Introduction
Squamous cell carcinoma (SCC) is a common skin
cancer originating from the upper layers of skin epidermis. The
incidence of SCC is relatively high, ranking second among the
non-melanoma skin cancers (1).
Numerous factors are implicated in the pathogenesis of SCC, of
which ultraviolet (UV) radiation is regarded as the most important
environmental risk factor. UV radiation induces DNA damage and
mutations of numerous susceptible genes, including the tumor
suppressor, p53 (2). In addition,
several intracellular signal regulators are involved in cancer
development and progression. Examples include epidermal growth
factor receptor, nuclear factor-κ-light-chain-enhancer of activated
B cells and the Wnt/β-catenin signaling pathways (3–7).
In canonical Wnt/β-catenin signaling, binding of Wnt
ligands to their cognate membrane receptors leads to the
inactivation of the β-catenin degradation complex, resulting in the
stabilization of cytoplasmic β-catenin. Once accumulated, β-catenin
locates to the nucleus and interacts with the Lef-1/TCF family of
DNA-binding proteins to generate a functional transcription factor
complex (8). The pivotal role of
Wnt/β-catenin signaling in cancer development has been previously
described (9–11). The importance of β-catenin in skin
cancer is supported by the fact that epidermis-specific ablation of
the β-catenin gene results in the loss of cancer initiating cells
and complete tumor regression (12). In addition, a previous genetic
study showed that chromosome loci harboring Wnt signaling genes are
frequently amplified in SCC (13),
indicating a potential importance of Wnt/β-catenin signaling in
cutaneous SCC.
Although the pivotal role of Wnt/β-catenin signaling
in SCC is well recognized, the downstream effectors of β-catenin
remain to be clearly elucidated. The present study identified Sox9
as a β-catenin-regulated gene and demonstrated that Sox9 has a
potential role in the regulation of SCC cells.
Materials and methods
Cell culture
The SCC12 human squamous cell carcinoma cell line
was provided by Professor Tae-Jin Yoon (Gyeongsang National
University, Jinju, South Korea) and maintained in Dulbecco's
modified Eagle's medium (DMEM), supplemented with 10% fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Human keratinocytes were isolated from skin specimens then
immortalized using a recombinant retrovirus expressing Simian virus
40 large T antigen (SV40T), as described in a previous report
(14). SV40T-transformed human
epidermal keratinocytes (SV-HEK) were maintained in
keratinocyte-serum free medium (Thermo Fisher Scientific, Inc.),
supplemented with bovine pituitary extract and recombinant human
epidermal growth factor (Thermo Fisher Scientific, Inc.) (14). Human dermal fibroblasts were
primary cultured and maintained in DMEM, supplemented with 10% FBS.
Human melanocytes were cultured in Medium 254 (Cascade Biologics,
Portland, OR, USA) and human melanocyte growth supplement (Cascade
Biologics) (15,16). Cells were maintained at 37°C in an
atmosphere 5% CO2 and 90% relative humidity.
Immunohistochemistry
All human skin samples were obtained under the
written informed consent of donors, in accordance with the Ethical
Committee approval process of the Institutional Review Board of
Chungnam National University School of Medicine. Paraffin sections
of skin specimens were dewaxed, rehydrated and washed three times
with phosphate-buffered saline (PBS). The tissue sections were
subsequently incubated with proteinase K (Dako, Carpinteria, CA,
USA) for 5 min at 37°C, and treated with H2O2
for 10 min at room temperature. The tissue sections were blocked in
0.1% Tween-20 and 1% bovine serum albumin (Sigma-Aldrich, St.
Louis, MO, USA) in PBS for 30 min. Following blocking, the tissue
sections were incubated at 4°C with the appropriate primary
antibodies as follows: Rabbit polyclonal anti-β-catenin (1:100
dilution; cat. no. sc-7199; Santa Cruz Biotechnologies, Inc., Santa
Cruz, CA, USA); mouse monoclonal anti-Sox9 (1:200 dilution; cat.
no. ab76997; Abcam, Cambridge, MA, USA) overnight. Subsequently,
the tissue sections were incubated with the following
peroxidase-conjugated secondary antibodies (dilution, 1:1,000):
Horseradish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit
immunoglobulin (Ig; cat. no. P0448) and HRP-conjugated polyclonal
rabbit anti-mouse Ig (cat. no. P0161; both Dako), visualized using
3,3′-diaminobenzidine solution from the Chemmate envision detection
kit (Dako) and photographed under a Diaphot inverted microscope
(Nikon Corporation, Tokyo, Japan).
