Introduction
Chronic hepatitis B is global public health issue.
Approximately 2 billion people worldwide have been infected with
the hepatitis B virus (HBV). Among them, more than 350 million
people have experienced chronic infections (1,2).
According to the World Health Organization reports, over one
million patients with chronic HBV die of liver failure, liver
cirrhosis or hepatocellular carcinoma (3). There is a high rate of chronic HBV
infection in China; it is estimated that there are over 120 million
people carrying the HBsAg antigen and 20 million with chronic
hepatitis B. Among them, there are almost 0.3 million mortalities
as a consequence of chronic HBV infection each year in China
(4).
The efficacy of antiviral treatment of patients with
chronic hepatitis B is affected by the virus itself and host
factors. Among host factors, host immune cells are important in
liver damage and the scavenging of invading viral cells. The immune
status of patients is one of the important factors impacting the
therapeutic response of patients to antiviral treatment. During
acute HBV infection, episodes of which are usually self-limited,
the cellular immune response against HBV is relatively strong.
However, in patients that become chronically infected, the T cell
response is much weaker (5).
Previous reports have indicated that the function of T cells in
chronic HBV infection is impaired and T cell dysfunction in the
host has been considered as the root cause of chronic HBV infection
(6,7).
It has been well-established that the host immune
response triggered by viral infection causes the liver damage
experienced by patients with chronic hepatitis B (8). However, the current therapeutic
strategies of chronic hepatitis B treatment, including
hepatoprotective drugs, and anti-viral and immune modulation
therapy, fail to completely eradicate HBV, even when treatments are
administered from an early stage. During the course of the immune
response to HBV, the specific cytotoxic T cells (CTL) determine the
outcome of the disease and are important in clearing the virus. The
T cell receptor (TCR) determines the CTL specificity and is the
feature of individual T cells. In peripheral blood, the TCR in 95%
T cells is composed of two different protein chains, α and β chains
(9). Additionally, these cells
actively participate in the specific immune response of the host
against the invading virus. Each chain is composed of two
extracellular domains: The variable (V) and constant (C) regions
(10). The TCR α-chain and β-chain
have complementarity determining regions, which can recognize the
major histocompatibility complex of viruses (11).
The diversity of TCRs in individual patients may be
important for the clinical outcomes of patients with HBV infection
(12,13). Thus, investigating the composition
of TCR α and β chains may further the understanding of the
pathogenesis and improve treatment strategies for HBV infection. In
humans, the genes which code for the α and β chains can be divided
into several regions according to the corresponding areas of the α
and β chain proteins, including the V, D, J and C regions. The
heterodimerical TCRs are highly diverse as a result of genomic
rearrangement of the V, D, J and C region segments of each T cell
(14). Although the genomic
rearrangement of these regions of TCRs in T cells has been
investigated during the past decade using high-throughput
sequencing (15,16), the effect of different treatments
on the genomic rearrangement of TCRs in patients with HBV remains
to be examined. In response to an effective treatment strategy, the
TCR gene may be adjusted to exert polymorphic expression of certain
genes in the β chain.
In the present study, peripheral blood lymphocytes
were isolated from patients with chronic hepatitis B prior and
subsequent to treatment, and the β chain V region gene diversity
was analyzed using high-throughput sequencing. The current study
also aimed to elucidate the gene patterns that may be closely
associated with specific mechanisms of cellular immune response to
the treatment in individual patients with HBV. Several genes with
abnormal expression levels were identified in the V and J segments
of the CD8+ T cell TCR of patients with chronic HBV
infection. These genomic abnormalities may be important during the
pathogenesis of HBV infection and progression of the chronic stage.
Furthermore, by comparing the expression profiles in patients with
or without routine treatment, the results of the current study
suggested that two gene variants, T cell receptor β variable 28
(TRBV28)_T cell receptor β joining 1–5 (TRBJ1.5) and TRBV6_TRBJ2.1,
in addition to their corresponding segments in the TCR of
CD8+ T cells, may be important sites whereby T cells can
be regulated and induced to eliminate the invading virus. The
expression levels of these genes were significantly altered
following treatment. The results of the present study may provide
evidence to demonstrate that the plasticity of the β chain V region
is an essential feature of the host immune response to virus
invasion and may provide insight into novel treatment strategies
for patients with HBV.
Materials and methods
Patients
The current study was conducted at the Beijing
You'an Hospital, Capital Medical University (Beijing, China).
Between March 2010 and December 2013, a total of 4 patients who had
been diagnosed with chronic hepatitis B and HBeAg+ were
enrolled. Routine treatment with nucleotide similitudes, interferon
and immunomodulators was administrated to all enrolled patients.
All procedures were performed in accordance with the guidelines set
by the Declaration of Helsinki and approved by the Clinical and
Animal Research Ethnic Committees of Capital Medical University
(Beijing, China). All participating individuals provided written
informed consent.
