Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney cancer in adults, accounting for ~90% of all renal tumors
and 3.9% of all cancer cases (1,2). RCC
is characterized by a lack of early-warning signs, protean clinical
manifestations, and resistance to radiotherapy and chemotherapy
(3). As a result, 25% of patients
present with advanced disease when initially diagnosed with RCC.
Furthermore, a third of patients who undergo resection of localized
disease exhibit recurrence (4).
The survival of patients with localized tumors who undergo radical
nephrectomy is markedly improved compared with patients with
regional and distant metastasis, thus demonstrating the importance
of early detection and treatment (5). Therefore, the detection of novel
biomarkers to aid early diagnosis is required.
Recently, numerous studies have highlighted the role
of a group of long non-coding RNAs (lncRNAs) in carcinogenesis, and
suggested that this class of molecules may be used as biomarkers in
cancer (6–11). LncRNAs (length, 200 nt to
~100,000nt) constitute a novel class of mRNA-like transcripts with
no protein-coding capacity (7). In
recent years, an increasing amount of lncRNAs have been identified
and observed to play crucial regulatory roles in various biological
processes, including embryonic development and carcinogenesis
(8), at transcriptional,
post-transcriptional and translational levels (9). For example, HOX transcript antisense
RNA and prostate cancer associated transcript 1, the first
identified lncRNAs, have been demonstrated to be critical in cancer
due to their interaction with the polycomb repressive complex 2, by
which they regulate numerous genes (10,11).
However, few studies have investigated the involvement of lncRNAs
in renal cancer progression.
The lncRNA urothelial carcinoma associated 1 (UCA1),
which was originally identified in bladder transitional cell
carcinoma (12), has been reported
to have a potential role in the progression of bladder cancer
(13–15). UCA1 has been reported to be
upregulated in several tumor tissues, including bladder, colon,
cervix, lung, breast, colorectal and brain cancer (13,16–19).
Furthermore, UCA1 promotes cell proliferation and transformation
(anchorage-independent growth), increases cell motility and
invasion, induces drug resistance, and reduces prognosis in cancer
(13,16,19).
However, the role of UCA1 in renal cancer remains to be elucidated.
The aim of the present study was to determine the expression of
UCA1 in renal tissues and cell lines, and the function of UCA1 with
regards to the regulation of cell growth, migration and apoptosis
in renal cancer. Further research is required to investigate the
roles and target genes of UCA1.
Materials and methods
Tumor tissues and cell lines
RCC tissue and adjacent normal tissue samples were
collected from 46 patients with RCC from Peking University Shenzhen
Hospital (Shenzhen, China). The tumor tissue samples contained
80–90% cancer cells, and the paired adjacent normal tissue samples
were obtained from ≥2 cm away from the tumor border and were
negative for tumor cells, as detected by microscopy. All fresh
tissue samples were immediately immersed in RNAlater (Qiagen GmbH,
Hilden, Germany) following surgical resection, frozen in liquid
nitrogen and stored at −80°C until further use. Informed consent
was obtained from participants for the use of their tissues in the
present study, and the study protocol was approved by the Ethics
Committees of Peking University Shenzhen Hospital. The diagnosis of
RCC was histopathologically confirmed and the data on all subjects
were obtained from medical records, pathology reports and personal
interviews. The clinicopathological information of the patients is
presented in Table I. Stage
classification was performed according to the 2010 American Joint
Committee on Cancer staging system (20).
 | Table IClinicopathological features of
patients with renal cell carcinoma. |
Table I
Clinicopathological features of
patients with renal cell carcinoma.
| Characteristic | Number of
cases |
|---|
| Mean age range
(years) | 51 (27–72) |
| Gender | |
| Male/female | 29/17 |
| Histological
type | |
| Clear
cell/papillary | 41/5 |
| pT-stage | |
| T1/T2/T3+T4 | 25/19/2 |
| Fuhrman grade | |
| I/II/III/IV | 15/20/8/3 |
| AJCC clinical
stages | |
| I/II/III+IV | 26/17/3 |
The human RCC cell lines: 786-O, ACHN, 769P and
Caki-2 cells, were obtained from the American Type Culture
Collection (Manassas, VA, USA). The human embryonic kidney 293T
(HEK-293T) cell line was purchased from the Type Culture Collection
of the Chinese Academy of Medical Sciences (Shanghai, China).
BamHI and EcoRI restriction enzymes were purchased
from Takara Bio, Inc., (Tokyo, Japan). All cell lines were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1%
antibiotics (100 units/ml penicillin and 100 mg/ml streptomycin
sulfates) and 1% glutamate (all Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified chamber containing 5%
CO2.
RNA extraction, reverse transcription and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Samples were homogenized using a motor-driven tissue
grinder (OSE-Y10; Tiangen Biotech Co., Ltd., Beijing, China) prior
to RNA extraction. Total RNA was extracted from the tissues (50–100
mg) and cell lines (1.5×106) using RNAiso Plus reagent
(Takara Bio, Inc.) for 5 min at room temperature, according to the
manufacturer's protocol. Following vigorous shaking by hand for 15
sec, the samples were centrifuged at 12,000 × g for 15 min at 4°C.
The concentration and quality of RNA was measured using the
NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific,
Inc., Pittsburgh, PA, USA) at 260 and 280 nm (A260/280 ratio). The
RNA samples with 260/280 ratios 1.8–2.0 were used for further
experiments. Total RNA was converted into cDNA using the
PrimeScript™ RT reagent kit (Takara Bio, Inc.) at 37°C for 15 min
followed by 85°C for 5 sec and was held at 4°C..
The expression levels of UCA1 in tissues and cell
lines were quantified using SYBR® Premix Ex Taq
II (Takara Bio, Inc.), according to the manufacturer's protocol, on
the Roche LightCycler® 480 Real-Time PCR system (Roche
Diagnostics, Basel, Switzerland). PCR cycling conditions were as
follows: 95°C for 15 min, followed by 40 cycles at 95°C for 15 sec
and 55°C for 30 sec, and final extension at 72°C for 30 sec.
Specific UCA1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
primers were purchased from Invitrogen (Thermo Fisher Scientific,
Inc.) (Table II). GAPDH was used
as the endogenous control to normalize the data. The expression
levels were presented as fold differences relative to GAPDH, based
on the following equation: Relative quantification
=2−ΔΔCq [ΔΔCq = (mean Cqtumor -
meanC-qcontrol) - (mean Cqnormal - mean
Cqcontrol) (21).