Western blot analysis
The cells were lysed in Proprep solution (Intron,
Daejeon, Korea). The total protein was quantified using a
bicinchoninic acid protein assay reagent (Pierce Biotechnology,
Inc., Rockford, IL, USA). The samples were separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (100 V for 1 h)
and transferred onto nitrocellulose membranes (Thermo Fisher
Scientific, Inc.). The membranes were then blocked using 5% skimmed
milk in Tris-buffered saline and Tween-20 (Sigma-Aldrich) for 1 h
at room temperature, and incubated (for 1 h at room temperature)
with mouse monoclonal anti-Sox9 (1:200 dilution), rabbit polyclonal
anti-β-catenin (1:100 dilution) and mouse monoclonal anti-β-actin
[1:1,000 dilution (cat. no. A2228), Sigma-Aldrich] primary
antibodies. Following primary antibody incubation, the membranes
were incubated for 1 h at room temperature with the following
secondary antibodies: Rabbit anti-mouse IgG (cat. no. ab97046) and
goat anti-rabbit IgG (cat. no. .ab6721; both Abcam) and were
visualized by enhanced chemiluminescence (Intron).
Production of recombinant adenovirus
The recombinant adenovirus expressing an 87-amino
acid N-terminally truncated β-catenin (Ad/ΔN87-β-cat) or Sox9
(Ad/Sox9) were previously described (17,18).
For knockdown experiments, the recombinant adenovirus expressing
microRNA (miR) was generated. The target sequences for β-catenin
and Sox9 were designed using BLOCK-iT™ RNAi Designer (Thermo Fisher
Scientific, Inc.). The double-stranded DNA oligonucle-otides were
synthesized and cloned into the parental vector, pcDNA6.2-GW
(Thermo Fisher Scientific, Inc.). In this vector system, the miR
sequence for the target gene is located downstream of emerald green
fluorescent protein (EmGFP) coding sequence, allowing the
identification of miR expressing cells by observing GFP under an
IX71 fluorescent microscope (Olympus Corporation, Tokyo, Japan).
This vector was termed pcDNA6.2-GW/EmGFP-miR. The expression
cassette for miR was moved into the pENT/CMV vector and then
adenovirus was generated. The miR sequences were as follows:
β-catenin, top strand:
5′-TGCTGTCTGCATGCCCTCATCTAATGGTTTTGGCCACTGACTGACCATTAGATGGGCATGCAGA-3′
and bottom strand:
5′-CCTGTCTGCATGCCCATCTAATGGTCAGTCAGTGGCCAAAACCATTAGATGAGGGCATGCAGAC-3′;
Sox9, top strand:
5′-TGCTGTGTTCTTGCTGGAGCCGTTGAGTTTTGGCCACTGACTGACTCAACGGCCAGCAAGAACA-3′
and bottom strand:
5′-CCTGTGTTCTTGCTGGCCGTTGAGTCAGTCAGTGGCCAAAACTCAACGGCTCCAGCAAGAACA-3′C.
For adenovirus transduction, SCC12 cells were incubated with 10
multiplicity of infection of adenovirus for 6 h at 37°C in an
atmosphere 5% CO2 and 90% relative humidity. The cells
were replenished with fresh medium and incubated for a further 2
days.
Colony forming assay
The cells were trypsinized and counted using a
hemocytometer. Following counting, ~1,000 cells were resuspended in
DMEM, supplemented with 10% FBS, and were seeded into 100 mm
culture dishes. The cells were incubated for 2–3 weeks and stained
with crystal violet (Sigma-Alrdrich).
Statistical analysis
The data were statistically analyzed using one-way
analysis of variance with SPSS software (v22.0; IBM SPSS, Chicago,
IL, USA). P<0.01 was considered to indicate a statistically
significant difference.
Results
In order to elucidate the importance of β-catenin in
cutaneous SCC, the present study first determined the expression of
β-catenin by immunohistochemistry. In normal skin, β-catenin
immunoreactivity was observed in all epidermal layers, with a
characteristic membrane staining pattern. In SCC, very intense
immunostaining of β-catenin was observed, and notably, many of the
SCC cells exhibited nuclear staining of β-catenin (Fig. 1A). These results suggested a
fundamental role of β-catenin signaling in cutaneous SCC. The
relative expression level of β-catenin in SCC12 cell line and
skin-comprising cells, including SV-HEK, fibroblasts and
melanocytes, was determined. Although it was not significant, the
total β-catenin level was marginally increased in the SCC12 cells
compared with the keratinocytes (Fig.
1B). To confirm the potential role of β-catenin, clonogenic
assays were performed as an in vitro tumorigenic test
(19). Following transduction of
recombinant adenovirus expressing miR specific for β-catenin, gene
knockdown was confirmed (Fig. 1C).