Inclusion criteria
All patients enrolled in the present study were
required to meet the following criteria: History of hepatitis B or
persistent HBsAg+ >6 months; copies of HBV-DNA
≥1×105/ml; HBeAg+; anti-HBe−;
abnormal alanine aminotransferase, aspartate aminotransferase or
γ-glutamyltransferase; liver abnormality not caused by another
medical issue; Knodell histology activity index ≥4 on histology;
fibrosis ≥S2; and necrosis ≥G2.
Exclusion criteria
The exclusion criteria for the candidates in the
current study were as follows: Co-infection with other viruses,
including hepatitis C or D virus, Epstein-Barr virus,
cytomegalovirus and human immunodeficiency virus; cancer diagnosis;
other liver diseases, including alcoholic hepatitis, nonalcoholic
fatty liver disease, drug-induced hepatitis and autoimmune
hepatitis; previous incidence of antiviral treatment; recent
steroid and immunosuppressant treatment; and intolerance to
antivirus treatment.
Peripheral blood mononuclear cell (PBMC)
isolation
PBMCs were separated according to the previous
reported procedure with minor modifications (17). Briefly, 5 ml venous blood was
collected from each patient following fasting and then mixed at a
ratio of 1:1 (vol/vol) with phosphate-buffered saline (PBS) at room
temperature. Subsequently, 2 ml Lymphocyte Separation Medium
(Corning Incorporated, Corning, NY, USA) was added to the mixture
and centrifuged at room temperature at 600 × g for 20 min. The
PBMCs were carefully aspirated (between the layers of plasma and
Lymphocyte Separation Medium) and transferred to a separate tube.
The pellets were washed twice with 5 ml Roswell Park Memorial
Institute 1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and then centrifuged following each wash at 400 × g for 10
min. All samples were stored at −80°C prior to use during
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) amplification of the TCR β
chain V area
The isolated PBMCs were washed three times with
ice-cold PBS then 1 ml TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was added to extract the total RNA according to
the manufacturer's instructions. An Oligotex kit (Qiagen, Inc.,
Valencia, CA, USA) was used to purify the mRNA from total RNA
according to the manufacturer's protocol. The purified mRNA was
converted to cDNA using SuperScript III Reverse Transcriptase,
DNase, buffers and dNTPs (all from Thermal Fisher Scientific,
Inc.). A degenerated V region primer (5′-GCIITKTIYTGGTAYMGACA-3′),
which was designed to cover the majority of the TRBV genes, and a
reverse primer (5′-GCACCTCCTTCCCATTCAC-3′) covering both TRBC genes
was used as previously reported (18,19).
Each 50 µl RT-qPCR reaction contained 2 µl cDNA, 12
µl ddH2O, 25 µl Premix Ex Taq (Takara Bio,
Inc., Otsu, Japan), 1 µl forward primer, 1 µl reverse
primer. The RT-qPCR cycling conditions were used as follows: 94°C
for 10 min; 40 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C
for 30 sec; and a final 10 min extension at 72°C and 4°C hold for
30 sec. The PCR reaction was performed using an ABI 7500 Real-Time
PCR System (Thermo Fisher Scientific, Inc.).
High-throughput sequencing analysis of
PCR products
PCR products (5 µl) were run on 1% agarose
gels pre-dyed with ethidium bromide (Sigma-Aldrich, St. Louis, MO,
USA) and purified using the QIAquick PCR Purification kit (Qiagen,
Inc.). Sequencing reactions and high-throughput sequencing were
performed using purified PCR products and the Ion PGM System for
Next-Generation Sequencing according to the manufacturer's
instructions (Thermo Fisher Scientific, Inc.).
Bioinformatics
DNA sequences were produced from each patient
corresponding to the samples provided prior and subsequent to
treatment. All sequences were analyzed by CLC Genomics Workbench
software, version 7.5 (Qiagen, Inc.) and compared with TRBV and
TRBJ sequences from the International ImMunoGeneTics database
(www.imgt.org). The sequence differences for each
patient from the samples with or without treatment were analyzed
and the changes in the V and J areas that were common in all
patients were identified.