RT-qPCR was performed in triplicate, including no-template
controls. The amplification of the appropriate product was
confirmed by melting curve analysis following amplification.
 | Table IISequences of siRNAs and primers. |
Table II
Sequences of siRNAs and primers.
| Name | Sequence,
5′-3′ |
|---|
| UCA1 | F:
TCGGCTTAGTGGCTGAAGA |
| R:
GGTCCATTGAGGCTGTAGAGT |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
| R:
GGCTGTTGTCATACTTCTCATGG |
| si-UCA1a | F:
UAGGAUCUGCAAUCAGAACUATT |
| R:
UAGUUCUGAUUGCAGAUCCUATT |
| si-UCA1b | F:
GCACCUUGUUAGCUACAUAAA |
| R:
UAUGUAGCUAACAAGGUGCCA |
| siRNA-NC | F:
UUCUCCGAACGUGUCACGUTT |
| R:
ACGUGACACGUUCGGAGATT |
| siRNA-GAPDH | F:
GUAUGACAACAGCCUCAAGTT |
| R:
CUUGAGGCUGUUGUCAUACTT |
| siRNA-NC-FAM | F:
UAUGUAGCUAACAAGGUGCCA |
| R:
UUCUCCGAACGUGUCACGUTT |
Ectopic expression and gene silencing of
UCA1 in cells
For the analysis of overexpression, PCR-amplified
UCA1, digested with BamHI and EcoRI restriction
enzymes, was subcloned into the pcDNA3.1 mammalian expression
vector (Invitrogen; Thermo Fisher Scientific, Inc.). Primers for
the amplification of the full-length UCA1 were as follows: forward
5′-CCGGAATTCTGACATTCTTCTGGACAATG-3′ and reverse
5′-CCGCTCGAGCTGACTCTTTTAGGAAGATTTCT-3′ (22). The UCA1 expression vector,
pcDNA3.1-UCA1, was verified by sequencing. For the knockdown study
of UCA1, two small interfering (si)RNAs targeting UCA1 (si-UCA1a
and si-UCA1b) were designed and purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). The negative control (siRNA-NC),
positive control (siRNA-GAPDH) and fluorescein amidite-labeled
siRNA-NC (siRNA-NC-FAM) used in the present study were designed and
purchased from Shanghai GenePharma Co., Ltd. The sequences of these
siRNAs are presented in Table
II.
Subsequently, the cells were transfected with
pcDNA3.1-UCA1/pcDNA3.1 empty vector (mock), si-UCA1a, si-UCA1b,
siRNA-NC, siRNA-GAPDH or siRNA-NC-FAM using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), which was mixed with Opti-MEM® I
Reduced Serum Medium (Gibco; Thermo Fisher Scientific, Inc.) 24 h
after plating. Fluorescence microscopy (Axio Imager 2; Carl Zeiss
AG, Oberkochen, Germany) and qPCR were used to assess and quantify
transfection efficiency.
Cell motility assay
A wound scratch assay and a Transwell migration
assay were performed to investigate the motility of RCC cells in
vitro. Approximately 3×105 786-O or ACHN cells were
seeded per 12-well dish and transfected after 24 h with 100 pmol of
either the siRNAs or 1 µg vectors, using
Lipofectamine® 2000. A total of 6 h post-transfection, a
sterile 200 µl pipette tip was used to scrape a clear line
through the cell layer. The cells were washed with
phosphate-buffered saline (PBS) and cultured in serum-free DMEM.
Images of the scratches were captured using a digital camera system
(DMIRB; Leica Microsystems GmbH, Wetzlar, Germany), 0 and 18 h
after the scratches were made. The experiments were performed in
triplicate and repeated ≥3 times.
For the Transwell migration assays, following
transfection with 100 pmol siRNAs and 1 µg vectors in
12-well plates for 24 h, 6×104 786-O and ACHN cells were
harvested and plated into the upper chambers (24-well insert; pore
size, 8 µm; Corning Incorporated, Corning, NY, USA) with 150
µl serum-free DMEM. DMEM (500 µl) supplemented with
10% FBS was added to the lower chambers. The cells were cultured in
a humidified chamber containing 5% CO2 at 37°C for 2
days, prior to being fixed with paraformaldehyde for 25 min and
stained with 0.1% crystal violet for 1 h. The chambers were washed
with PBS three times and dried prior to images being captured with
a digital camera system. The experiments were performed in
triplicate and repeated ≥3 times.
Cell proliferation assay
Cell proliferation assay was performed using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich, St. Louis, MO, USA). Approximately 5×103
786-O and ACHN cells were plated into a 96-well plate and
transfected with either 13.3 pmol siRNA or 125 ng vectors per well.
At 0, 24, 48 or 72 h post-transfection, 20 µl MTT was added
into five wells and incubated for 6 h. The MTT was subsequently
discarded and 120 µl dimethyl sulfoxide (Sigma-Aldrich) was
added to dissolve the purple formazan crystals. Following agitation
for 30 min at room temperature, the optical value of each well was
measured using an enzyme-linked immunosorbent assay microplate
reader (iMark 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at a wavelength of 490 nm (reference wavelength, 630 nm).
Cell apoptosis assay
The apoptotic rate of RCC cells was determined by
flow cytometry (EPICS® Xl-4, Beckman Coulter, Inc.,
Brea, CA, USA). Approximately 3×105 786-O or ACHN cells
were plated in 6-well plates for 24 h and were then transfected
with 200 pmol siRNAs (si-UCA1 or si-NC) and 2 µg
(pcDNA3.1-UCA1 or mock) vectors. At 48 h post-transfection, the
cells in each well, including the floating cells, were pelleted by
centrifugation at 200 × g The cells were washed twice with 4°C PBS,
resuspended in 100 µl 1X binding buffer and stained with 5
µl propidium iodide and 5 µl Annexin V-fluorescein
isothiocyanate (Invitrogen; Thermo Fisher Scientific, Inc.) for 15
min at room temperature. Within 1 h, each cell sample was added and
mixed with 400 µl 1X binding buffer prior to flow cytometry.
Each experiment was conducted at least three times.
Statistical analysis
Relative UCA1 expression data were presented as the
mean ± standard error of the mean (Fig. 1). All remaining data were expressed
as the mean ± standard deviation of three independent experiments.