Knockdown of β-catenin markedly reduced the colony-forming
potential of SCC12 cells (Fig.
1D), supporting the pivotal role of β-catenin in the
tumorigenesis of SCC.
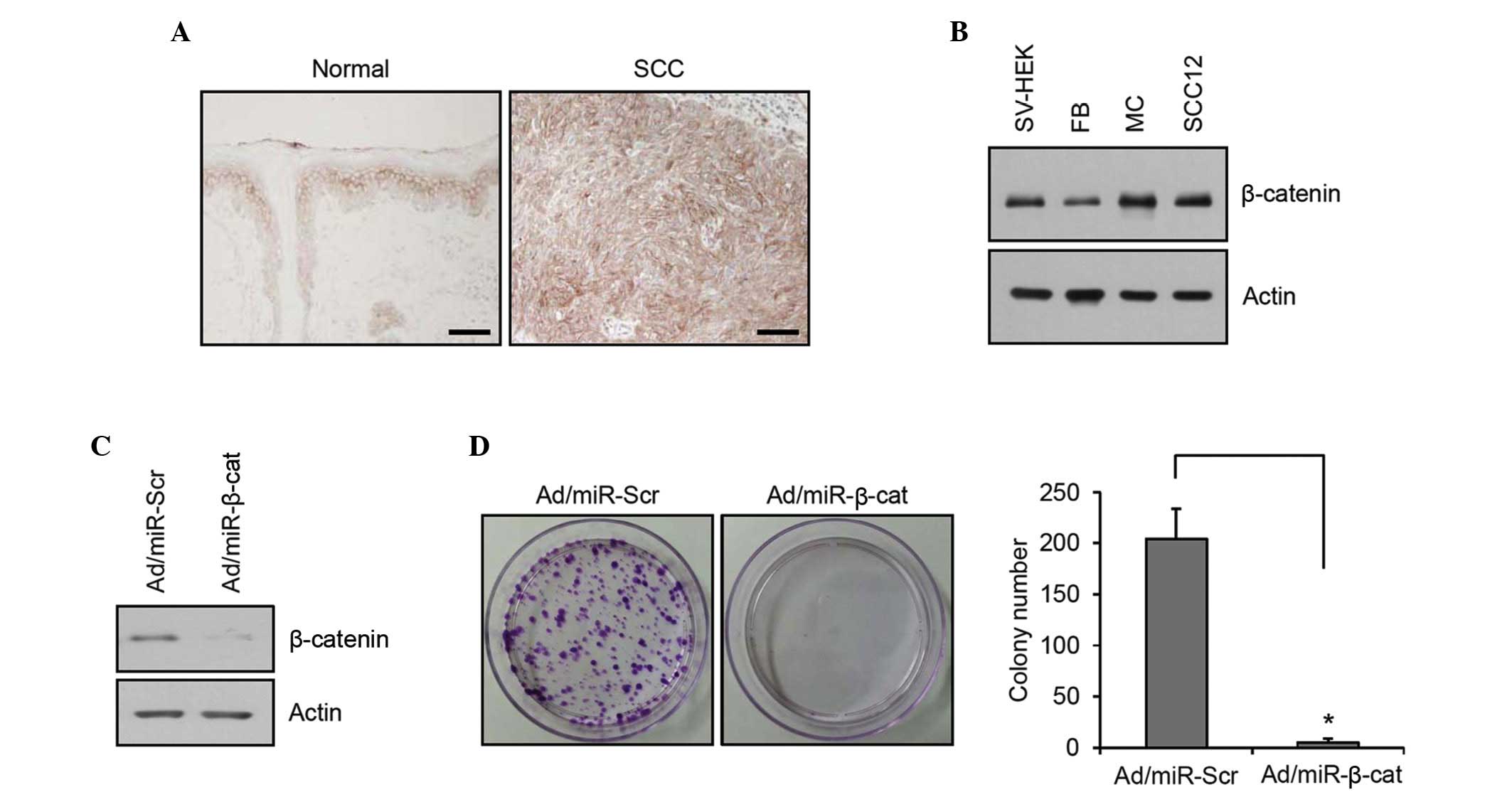 | Figure 1Expression and role of β-catenin in
SCC. (A) Immunohistochemistry analysis of β-catenin expression in
normal skin and cutaneous SCC. In SCC, intense immunoreactivity was
observed in both the membrane and nuclei (scale bar, 100
µm). (B) The protein expression of β-catenin in cultured
skin cells was determined by western blotting. Actin was used as a
loading control. (C) The SCC12 cells were transduced with
Ad/miR-β-cat. Western blotting revealed effective knockdown of
β-catenin. Ad/miR-Scr was used for negative control and actin was
used as a loading control. (D) A colony-forming assay was performed
after Ad transduction. The SCC12 cells were cultured for 2 days,
treated with trypsin/EDTA, re-suspended and seeded at a low
concentration. The cells were cultured for 2 weeks and stained with
crystal violet. Colonies were counted. The data are expressed as
the mean± standard deviation (n=3; *P<0.01). SCC,
squamous cell carcinoma; SV-sHEK, simian virus 40 large T
antigen-transformed human epidermal keratinocytes; FB, dermal
fibroblasts; MC, melanocytes; miR, microRNA; Ad, adenovirus; Scr,