Results
Quality of PCR products
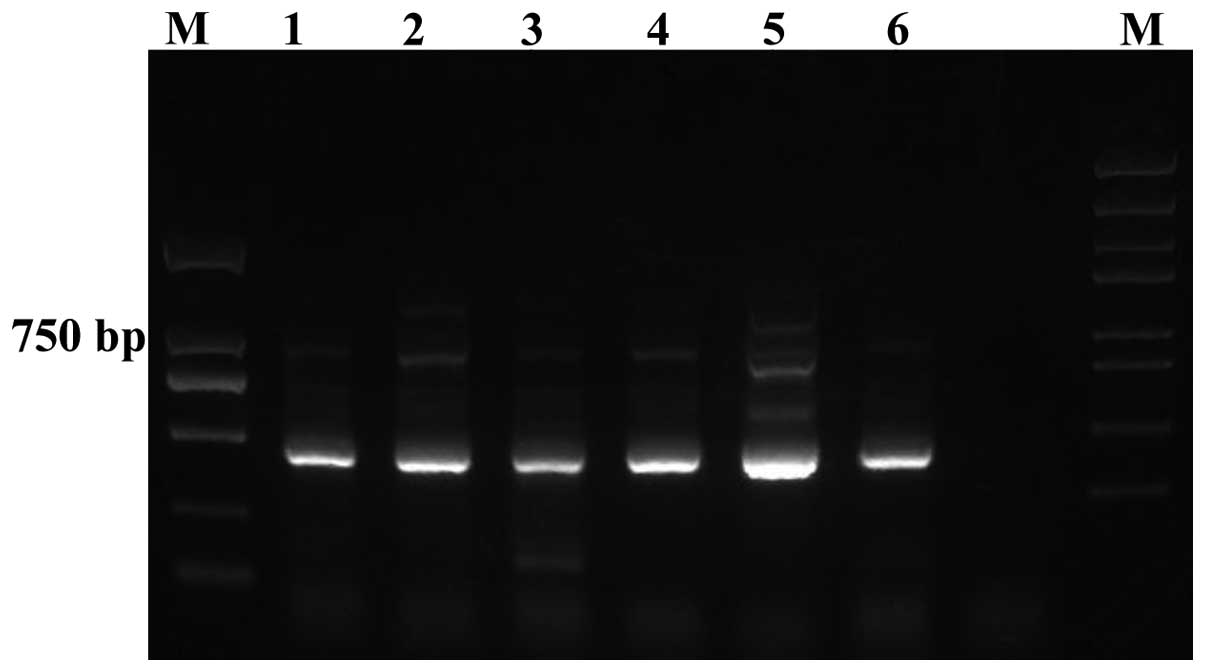
The quality of the amplified PCR products was tested
by running on an agarose gel, which demonstrated clear bands with
400–500 bp size (Fig. 1). Agilent
2100 Bioanalyzer System (Agilent Technologies, Inc., Santa Clara,
CA, USA) was used to determine the quality of the PCR products from
the patient samples prior and subsequent to treatment.
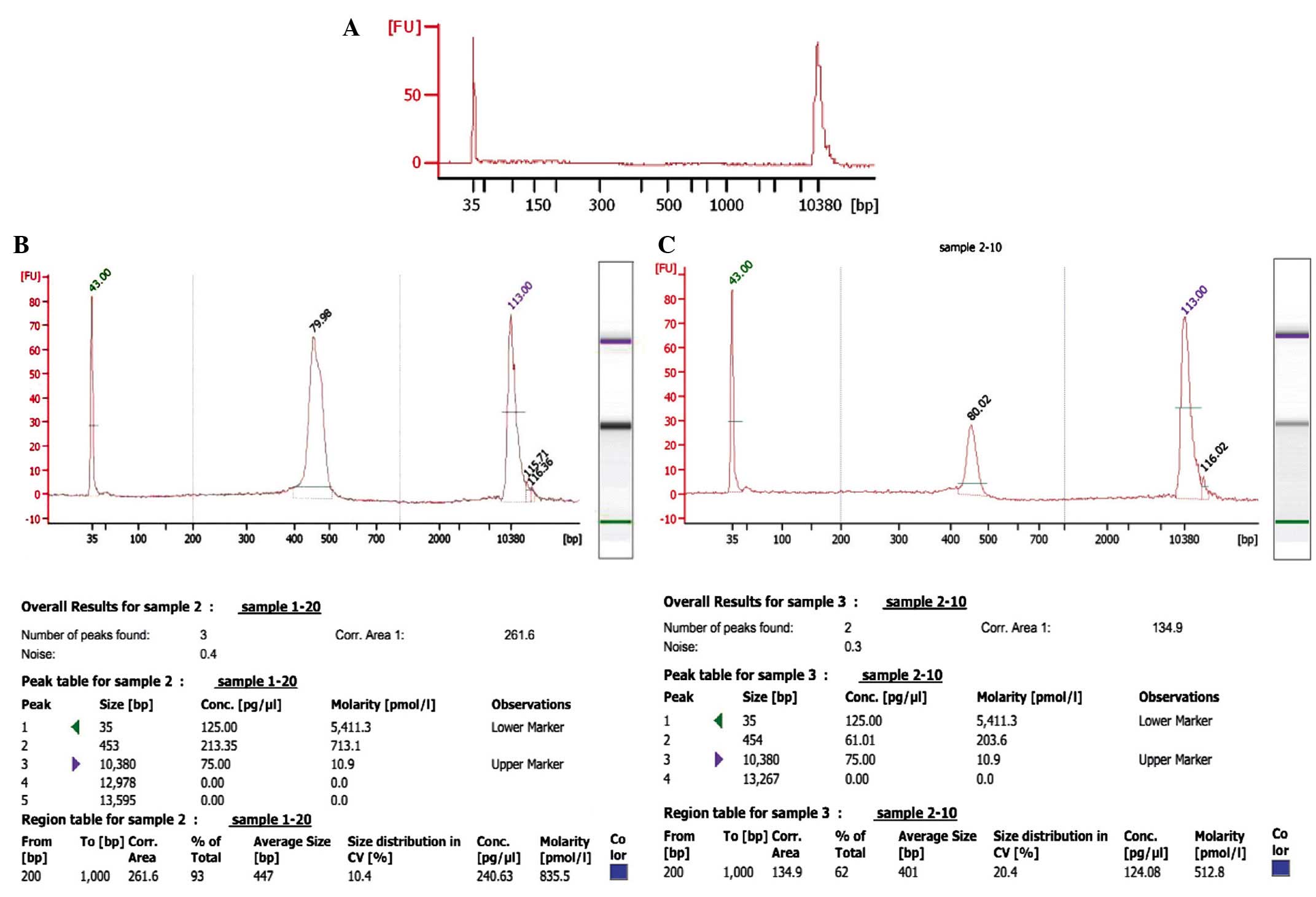
Representative data of the samples from one patient (prior and