Statistical analyses were performed using SPSS 19.0 statistical
software (IBM SPSS, Armonk, NY, USA). Statistical significance was
determined using Student's t-test. A paired t-test was used to
compare the expression levels of UCA1 in matched tumor and adjacent
normal samples. P<0.05 was considered to indicate a
statistically significant difference.
Results
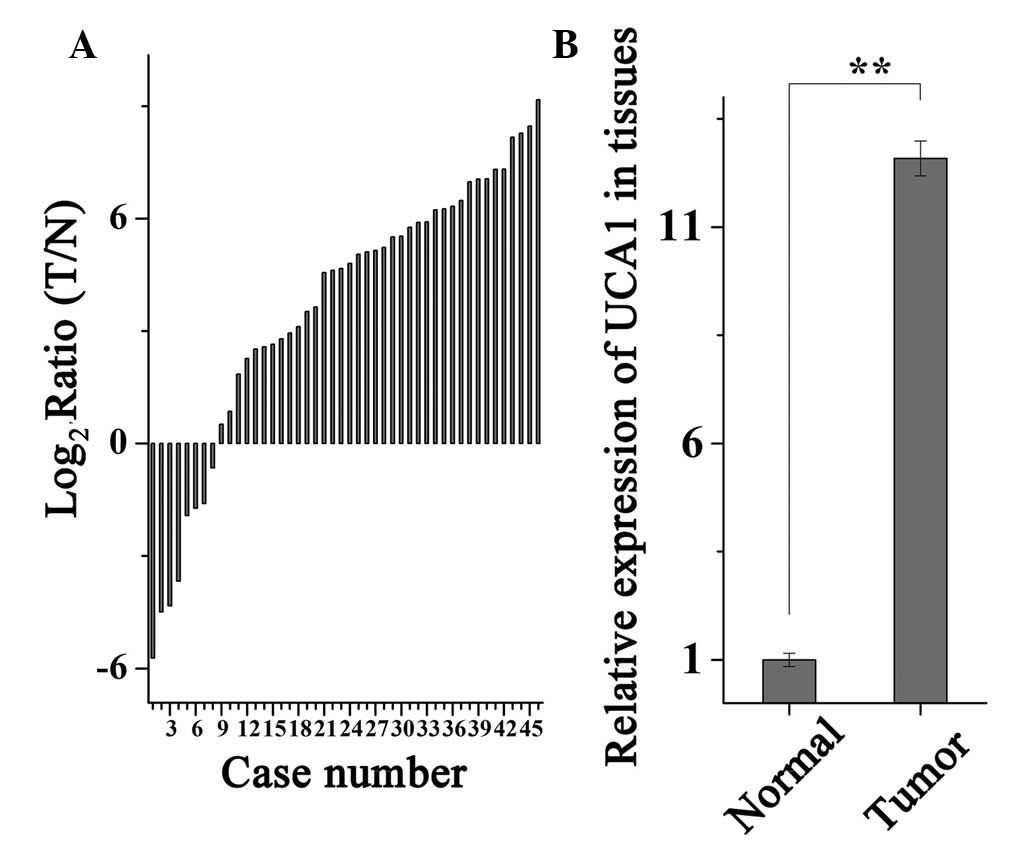
UCA1 is upregulated in RCC tissues and
cell lines
To determine the mRNA expression levels of UCA1,
RT-qPCR was performed on 46 paired RCC tissue and adjacent normal
tissue samples. Relative expression levels of UCA1
(log2; T/N) are presented in Fig. 1A. The expression levels of UCA1
were significantly higher in RCC tissues, as compared with in
adjacent normal tissues (P<0.01), as demonstrated in Fig. 1B.
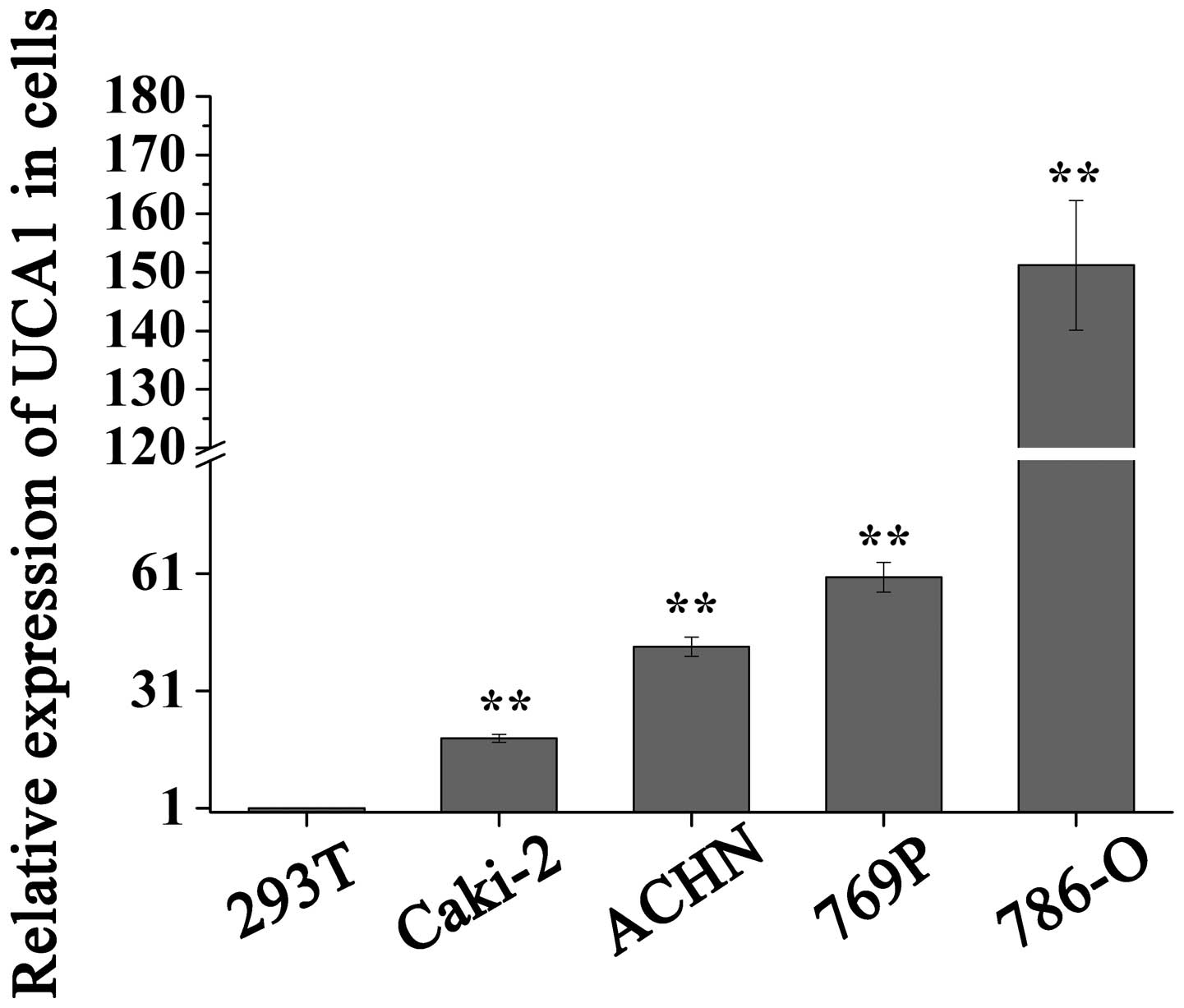
In addition, the present study analyzed the
expression levels of UCA1 in four RCC cell lines (786-O, ACHN, 769P
and Caki-2) and one normal kidney human embryonic kidney cell line
(HEK-293T). As presented in Fig.