scrambled; β-cat, β-catenin. |
The present study next attempted to identify the
genes regulated by β-catenin. To this end, the present study
transduced the recombinant adenovirus expressing N-terminal
87-amino acid truncated β-catenin (Ad/ΔN87-β-cat), a constitutively
active form of β-catenin (20). It
was found that Sox9 was induced by the overexpression of the
constitutively active form of β-catenin. Conversely, knockdown of
β-catenin resulted in downregulation of Sox9 (Fig. 2A). These results suggested that
Sox9 is a β-catenin-regulated gene in SCC cells and has a potential
role in tumorigenesis. Therefore, the expression of Sox9 was next
determined in cutaneous SCC. Immunohistochemistry and western
blotting showed that the expression of Sox9 was increased in
cutaneous SCC tissue compared with normal skin tissue (Fig. 2B and C). Consistent with these
data, the relative protein expression level of Sox9 was
significantly increased in SCC12 cells compared with keratinocytes
cultured in vitro (Fig.
2D).
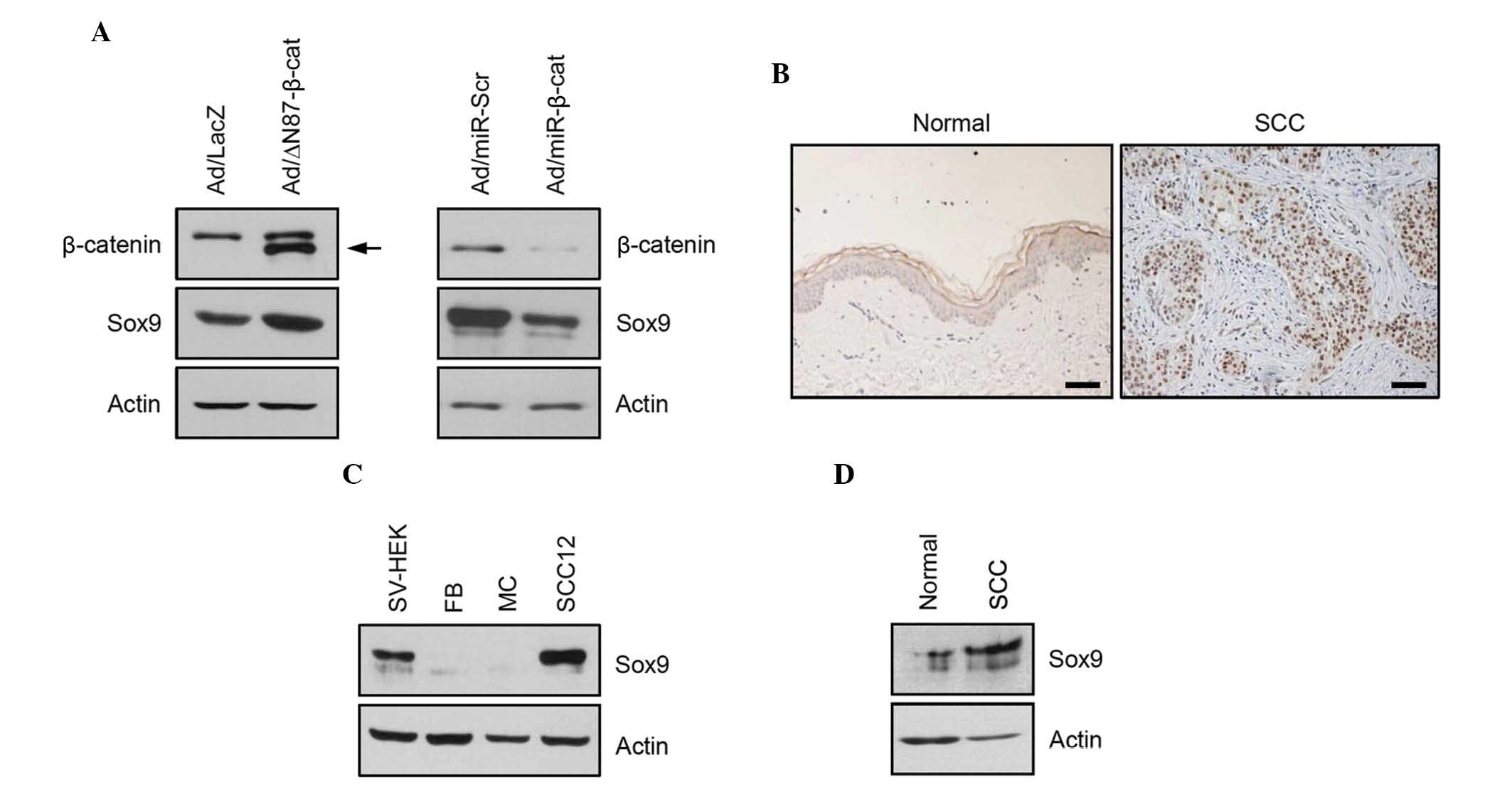 | Figure 2Identification of Sox9 as a
β-catenin-regulated gene in SCC12 cells. (A) SCC12 cells were
transduced with Ad expressing a constitutively active form of
β-catenin (ΔN87) and/or miR specific for β-catenin. Exogenous
N-terminal truncated β-catenin (arrow) was detected. Following the