subsequent to treatment) is presented in Fig. 2. As demonstrated, the peak value of
the sample prior to treatment was on 454 bp, whereas following
treatment while the one of sample after treatment was on the 453
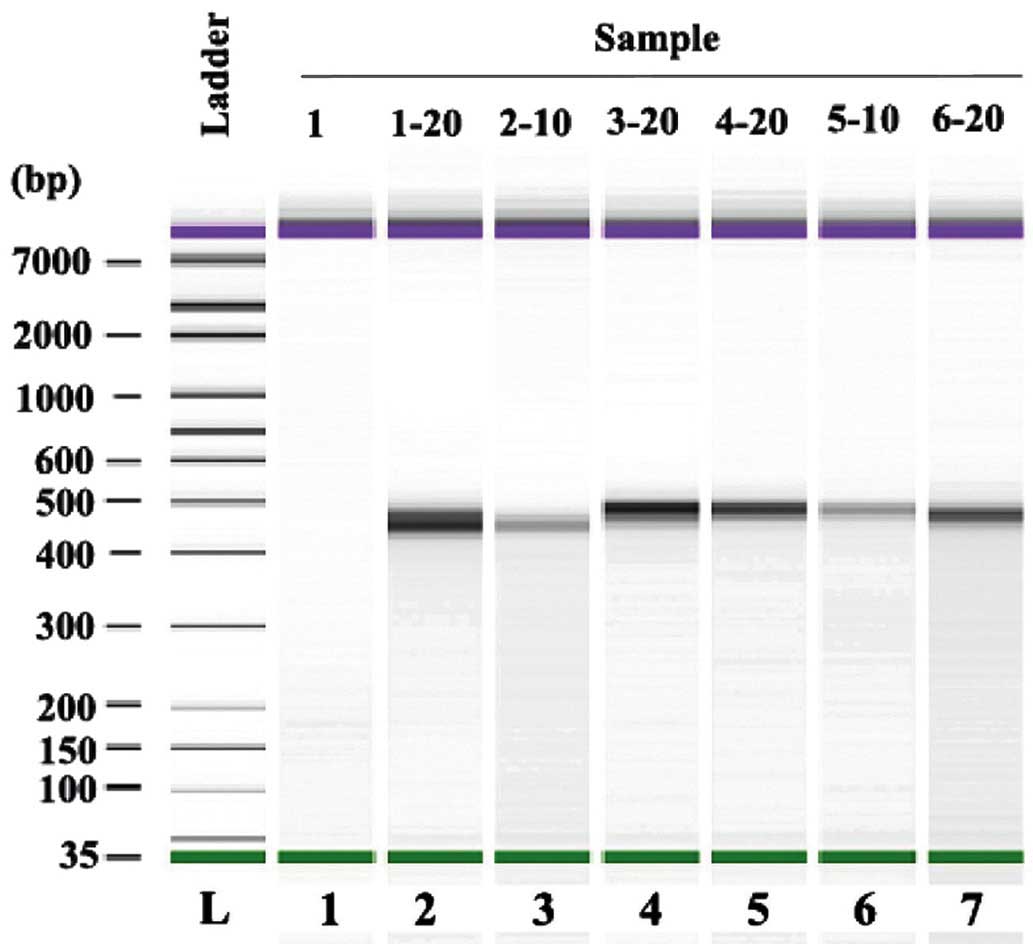
bp. The data was further analyzed using a mimic diagram to
demonstrate the sizes of the bands with using the Agilent 2100
Bioanalyzer system (Fig. 3). The
results from Agilent 2100 analysis were consistent with the results
demonstrated in Fig. 1.