2, the expression levels of UCA1 were significantly higher in
786-O, 769P, ACHN and Caki-2 cells (P<0.01), as compared with
the HEK-293T cells, which is consistent with the expression pattern
of UCA1 in RCC tissues. In the functional experiments, two human
RCC cell lines were selected. 786-O and 769P cells exhibited the
highest expression levels of UCA1; however, ACHN exhibited more
successful growth in the laboratory than 769P cells. Therefore, in
the present study, 786-O and ACHN cells were selected for further
experiments.
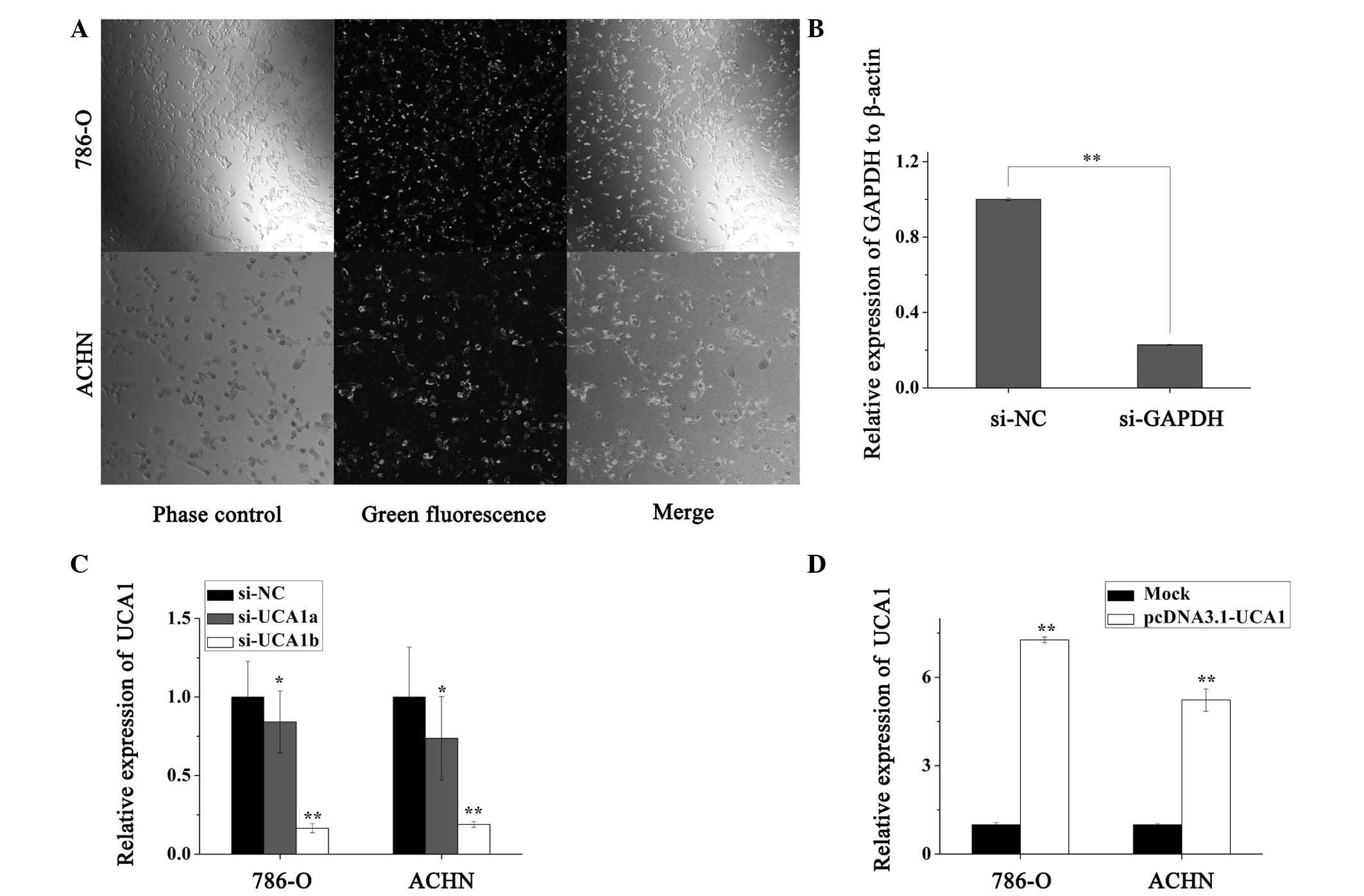
Validation of cell transfection
efficiency
The transfection efficiency was >90%, when the
cells were transfected with FAM-conjugated siRNA-NC, as detected by
fluorescence microscopy (Fig. 3A).