activation and/or repression of β-catenin, the expression of Sox9
was determined by western blotting. Ad expressing LacZ and/or
miR-scr were used as negative controls. Actin was used as a loading
control. (B) Immunohistochemical analysis of the expression of Sox9
in normal skin and cutaneous SCC tissues. In SCC, intense
immunoreactivity is seen in the nucleus (scale bar, 100 µm).
(C) The expression of Sox9 in normal skin tissue and SCC tissue was
confirmed by western blotting. Actin was used as a loading control.
(D) The expression levels of Sox9 in cultured skin cells were
confirmed by western blotting. Actin was used as a loading control.
Ad, adenovirus; miR, microRNA; scr, scrambled; SCC, squamous cell
carcinoma; SV-sHEK, simian virus 40 large T antigen-transformed
human epidermal keratinocytes; FB, dermal fibroblasts; MC,
melanocytes. |
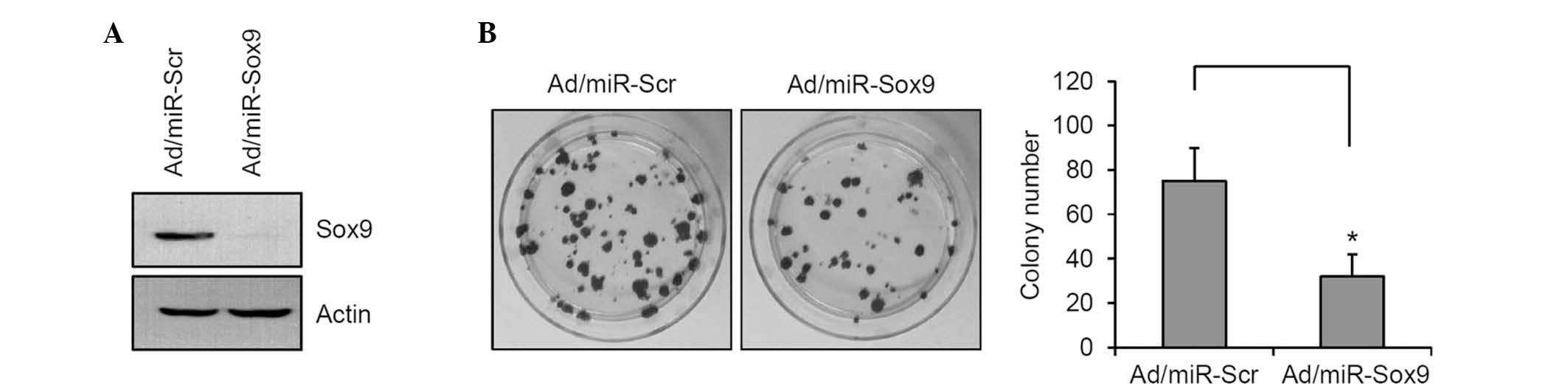
The expression of Sox9 was reduced in SCC12 cells
using the recombinant adenovirus expressing miR specific for Sox9
(Fig. 3A). The colony-forming
potential of SCC12 cells was determined, and the data showed that
knockdown of Sox9 significantly decreased the colony number
(Fig. 3B). These results suggested
that Sox9 is a potential transcription factor that can increase
tumorigenicity of SCC cells.