Sequence analysis of PCR products
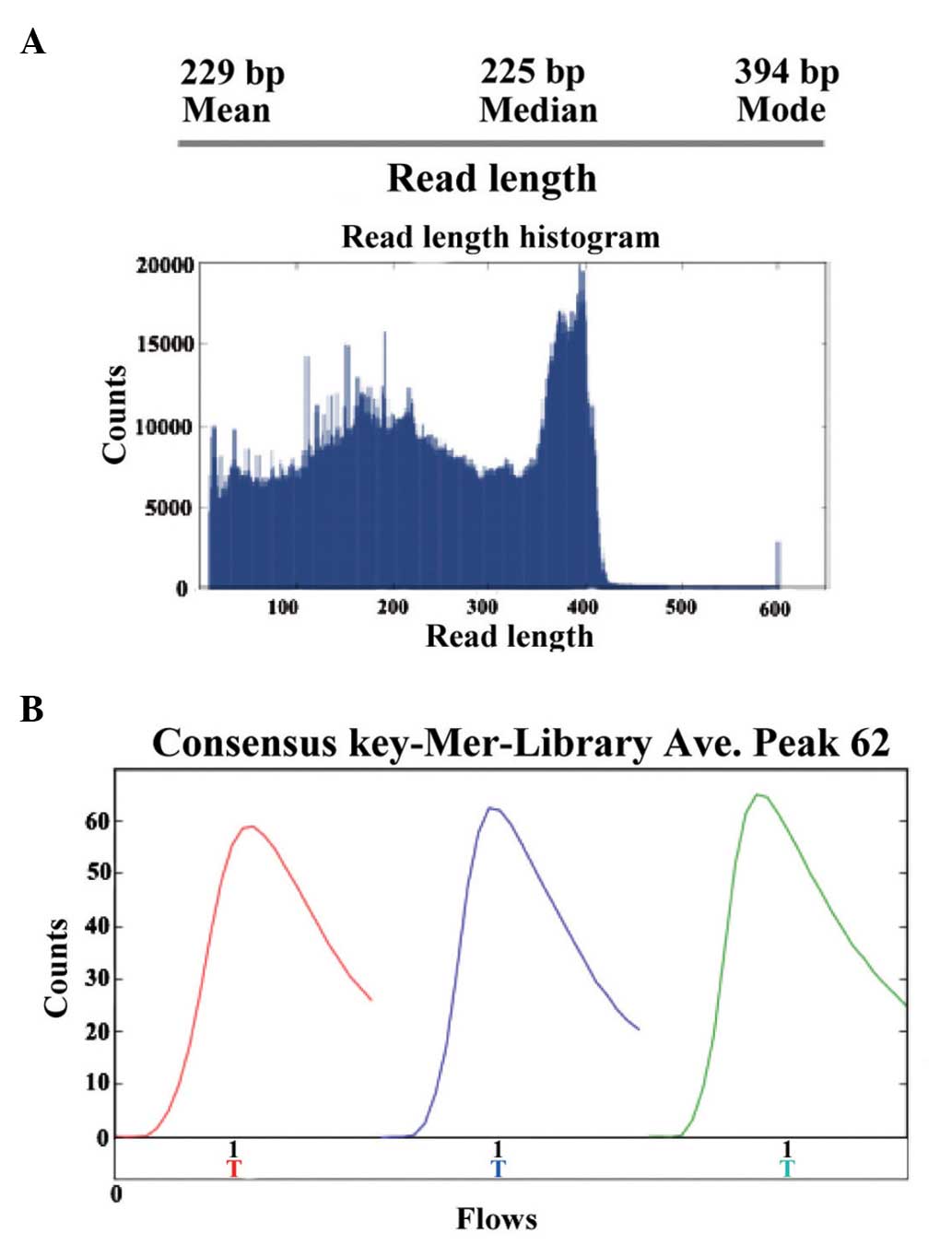
The present study performed high-throughput
sequencing to analyze the genomic expression patterns of samples in
patients prior and subsequent to treatment for HBV. Ion sphere
particle (ISP) identification summaries of representative samples
are presented in Fig. 4 and
Table I. In total, 3,829,994 reads
were identified and a relatively stronger red color suggested the
relative stable sequencing process. In a further library summary,
the average size of all reads was 225 bp (Fig. 5 and Table II). The differential genomic
expression of TCR β chains is presented in Table III. By comparing the expression
profile in the samples prior and subsequent to treatment, the
results of the present study demonstrated there to be markedly
differential expression of the following TCR recombinations:
TRBV10_TRBJ2.1; TRBV10_TRBJ2.2; TRBV10_TRBJ2.7; TRBV12_TRBJ1.1;
TRBV27_TRBJ2.2; TRBV28_TRBJ1.5; and TRBV6_TRBJ2. Among them,
TRBV28_TRBJ1.5 and TRBV6_TRBJ2.1 were markedly different.
 | Table IISP identification summaries of
representative samples. |
Table I
ISP identification summaries of
representative samples.