The control experiment was performed by transfecting the cells with
si-GAPDH and si-NC (Fig. 3B). As
presented in Fig. 3C, to quantify
the efficiency of the siRNA, qPCR was performed. The results
indicated that UCA1 was significantly downregulated by 26.3 and
81.1% in ACHN cells (P<0.05), and by 15.8 and 83.4% in 786-O
cells (P<0.05) by si-UCA1a and si-UCA1b, respectively, as
compared with the si-NC group. Therefore, knockdown of UCA1
expression was performed by transfecting the cells with si-UCA1b,
in order to investigate the biological functions of UCA1 in RCC
cells. To verify the overexpression efficiency, qPCR was performed
following transfection with pcDNA3.1-UCA1 or a mock vector. UCA1
was significantly upregulated, 5.23 and 7.27 times compared with
the mock group (P<0.01), in ACHN and 786-O cells respectively
(Fig. 3D).
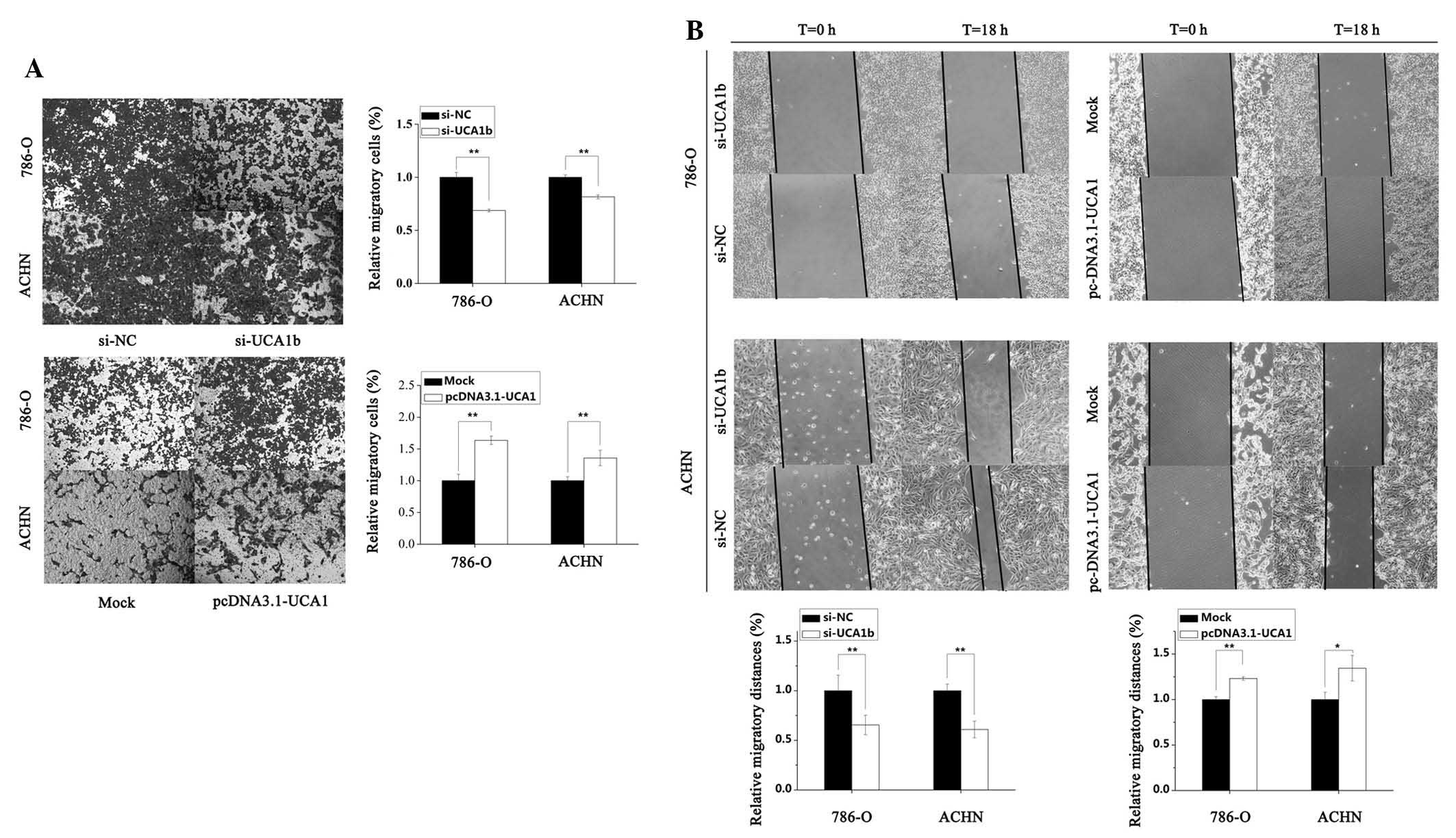
UCA1 increases RCC cell motility
To investigate the effects of UCA1 on cell motility
in 786-O and ACHN RCC cells, Transwell migration and cell scratch
assays were conducted. As presented in Fig. 4A, the results of the Transwell
migration assay demonstrated that the motility of cells transfected
with si-UCA1b was significantly reduced by 30.5% (P<0.01) in
786-O cells and by 18.4% (P<0.01) in ACHN cells, as compared
with the si-NC group. Conversely, the motility of cells transfected
with pcDNA3.1-UCA1 was significantly increased by 63.6% (P<0.01)
and 35.7% (P<0.01) in 786-O and ACHN cells, as compared with the
mock group, respectively. Similar results were exhibited for the
cell scratch assay. Downregulation of UCA1 by transfection of the
cells with si-UCA1b significantly suppressed the migration of 786-O
and ACHN cells by 34.5% (P<0.01) and 39.0% (P<0.01), as
compared with the si-NC group. Conversely, upregulation of UCA1 by
transfection with pcDNA3.1-UCA1 promoted migration by 23.1%
(P<0.01) and 34.5% (P<0.05) in 786-O and ACHN cells
respectively, as compared with the mock group (Fig. 4B).