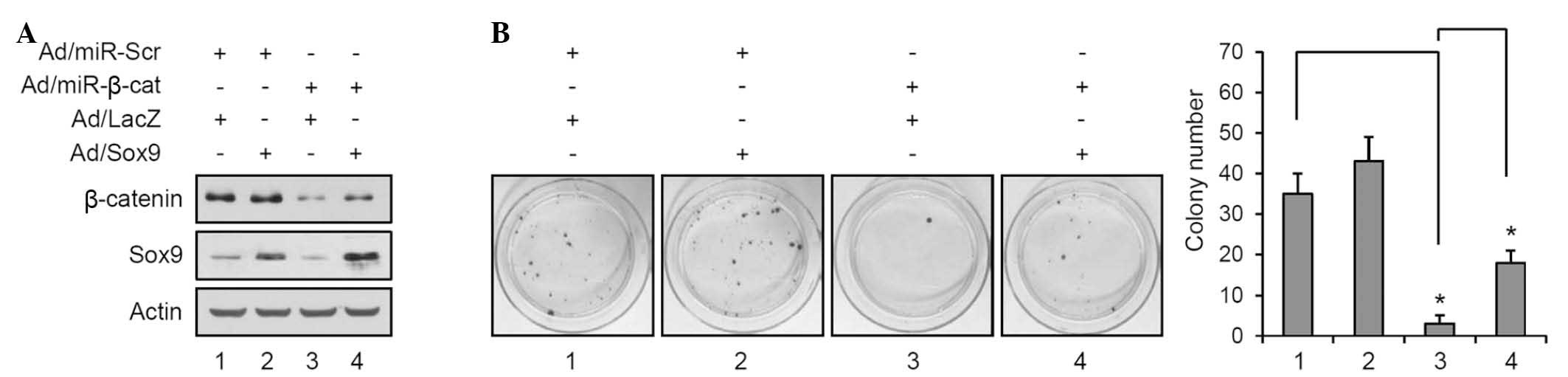
To further confirm the association between β-catenin
and Sox9, β-catenin levels were reduced and the expression of Sox9
was simultaneously increased using the recombinant adenovirus
(Fig. 4A). Overexpression of Sox9
led to a slight increase of colony number in the control miR
transduced group, however, it was not significant (lane 1 vs. 2;
Fig. 4B). Consistent with previous
data, knockdown of β-catenin resulted in a marked decrease of
colony formation; however overexpression of Sox9 partially restored
the colony-forming potential (lane 3 vs. 4; Fig. 4B). These results strengthened the
notion that Sox9 is a β-catenin-downstream transcription factor and
is involved in the development of SCC.
Discussion
In the present study, it was found that Sox9 was
regulated by β-catenin in SCC12 cells. When β-catenin signaling was
activated by the overexpression of stabilized β-catenin
(ΔN87-β-Cat), Sox9 expression was increased. By contrast, knockdown
of β-catenin using specific miR resulted in the downregulation of
Sox9. Sox9 demonstrated tumorigenicity in terms of colony-forming
activity. Therefore, the present data suggested that
β-catenin-regulated Sox9 is one of the effector molecules that can
positively affect the development of SCC.
Sox9 is a member of high mobility group box
transcription factor family and serves a critical role in various
biological events, including embryonic development, cell fate
determination and lineage commitment (21,22).
Germline mutation for Sox9 causes campomelic dysplasia, a disorder
characterized by numerous skeletal abnormalities, defects in
central nervous system and XY sex reversal (23,24).
A potential role of Sox9 for cancer development has been previously
recognized. For example, Sox9 supports breast tumor cell
proliferation and directly contributes to the poor clinical
outcomes associated with invasive breast cancer (25,26).
In other examples, high levels of Sox9 were detected in colorectal
cancer and Sox9 exhibits several properties, including the ability
to promote proliferation, inhibit senescence and collaborate with
other oncogenes in neoplastic transformation (27,28).
Based on these findings, it is plausible that Sox9 can also affect
cutaneous SCC development. The present study demonstrated that Sox9
was increased in cutaneous SCC, and that Sox9 enhanced the
colony-forming activity of SCC12 cells. These data supported the
notion that Sox9 is an important pro-oncogenic protein in SCC
development.
The putative association between β-catenin and Sox9
has been demonstrated several times in other systems. For example,
the Wnt signaling pathway promotes chondrocyte differentiation in a
Sox9-dependent manner (29).