A, Addressable
wells
(n= 11,304,141) | Count | Percentage |
|---|
| With ISPs | 8,917,942 | 78.9% |
| Live | 8,867,906 | 99.4% |
| Test fragment | 20,441 | 9.2% |
| Library | 8,847,465 | 99.8% |
|
B, Library
ISPs
(n= 8,847,465) | Count | Percentage |
|
| Filtered:
Polyclonal | 2,634,948 | 29.8% |
| Filtered: Low
quality | 2,380,081 | 26.9% |
| Filtered: Primer
dimer | 2,442 | 00.0% |
| Final Library
ISPs | 3,829,994 | 43.3% |
 | Table IIBarcode of loading samples from
patients. |
Table II
Barcode of loading samples from
patients.
| Barcode | Sample | Bases (bp) | ≥Q20 | Reads | Mean read length |
|---|
| No barcode | None | 12,567,420 | 9,050,676 | 90,123 | 139 |
| IonXpress.017 | None | 135,103,823 | 113,524,381 | 526,527 | 256 |
| IonXpress.019 | None | 127,371,212 | 109,752,867 | 568,263 | 224 |
| IonXpress.021 | None | 159,903,935 | 133,142,164 | 685,596 | 233 |
| IonXpress.023 | None | 166,661,460 | 141,148,845 | 727,848 | 228 |
| IonXpress.025 | None | 133,847,320 | 113,599,121 | 618,022 | 216 |
| IonXpress.027 | None | 143,665,632 | 122,754,662 | 607,994 | 236 |
 | Table IIIRepresentative upregulated genes in
TCR β chain (V and J area) after routine treatment. |
Table III
Representative upregulated genes in
TCR β chain (V and J area) after routine treatment.
| TRBJ |
TRBV
|
|---|
| 10-1-01 | 10-3-01 | 12-4-01 | 19-01 | 27-01 | 28-01 | 6-3-01 | 6-4-01 | 6-5-01 | 6-6-01 | 7-2-01 | 7-8-01 |
|---|
| 1-1 | 14.0562 | 63.6778 | 164.0466 | 70.7131 | 73.7088 | 41.2304 | 40.7399 | −0.2857 | 10.4917 | 27.2504 | 47.9113 | 1.0774 |
| 1-2 | 0.0923 | 35.6464 | 164.7710 | 84.8584 | 37.2387 | 120.0921 | 21.6486 | 3.9117 | 13.5238 | 21.6501 | 39.5857 | 0.0654 |
| 1-3 | −0.0032 | 7.7549 | 14.4925 | −4.6894 | 1.6685 | 4.2581 | 4.2526 | 0.3093 | −0.0709 | 7.1806 | 13.8860 | 0.1899 |
| 1-4 | 0.6782 | −6.8893 | 29.5795 | 6.0467 | 10.4934 | −1.8589 | 0.6628 | 0.4673 | 14.6283 | 6.6969 | 11.8412 | 0.3314 |
| 1-5 | −24.7328 | 26.0093 | 30.1689 | −112.0634 | 52.4666 | 90.3730 | −67.7313 | 7.4276 | 5.2049 | 23.2271 | 160.5450 | 1.3626 |
| 1-6 | 0.6701 | −2.6944 | 22.4786 | 29.6578 | −3.7970 | −72.7041 | 4.4369 | −17.3607 | 15.0942 | 14.3288 | 25.8857 | 2.9350 |
| 2-1 | 16.7451 | 73.6994 | 172.5381 | 395.2790 | −32.2191 | 61.0448 | 90.3862 | 53.8860 | 11.8641 | 59.1640 | 152.8131 | 0.6849 |
| 2-2 | −7.2258 | 40.9557 | 93.3107 | −190.8478 | 38.6384 | −53.9616 | 34.6702 | −28.1767 | 16.8347 | 23.5272 | 38.9658 | 0.4695 |
| 2-2P | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 2-3 | 7.9254 | 124.7426 | 210.6778 | −146.9102 | −580.9940 | 201.5457 | 48.9872 | −19.2801 | 9.9758 | 32.9447 | 139.6743 | −0.4528 |
| 2-4 | 6.2481 | −1.7408 | 14.5234 | 20.8885 | −13.7698 | −19.9982 | 3.1636 | −1.9191 | 3.3963 | 1.5966 | −12.1429 | −0.0665 |
| 2-5 | 4.0523 | 26.6260 | 63.1694 | 28.1325 | −0.3912 | 81.8239 | 38.3662 | 17.3466 | 6.8584 | 20.2083 | 49.4611 | −0.2882 |
| 2-6 | 0.0000 | 28.3354 | 28.9562 | −104.5056 | 11.1357 | 65.0562 | 13.9030 | −1.3684 | 1.3235 | 9.2550 | 9.5562 | 0.0851 |
Discussion
Chronic HBV infections are highly prevalent and
create a substantial burden to healthcare systems (20). HBV infection is a major cause of
morbidity and mortality worldwide, particularly in developing
countries (21).