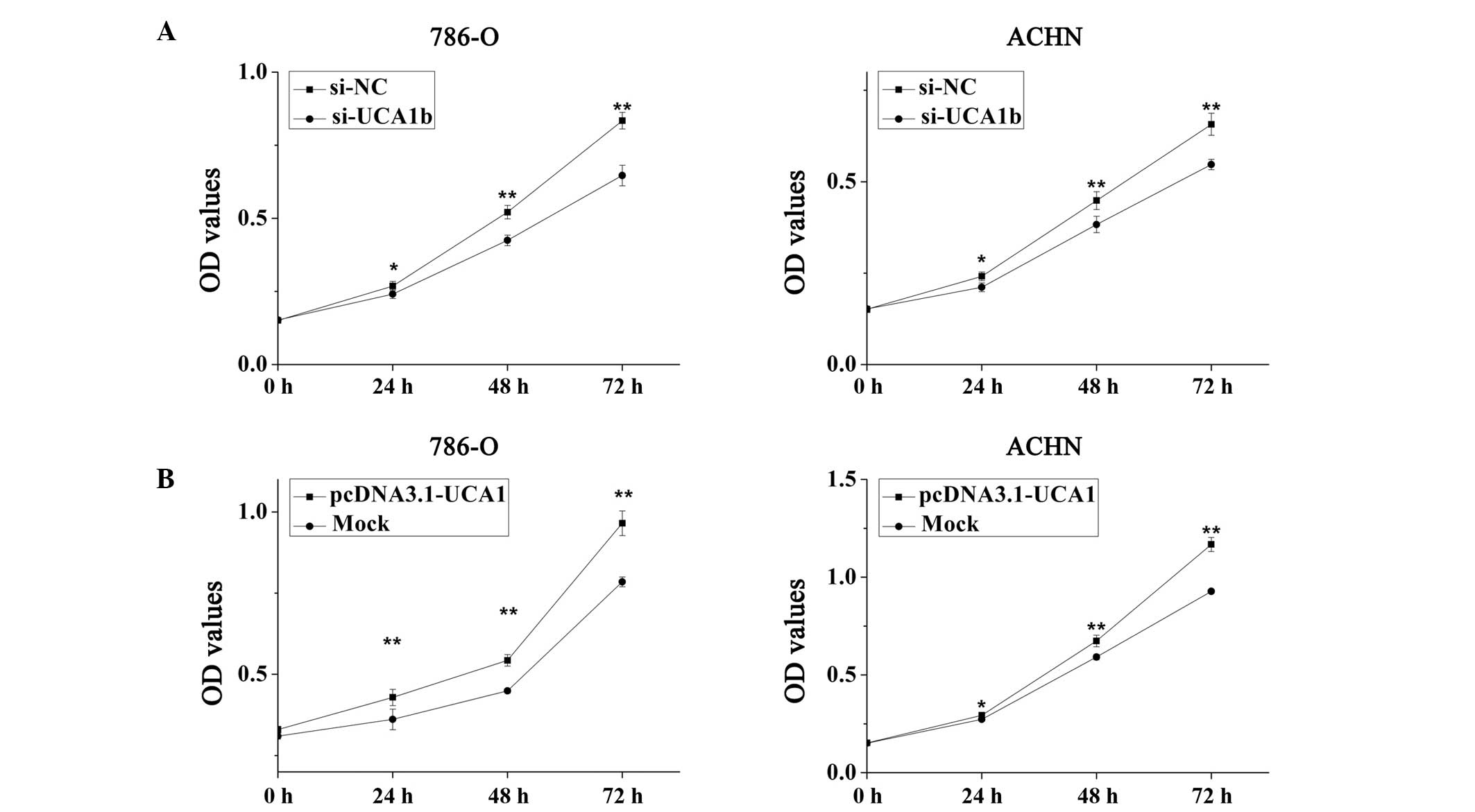
UCA1 promotes cell proliferation
A cell proliferation assay was performed using MTT
in vitro. The results of the MTT assay suggested that
upregulation of UCA1 promoted cell proliferation, whereas
downregulation of UCA1 attenuated cell proliferation. As presented
in Fig. 5A, the cell proliferation
of 786-O and ACHN cells was significantly reduced by 10.31%
(P<0.05), 18.54% (P<0.01) and 22.48% (P<0.01), and 12.43%
(P<0.05), 14.62% (P<0.01) and 16.70% (P<0.01), following
transfection with si-UCA1b for 24, 48 and 72 h respectively, as
compared with the si-NC group. Conversely, the proliferation of
786-O cells was significantly increased by 18.73% (P<0.01),
20.94% (P<0.01) and 22.98% (P<0.01), and the proliferation of
ACHN cells was significantly increased by 7.70% (P<0.05), 13.91%
(P<0.01) and 25.87% (P<0.01), at 24, 48 and 72 h following
transfection with pcDNA3.1-UCA1 respectively, as compared with the
mock group (Fig. 5B).
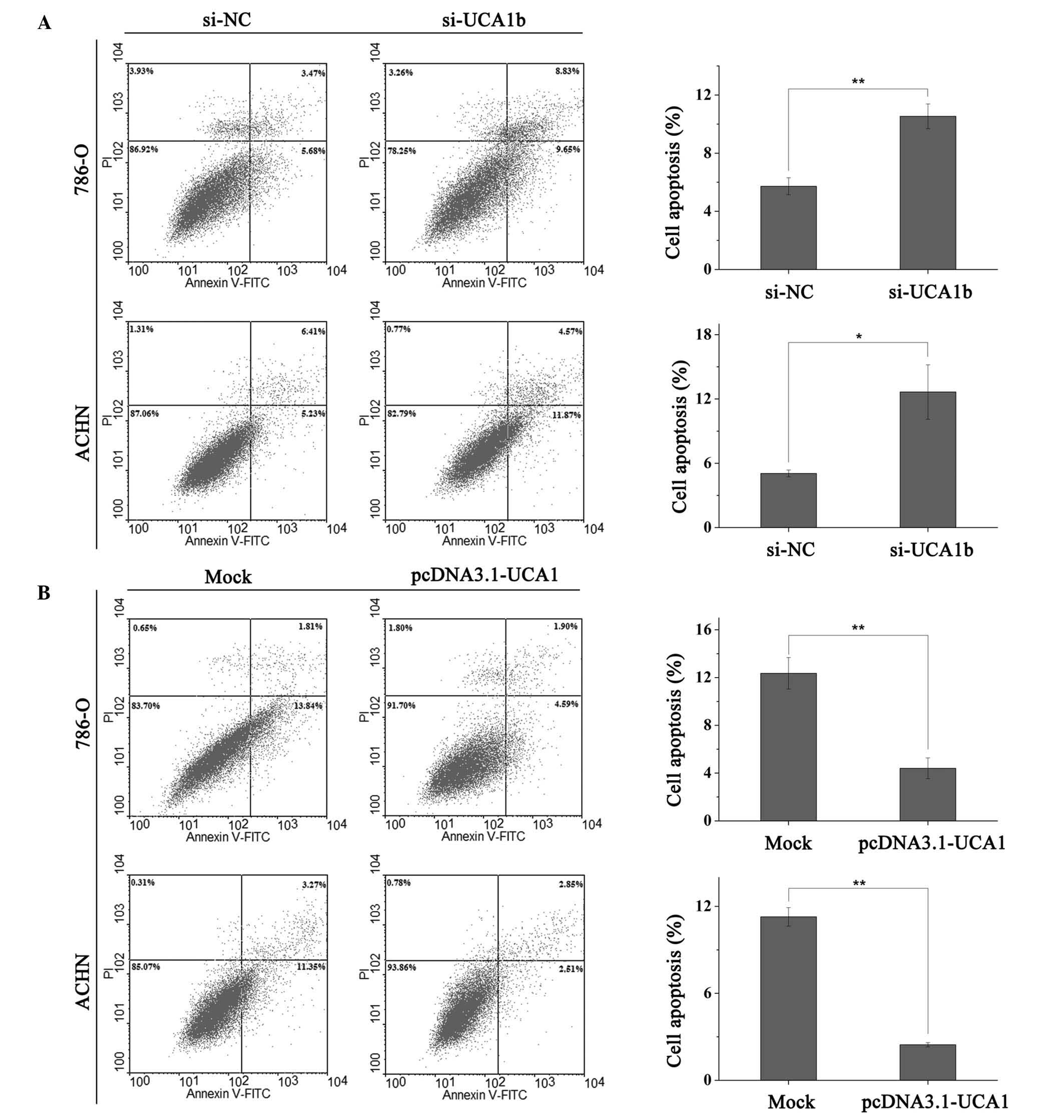
UCA1 suppresses apoptosis
Apoptotic rate was quantified by flow cytometry
(EPICS® Xl-4, Beckman Coulter, Inc.). UCA1 had a
negative effect on apoptosis, indicating that ectopic expression of
UCA1 suppressed apoptosis, whereas downregulation of UCA1 increased
apoptosis of RCC cells. Following transfection (48 h) with siRNAs
(si-NC or si-UCA1b) or vectors (pcDNA3.1-UCA1 and mock), 786-O and
ACHN cells were collected for apoptotic quantification. The early
apoptotic rate of 786-O cells transfected with si-NC or si-UCA1b
was 5.72 vs. 10.53% (P<0.01) and the apoptotic rate of ACHN
cells was 5.05 vs. 12.65% (P<0.05; Fig. 6A). As presented in Fig. 6B, upregulation of UCA1 by
transfection with pcDNA3.1-UCA1 inhibited the apoptotic rate of
786-O and ACHN cells from 12.35 to 4.4% (P<0.01) and from 11.27
to 2.45% (P<0.01) respectively, as compared with mock
transfection.
Discussion
RCC is a type of cancer associated with poor
prognosis, since ~25% of patients are initially diagnosed with
advanced RCC, and up to 33.3% of patients with clinically localized
disease develop recurrence post-surgery (4). It is therefore critical to identify
novel biomarkers for the early diagnosis and monitoring of RCC, and
as potential therapeutic targets for the treatment of RCC. LncRNAs
have recently received attention and have been observed to be
dysregulated in numerous types of disease, particularly in cancer
(23). Dysregulated lncRNA
expression is important in diverse cellular processes, including
cell migration, invasion, proliferation and apoptosis (7,24).
Although several lncRNAs have been demonstrated as crucial in RCC,
including onco-lncRNAs (metastasis associated lung adenocarcinoma