Another example shows that Sox9 protein is expressed in the
intestinal epithelium in a pattern characteristic of Wnt/β-catenin
targets. Additionally, inhibition of β-catenin signaling by the
over-expression of dominant negative TCF4 resulted in a marked
decrease of Sox9 in human colon carcinoma cells (30). Finally, conditional inactivation of
adenomatous polyposis coli, thereby activating Wnt/β-catenin
signaling in mouse colon, is associated with the induction of Sox9
expression and initiation of crypt budding (31). In the present study, activation and
inactivation of β-catenin signaling clearly affected Sox9
expression in a positive manner. Therefore, the present study
suggested that Sox9 is an authentic β-catenin-downstream
transcription factor and exerts its effect as a positive regulator
for cutaneous SCC.
In conclusion, the present study demonstrated that
Sox9 is a functional downstream effector of β-catenin in cutaneous
SCC. These findings provided novel insights into the association
between β-catenin and Sox9 in SCC, and may assist with developing
novel therapeutic targets for skin cancer in the future.
Acknowledgments
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education, Science and Technology (grant
no. 2012R1A1A1041389).
References
|
1
|
Alam M and Ratner D: Cutaneous
squamous-cell carcinoma. N Engl J Med. 344:975–983. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brash DE, Rudolph JA, Simon JA, Lin A,
McKenna GJ, Baden HP, Halperin AJ and Pontén J: A role for sunlight
in skin cancer: UV-induced p53 mutations in squamous cell
carcinoma. Proc Natl Acad Sci USA. 88:10124–10128. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kolev V, Mandinova A, Guinea-Viniegra J,
Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF and Dotto
GP: EGFR signalling as a negative regulator of Notch1 gene
transcription and function in proliferating keratinocytes and
cancer. Nat Cell Biol. 10:902–911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Hogerlinden M, Rozell BL,
Ahrlund-Richter L and Toftgård R: Squamous cell carcinomas and
increased apoptosis in skin with inhibited Rel/nuclear
factor-kappaB signaling. Cancer Res. 59:3299–3303. 1999.PubMed/NCBI
|
|
5
|
Brasanac D, Boricic I, Todorovic V,
Tomanovic N and Radojevic S: Cyclin A and beta-catenin expression
in actinic keratosis, Bowen's disease and invasive squamous cell
carcinoma of the skin. Br J Dermatol. 153:1166–1175. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lyakhovitsky A, Barzilai A, Fogel M, Trau
H and Huszar M: Expression of e-cadherin and beta-catenin in
cutaneous squamous cell carcinoma and its precursors. Am J
Dermatopathol. 26:372–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doglioni C, Piccinin S, Demontis S, Cangi
MG, Pecciarini L, Chiarelli C, Armellin M, Vukosavljevic T,
Boiocchi M and Maestro R: Alterations of beta-catenin pathway in
non-melanoma skin tumors: Loss of alpha-ABC nuclear reactivity
correlates with the presence of beta-catenin gene mutation. Am J
Pathol. 163:2277–2287. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hisken J and Behrens J: The Wnt signaling
pathway. J Cell Sci. 113:3545–3546. 2000.
|
|
9
|
Zhang Y, Liu B, Zhao Q, Hou T and Huang X:
Nuclear local-izaiton of β-catenin is associated with poor survival
and chemo-/radioresistance in human cervical squamous cell cancer.
Int J Clin Exp Pathol. 7:3908–3917. 2014.
|
|
10
|
Li P, Cao Y, Li Y, Zhou L, Liu X and Geng
M: Expression of Wnt-5a and β-catenin in primary hepatocellular
carcinoma. Int J Clin Exp Pathol. 7:3190–3195. 2014.
|
|
11
|
Cui J, Xi H, Cai A, Bian S, Wei B and Chen
L: Decreased expression of Sox7 correlates with the upregulation of
the Wnt/β-catenin signaling pathway and the poor survival of
gastric cancer patients. Int J Mol Med. 34:197–204. 2014.PubMed/NCBI
|
|
12
|
Malanchi I, Peinado H, Kassen D, Hussenet
T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W and
Huelsken J: Cutaneous cancer stem cell maintenance is dependent on
beta-catenin signalling. Nature. 452:650–653. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Popp S, Waltering S, Herbst C, Moll I and
Boukamp P: UV-B-type mutations and chromosomal imbalances indicate
common pathways for the development of Merkel and skin squamous
cell carcinomas. Int J Cancer. 99:352–360. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Choi HI, Sohn KC, Hong DK, Lee Y, Kim CD,
Yoon TJ, Park JW, Jung S, Lee JH and Lee YH: Melanosome uptake is
associated with the proliferation and differentiation of
keratinocytes. Arch Dermatol Res. 306:59–66. 2014. View Article : Google Scholar
|
|
15
|
Je YJ, Choi DK, Sohn KC, Kim HR, Im M, Lee
Y, Lee JH, Kim CD and Seo YJ: Inhibitory role of Id1 on
TGF-β-induced collagen expression in human dermal fibroblasts.
