH, Chiu WT, Tan C, Xie K, Wang J and Huang S: FoxM1 promotes
breast tumorigenesis by activating PDGF-A and forming a positive
feedback loop with the PDGF/AKT signaling pathway. Oncotarget.
6:11281–11294. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Consolaro F, Basso G, Ghaem-Magami S, Lam
EW and Viola G: FOXM1 is overexpressed in B-acute lymphoblastic
leukemia (B-ALL) and its inhibition sensitizes B-ALL cells to
chemotherapeutic drugs. Int J Oncol. 47:1230–1240. 2015.PubMed/NCBI
|
|
26
|
Barger CJ, Zhang W, Hillman J, Stablewski
AB, Higgins MJ, Vanderhyden BC, Odunsi K and Karpf AR: Genetic
determinants of FOXM1 overexpression in epithelial ovarian cancer
and functional contribution to cell cycle progression. Oncotarget.
6:27613–27627. 2015. View Article : Google Scholar : PubMed/NCBI
|