transcript 1 and KCNQ1 opposite strand/antisense transcript 1) and
tumor suppressor lncRNAs (growth arrest specific 5, H19 and
maternally expressed 3) (25), the
present study focused on the effects of UCA1 on the onset and
development of RCC.
In the present study, the upregulation of UCA1 in 46
paired RCC tissues and four RCC cell lines was compared with
adjacent normal tissues and HEK-293T cells using qPCR. To
investigate the function of UCA1 in RCC cell lines (786-O and
ACHN), primers were designed and constructed to confirm that
si-UCA1b silenced the expression of UCA1 and that the
overexpression vector pcDNA3.1-UCA1 upregulated the expression of
UCA1. These models of up- and downregulation were then examined
using various assays, including Transwell and scratch migration,
MTT and apoptosis assays. The results demonstrated that suppression
of UCA1 inhibited migration and cell proliferation, and induced
cell apoptosis, whereas overexpression of UCA1 promoted migration
and cell proliferation, and reduced cell apoptosis. These outcomes
suggested that UCA1 may function as an oncogene in RCC by
regulating cellular migration, proliferation and apoptosis.
Although aberrant UCA1 expression has been observed,
and its function on the cell biology of migration, invasion,
proliferation, apoptosis, glycolysis (26) and senescence (27) has been described in certain types
of cancer, the signaling pathway involving UCA1 remains to be
elucidated. LncRNAs function via chromatin modification,
transcription initiation, co- and post-transcriptional regulation
or as competitive endogenous RNAs to microRNAs (8,28).
In bladder cancer, the UCA1 gene is localized in the cytoplasm and
its expression is correlated with the progression of bladder cancer
(14). In a microarray assay,
UCA1a-overexpressing bladder cancer cells were shown to exhibit
alterations in the gene expression of platelet-derived growth
factor β polypeptide, Fas and ATM serine/threonine kinase, which
regulate cell apoptosis and tumorigenesis (29). Another study also applied
microarray analysis and qPCR to identify and confirm the
dysregulated genes affected by ectopic UCA1 expression, including
upregulated wingless-type MMTV integration site family member 6,
cytochrome P450, 1A1 and aurora kinase C, and downregulated
methyl-CpG binding domain protein 3 and serine/arginine-rich
protein-specific kinase 1. These genes have already been detected
in tumorigenesis, embryonic development and cisplatin resistance
(13). However, the molecular
mechanism remains to be elucidated and further studies are required
to investigate how UCA1 affects these genes and exerts its
function.