Biochem Biophys Res Commun. 444:81–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JS, Kim DH, Choi DK, Kim CD, Ahn GB,
Yoon TY, Lee JH and Lee JY: Comparison of gene expression profiles
between keratinocytes, melanocytes and fibroblasts. Ann Dermatol.
25:36–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sohn KC, Shi G, Jang S, Choi DK, Lee Y,
Yoon TJ, Park H, Hwang C, Kim HJ, Seo YJ, et al: Pitx2, a
beta-catenin-regulated transcription factor, regulates the
differentiation of outer root sheath cells cultured in vitro. J
Dermatol Sci. 54:6–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi G, Sohn KC, Li Z, Choi DK, Park YM,
Kim JH, Fan YM, Nam YH, Kim S, Im M, et al: Expression and
functional role of Sox9 in human epidermal keratinocytes. PLoS One.
8:e543552013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Franken NA, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:2315–2319. 2006. View Article : Google Scholar
|
|
20
|
Gat U, DasGupta R, Degenstein L and Fuchs
E: De Novo hair follicle morphogenesis and hair tumors in mice
expressing a truncated beta-catenin in skin. Cell. 95:605–614.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lefebvre V, Dumitriu B, Penzo-Méndez A,
Han Y and Pallavi B: Control of cell fate and differentiation by
Sry-related high-mobility-group box (Sox) transcription factors.
Int J Biochem Cell Biol. 39:2195–2214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sarkar A and Hochedlinger K: The sox
family of transcription factors: Versatile regulators of stem and
progenitor cell fate. Cell Stem Cell. 12:15–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Foster JW, Dominguez-Steglich MA, Guioli
S, Kwok C, Weller PA, Stevanović M, Weissenbach J, Mansour S, Young
ID, Goodfellow PN, et al: Campomelic dysplasia and autosomal sex
reversal caused by mutations in an SRY-related gene. Nature.
372:525–530. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schafer AJ, Foster JW, Kwok C, Weller PA,
Guioli S and Goodfellow PN: Campomelic dysplasia with XY sex
reversal: Diverse phenotypes resulting from mutations in a single
gene. Ann NY Acad Sci. 785:137–149. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chakravarty G, Moroz K, Makridakis NM,
Lloyd SA, Galvez SE, Canavello PR, Lacey MR, Agrawal K and Mondal
D: Prognostic significance of cytoplasmic SOX9 in invasive ductal
carcinoma and metastatic breast cancer. Exp Biol Med (Maywood).
236:145–155. 2011. View Article : Google Scholar
|
|
26
|
Wang H, He L, Ma F, Regan MM, Balk SP,
Richardson AL and Yuan X: SOX9 regulates low density lipoprotein
receptor-related protein 6 (LRP6) and T-cell factor 4 (TCF4)
expression and Wnt/β-catenin activation in breast cancer. J Biol
Chem. 288:6478–6487. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matheu A, Collado M, Wise C, Manterola L,
Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, et
al: Oncogenicity of the developmental transcription factor Sox9.
Cancer Res. 72:1301–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Panza A, Pazienza V, Ripoli M, Benegiamo
G, Gentile A, Valvano MR, Augello B, Merla G, Prattichizzo C,
Tavano F, et al: Interplay between SOX9, β-catenin and PPARγ
activation in colorectal cancer. Biochim Biophys Acta.
1833:1853–1865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yano F, Kugimiya F, Ohba S, Ikeda T,
Chikuda H, Ogasawara T, Ogata N, Takato T, Nakamura K, Kawaguchi H
and Chung UI: The canonical Wnt signaling pathway promotes
chondrocyte differentiation in a Sox9-dependent manner. Biochem
Biophys Res Commun. 333:1300–1308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blache P, van de Wetering M, Duluc I,
Domon C, Berta P, Freund JN, Clevers H and Jay P: SOX9 is an
intestine crypt transcription factor, is regulated by the Wnt
pathway, and represses the CDX2 and MUC2 genes. J Cell Biol.
166:37–47. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feng Y, Sentani K, Wiese A, Sands E, Green
M, Bommer GT, Cho KR and Fearon ER: Sox9 induction, ectopic Paneth
cells, and mitotic spindle axis defects in mouse colon adenomatous
epithelium arising from conditional biallelic Apc inactivation. Am
J Pathol. 183:493–503. 2013. View Article : Google Scholar : PubMed/NCBI
|