The natural process of HBV infection is determined
by the interaction between virus replication and the host immune
response. Host immune responses are important for the successful
elimination of viruses (22). T
cell immune responses are central for determining the clinical
outcome of infection (19). It is
well-established that there is a significant correlation between
the strength of the T cell response and virus clearance (23). A previous study also suggested that
investigating variable gene expression patterns of TCR may provide
important information regarding the host immune response against
invading HBV (24). However,
little is understood regarding whether genomic rearrangement of TCR
occurs in response to treatment of the host infected with HBV. As T
cell dysfunction may be an important cause of the chronic
deterioration in HBV patients, the present study hypothesized that
effective routine treatment may regulate the TCR expression
profiles by regulating genomic rearrangement. High-throughput
sequencing technology is widely used in biology and medicine to
investigate important biological questions in rare diseases.
Single-cell sequencing is also used in cancer research (25). With the advent of
next-generation-sequencing technologies, it has now become feasible
to directly sequence a numerous TCR β chains in parallel. The
current study used Ion Torrent high-throughput sequencing
technology to investigate the effect of treatment for chronic
hepatitis B on the TCR β chain V region genomic recombination
pattern. The present study aimed to provide evidence to further
understand the host immune mechanisms in patients infected with HBV
and how treatment influences the outcome of HBV infection. In
total, 3,829,994 reads were sequenced and analyzed. The sizes of
the majority of the sequence PCR products were 100–400 bp.
Furthermore, the results of the current study demonstrated that the
ratio for library reads is 43.3%. These results are consistent with
the recognized advantages of high-throughput and higher quality
next-generation-sequencing technology. By comparing the expression
profile in TCR β chains of patients prior and subsequent to
treatment, to the best of our knowledge, the present study is the
first to demonstrate alterations to several genes at the V and J
regions following treatment for HBV. Only TRBV28_TRBJ1.5 and
TRBV6_TRBJ2.1 were significantly different following treatment
compared with the samples from the same patient prior to treatment.
TRBV families, including TRBV28 and TRBV6, have been previously
demonstrated to be important in abnormal immune responses of T
cells in certain diseases, including psoriasis (26) and myasthenia gravis (27). The observations of the current
study provide useful evidence demonstrating that these genes are
closely involved in the response of TCR to HBV in patients.
In summary, the present study demonstrated rapid,
high-throughput analysis of the expression TCR-β chain genomic
recombinants. The results of the current study may support future
research into the T cell response in patients with HBV.
Acknowledgments
The current study was partially supported by the
National Natural Science Foundation of China (grant no.
81400901).
References
|
1
|
Hollinger FB and Liang TJ: Hepatitis B
Virus. Fields Virology. 4th edition. Knipe DM, Howley PM, Griffin
DE, et al: Lippincott Williams & Wilkins; Philadelphia: pp.
2971–3036. 2001
|
|
2
|
Chisari FV and Ferrari C: Viral Hepatitis.
Viral Pathogenesis. Nathanson N, Ahmed R, Gonzales-Scarano F,
Griffin DE, Holmes KV, Murphy FA and Robinson HL: Lippincott -
Raven; Philadelphia: pp. 745–778. 1997
|
|
3
|
World Health Organization: Introduction of
hepatitis B vaccine into childhood immunization services. WHO;
Geneva: 2001
|
|
4
|
Liang X, Bi S, Yang W, Wang L, Cui G, Cui
F, Zhang Y, Liu J, Gong X, Chen Y, et al: Epidemiological
serosurvey of hepatitis B in China - declining HBV prevalence due
to hepatitis B vaccination. Vaccine. 27:6550–6557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stoop JN, van der Laan LJ, Kuipers EJ,
Kusters JG and Janssen HL: Regulatory s contribute to the impaired
immune response in patients with chronic hepatitis B virus
infection. Hepatology. 41:771–778. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boni C, Fisicaro P, Valdatta C, Amadei B,
Di Vincenzo P, Giuberti T, Laccabue D, Zerbini A, Cavalli A,
Missale G, et al: Characterization of hepatitis B virus
(HBV)-specific T-cell dysfunction in chronic HBV infection. J
Virol. 81:4215–4225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sobao Y, Tomiyama H, Sugi K, Tokunaga M,
Ueno T, Saito S, Fujiyama S, Morimoto M, Tanaka K and Takiguchi M:
The role of hepatitis B virus-specific memory CD8 T cells in the
control of viral replication. J Hepatol. 36:105–115. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Busca A and Kumar A: Innate immune
responses in hepatitis B virus (HBV) infection. Virol J. 11:222014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kimura N, Toyonaga B, Yoshikai Y, Du RP
and Mak TW: Sequences and repertoire of the human T cell receptor
alpha and beta chain variable region genes in thymocytes. Eur J
Immunol. 17:375–383. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janeway CA Jr, Travers P, Walport M and
Shlomchik MJ: Immunobiology: The Immune System In Health and
Disease. 5th edition. Garland Science; New York, NY: 2001
|
|
11
|
Marrack P, Scott-Browne JP, Dai S, Gapin L
and Kappler JW: Evolutionarily conserved amino acids that control
TCR-MHC interaction. Annu Rev Immunol. 26:171–203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiong Y, Tan Y and Song YG: Analysis of T
cell receptor Vβ diversity in peripheral CD4+ and
CD8+ T lymphocytes obtained from patients with chronic
severe hepatitis B. Hepat Mon. 14:e159002014. View Article : Google Scholar
|
|
13
|
Yang J, Chen J, He J, Xie Y, Zhu Y, Cao H
and Li L: Profiling the repertoire of T-cell receptor beta-chain
variable genes in peripheral blood lymphocytes from subjects who
have recovered from acute hepatitis B virus infection. Cell Mol
Immunol. 11:332–342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Davis MM, Boniface JJ, Reich Z, Lyons D,
Hampl J, Arden B and Chien Y: Ligand recognition by alpha beta T
cell receptors. Annu Rev Immunol. 16:523–544. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Welsh RM, Selin LK and Szomolanyi-Tsuda E:
Immunological memory to viral infections. Annu Rev Immunol.
22:711–743. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lanzavecchia A and Sallusto F: Antigen
decoding by T lymphocytes: From synapses to fate determination. Nat
Immunol. 2:487–492. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mylona EE, Mouktaroudi M, Crisan TO,
Mylona EE, Mouk-taroudi M, Crisan TO, Makri S, Pistiki A, Georgitsi
M, Savva A, Netea MG, van der Meer JW, Giamarellos-Bourboulis EJ
and Joosten LA: Enhanced interleukin-1β production of PBMCs from
patients with gout after stimulation with Toll-like receptor-2
ligands and urate crystals. Arthritis Res Ther. 14:R1582012.
View Article : Google Scholar
|
|
18
|
Krell PF, Reuther S, Fischer U, Keller T,
Weber S, Gombert M, Schuster FR, Asang C, Stepensky P, Strahm B, et
al: Next-generation-sequencing-spectratyping reveals public T-cell
receptor repertoires in pediatric very severe aplastic anemia and
identifies a β chain CDR3 sequence associated with
hepatitis-induced pathogenesis. Haematologica. 98:1388–1396. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou D, Srivastava R, Grummel V, Cepok S,
Hartung HP and Hemmer B: High throughput analysis of TCR-beta
rearrangement and gene expression in single T cells. Lab Invest.
86:314–321. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
El-Shabrawi M and Hassanin F: Treatment of
hepatitis B and C in children. Minerva Pediatr. 66:473–489.
2014.PubMed/NCBI
|
|
21
|
Chatterjee R and Mitra A: An overview of
effective therapies and recent advances in biomarkers for chronic
liver diseases and associated liver cancer. Int Immunopharmacol.
24:335–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Freeman AJ, Marinos G, Ffrench RA and
Lloyd AR: Immunopathogenesis of hepatitis C virus infection.
Immunol Cell Biol. 79:515–536. 2001. View Article : Google Scholar
|
|
23
|
Bauer T, Sprinzl M and Protzer U: Immune
control of hepatitis B virus. Dig Dis. 29:423–433. 2011.PubMed/NCBI
|
|
24
|
Soroosh P, Shokri F, Azizi M and
Jeddi-Tehrani M: Analysis of T-cell receptor beta chain variable
gene segment usage in healthy adult responders and nonresponders to
recombinant hepatitis B vaccine. Scand J Immunol. 57:423–431. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Soon WW, Hariharan M and Snyder MP:
High-throughput sequencing for biology and medicine. Mol Syst Biol.
9:6402013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yin G, Li X, Li J, Cui H, Hou R, Zhang J,
Dong F, Liang H and Zhang K: Screening of differentially expressed
genes and predominant expression of β variable region of T cell
receptor in peripheral T cells of psoriatic patients. Eur J
Dermatol. 21:938–944. 2011.PubMed/NCBI
|
|
27
|
Marino M, Maiuri MT, Di Sante G, Scuderi
F, La Carpia F, Trakas N, Provenzano C, Zisimopoulou P, Ria F,
Tzartos SJ, et al: T cell repertoire in DQ5-positive MuSK-positive
myasthenia gravis patients. J Autoimmun. 52:113–121. 2014.
View Article : Google Scholar : PubMed/NCBI
|