In cancer development, UCA1 has been shown to be
upregulated by transcription factor CCAAT/enhancer binding protein
α (30) and hypoxia (31). In addition, overexpression of UCA1
may increase chemoresistance in bladder cancer by regulating Wnt
signaling (32), and knockdown of
UCA1 may accelerate cell cycle progression in bladder cancer via
cAMP response element-binding protein and the
phosphatidylinositol-4,5-bisphosphate 3-kinase dependent signaling
pathway (15). UCA1 may also be
involved in activation of the Akt signaling pathway by Ets-2,
regulating apoptosis in bladder cancer cells (33). Aerobic glycolysis is critical for
most cancer cells to generate the energy required for cellular
processes, instead of the more efficient mitochondrial oxidative
phosphorylation; this phenomenon is known as the Warburg effect
(34). UCA1 promotes glycolysis by
upregulating hexokinase 2 via activation of mammalian target of
rapamycin-signal transducer and activator of transcription 3 and
suppression of the microRNA-143 signaling pathway (26). The role of UCA1 and the
UCA1-mediated signaling pathway in the onset and development of RCC
remains to be elucidated; therefore, further studies are required
to investigate and identify target genes of UCA1.
In clinical settings, UCA1 in urine may be used as a
noninvasive biomarker, with a high level of sensitivity and
specificity, for the early diagnosis and post-operative follow-up
of transitional cell carcinoma (12,35,36).
In addition to bladder cancer, increased expression of UCA1 has
been associated with the later stages and metastasis of melanoma
(17). In colorectal cancer, a
high level of UCA1 expression is notably correlated with a larger
tumor size, less differentiated histology, greater tumor depth and
a markedly poorer prognosis, as compared with in patients with low
UCA1 expression (18).
In conclusion, UCA1 expression was upregulated in
RCC tissues and cell lines. Due to the limited number of RCC
samples used in the present study, no correlation was observed
between the UCA1 expression and clinicopathological
characteristics. Ectopic overexpression and gene silencing of UCA1
in RCC cell lines had opposite effects on cellular proliferation,
migration and apoptosis, and the results suggested that UCA1 may
function as an oncogene in RCC. The results of the present study
indicated that UCA1 may be considered a promising biomarker for
diagnosis and a potential therapeutic target in RCC. Further
research is required to investigate the roles and target genes of
UCA1.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81101922),
the Science and Technology Fund Project of Shenzhen (grant nos.
JCYJ20130402114702124 and JCYJ20150403091443329) and the fund of
Guangdong Key Medical Subject.
References
|
1
|
Tavani A and La Vecchia C: Epidemiology of
renal-cell carcinoma. J Nephrol. 10:93–106. 1997.PubMed/NCBI
|
|
2
|
National Cancer Institute: Surveillance,
Epidemiology, and End Results Program. SEER stat fact sheets:
Kidney and renal pelvis cancer. Available at: http://seer.cancer.gov/statfacts/html/kidrp.html.
Accessed 30, 2014.
|
|
3
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J. 353:2477–2490. 2005. View Article : Google Scholar
|
|
5
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wu Z, Fu X and Han W: Long noncoding
RNAs: Insights from biological features and functions to diseases.
Med Res Rev. 33:517–553. 2013. View Article : Google Scholar
|
|
10
|
Takagi M, Absalon MJ, McLure KG and Kastan
MB: Regulation of p53 translation and induction after DNA damage by
ribosomal protein L26 and nucleolin. Cell. 123:49–63. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie XJ, Li X, Wang F and Chen W: Cellular
localization and tissue expression pattern of UCA1, a non-coding
RNA. Nan Fang Yi Ke Da Xue Xue Bao. 30:57–60. 2010.In Chinese.
PubMed/NCBI
|
|
15
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsang WP, Wong TW, Cheung AH, Co CN and
Kwok TT: Induction of drug resistance and transformation in human
cancer cells by the noncoding RNA CUDR. RNA. 13:890–898. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han Y, Yang YN, Yuan HH, Zhang TT, Sui H,
Wei XL, Liu L, Huang P, Zhang WJ and Bai YX: UCA1, a long
non-coding RNA up-regulated in colorectal cancer influences cell
proliferation, apoptosis and cell cycle distribution. Pathology.
46:396–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7th edition.
Springer; New York, NY: pp. 479–89. 2010
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Wang T, Yuan J, Feng N, Li Y, Lin Z, Jiang
Z and Gui Y: Hsa-miR-1 downregulates long non-coding RNA urothelial
cancer associated 1 in bladder cancer. Tumour Biol. 35:10075–10084.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Li X, Wu S, Xue M and Chen W: Long
non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase
2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci.
105:951–955. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kumar PP, Emechebe U, Smith R, Franklin S,
Moore B, Yandell M, Lessnick SL and Moon AM: Coordinated control of
senescence by lncRNA and a novel T-box3 co-repressor complex.
Elife. 3:e028052014. View Article : Google Scholar
|
|
28
|
Yang L, Froberg JE and Lee JT: Long
noncoding RNAs: Fresh perspectives into the RNA world. Trends
Biochem Sci. 39:35–43. 2014. View Article : Google Scholar :
|
|
29
|
Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X
and Li X: Long non-coding RNA UCA1a(CUDR) promotes proliferation
and tumorigenesis of bladder cancer. Int J Oncol. 41:276–284.
2012.PubMed/NCBI
|
|
30
|
Xue M, Li X, Wu W, Zhang S, Wu S, Li Z and
Chen W: Upregulation of long non-coding RNA urothelial carcinoma
associated 1 by CCAAT/enhancer binding protein α contributes to
bladder cancer cell growth and reduced apoptosis. Oncol Rep.
31:1993–2000. 2014.PubMed/NCBI
|
|
31
|
Xue M, Li X, Li Z and Chen W: Urothelial
carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted
long noncoding RNA that enhances hypoxic bladder cancer cell
proliferation, migration, and invasion. Tumour Biol. 35:6901–6912.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu W, Zhang S, Li X, Xue M, Cao S and Chen
W: Ets-2 regulates cell apoptosis via the Akt pathway, through the
regulation of urothelial cancer associated 1, a long non-coding
RNA, in bladder cancer cells. PLoS One. 820132013.
|
|
34
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Srivastava AK, Singh PK, Rath SK, Dalela
D, Goel MM and Bhatt ML: Appraisal of diagnostic ability of UCA1 as
a biomarker of carcinoma of the urinary bladder. Tumour Biol.
35:11435–11442. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Hao H, Zhang CJ, Yang XY, He Q
and Lin J: Evaluation of novel gene UCA1 as a tumor biomarker for
the detection of bladder cancer. Zhonghua Yi Xue Za Zhi.
92:384–387. 2012.In Chinese. PubMed/NCBI
|