Introduction
Mesenchymal stem cells (MSCs) are multipotent
stromal cells with the capability to differentiate into several
cell types (1). MSCs have been
isolated from various sources, including bone marrow, umbilical
cord blood, muscle and adipose tissues. These cells have been shown
to be able to differentiate into chondrocytes, adipocytes,
myoblasts and osteoblasts in vitro, and are useful
candidates for cell replacement therapy (2–4).
Dental stem cells are a particular type of MSCs and
have been isolated from dental pulp, periodontal ligament, apical
papilla and dental follicle. Each of these sources yields a
different type of dental stem cell (5–8).
Dental pulp stem cells (DPSCs), including exfoliated deciduous
teeth cells (SHEDs), have the capacity for self-renewal and
multi-lineage differentiation with the ability to differentiate
into osteoblasts, hepatocytes, adipocytes, neural cells,
chondrocytes, myocytes and odontoblasts (6,9–19).
Dental stem cells express specific markers that are only expressd
by MSCs and embryonic stem cells, including STRO-1, CD106, OCT4 and
NANOG (5–8,20,21).
Compared with other dental pulp stem cells, SHEDs
have certain advantages: First, SHEDs exhibit a greater
proliferative rate than other types of dental stem cell (6,15,22);
furthermore, SHEDs have been shown to exhibit a high capacity for
bone/dentin formation in single colony-derived SHED clones when
injected to immunodeficient mice (6).
However, SHEDs have remained to be fully
characterized. Therefore, the present study was performed to
examine the proliferative abilities, surface marker and specific
gene expression as well as the multi-lineage differentiation
ability of SHEDs. These cells may represent a valuable resource for
tissue engineering and cell replacement therapy.
Materials and methods
Subjects and sample collection
Normal human deciduous incisors were collected from
6- to 8-year-old individuals at the dental clinic of Liaocheng
People's Hospital with strict adherence to the guidelines approved
by the Ethics Committee of Liaocheng People's Hospital (Liaocheng,
China). Consent of the guardians of the individuals was obtained
prior to sample extraction. Furthermore, each subject was examined
to exclude systemic and oral infections, and only samples from
disease-free subjects were used in the present study.
Cell culture and isolation
SHEDs were isolated and cultured according to the
guidelines from established protocols with certain adjustments
(23). Under aseptic conditions,
the dental pulp cavity of the crown was opened using drills. The
pulp was extracted with a broach, immediately placed in Dulbecco's
modified Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and transported to the
laboratory. In the sterile cell culture hood, the pulp was washed
three times with phosphate-buffered saline (PBS). The pulp tissue
was then cut into small pieces (≤0.5 mm) in 10-cm culture dishes.
Sterile coverslips were used to cover the tissue blocks to keep
them in place. The cells were cultured in maintenance medium
comprising DMEM (Gibco) supplemented 10% fetal bovine serum
(Gibco), 100 U/ml penicillin (Gibco) and 100 U/ml streptomycin
(Gibco) at 37°C in a humidified atmosphere with 5% CO2.
The culture medium was replaced every three days. Upon reaching 90%
confluence, the cells were digested with TrypLE (Gibco) and
passaged. Cells at passages 3–5 were used in the subsequent
experiments.
Flow cytometry
The phenotypic characteristics of SHEDs were
assessed by flow cytometry. Cells in the logarithmic growth phase
were trypsinized, collected and washed twice with PBS. The cell
concentration was adjusted to 1×107 cells/ml and 100
µl of the suspension was transferred into a fresh Eppendorf
tube. Cells were blocked with 50 µl normal goat serum
(Gibco) for 15–20 min at room temperature. The cells were incubated
with mouse anti-human fluorescein isothiocyanate-labeled monoclonal
anti-CD90 (dilution, 1:50; cat. no. 555595), mouse anti-human
phyeoerythrin (PE)-labeled monoclonal anti-CD73 (dilution, 1:50;
cat. no. 561014) and mouse anti-human PE-labeled monoclonal
anti-CD34 (dilution, 1:50; cat. no. 348057) (all purchased from BD
Biosciences, Franklin Lakes, NJ, USA), and kept at 4°C for 20 min.
Excess antibody was removed by washing twice with PBS. Finally, 500
µl fixative solution was added. Cells were analyzed using a
BD FACSCalibur (BD Biosciences) and quantification was performed
using Cell Quest software (version 3.3; BD Biosciences). FITC- or
PE-labeled immunoglobulin G-stained cells were used as negative
controls.
RNA isolation and polymerase chain
reaction (PCR) analysis
Total RNA was obtained from 1×107 cells
using TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
The residual DNA was removed with DNase I (Invitrogen). The
activity of DNase I was inhibited by addition of 2.5 mM ethylene
diamine tetraacetic acid (Sigma-Aldrich, St. Louis, MO, USA) and
incubation at 65°C for 10 min. The quantity of total RNA was
determined using SuperScript III Reverse Transcriptase (Invitrogen)
according to the manufacturer's instructions. One microgram of
total RNA (DNase I-treated) was used as a template to synthesize
first-strand complementary DNA using SuperScript® III
Reverse Transcriptase (Invitrogen). Primers listed in Table I were designed according to the
cDNA sequences from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The synthesized
cDNA was then subjected to PCR amplification. For real-time PCR,
the following thermocycling conditions were used: 34 cycles of
denaturation at 95°C for 30 sec, primer annealing at 60°C for 20
sec and extension at 72°C for 30 sec. The PCR products were
separated by electrophoresis in 1.5% agarose gels stained with
ethidium bromide (Sigma-Aldrich) and visualized using a Bio-Rad
ChemiDoc MP Imaging System (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
 | Table IOligonucleotide primer sequences
utilized for polymerase chain reaction analysis. |
Table I
Oligonucleotide primer sequences
utilized for polymerase chain reaction analysis.
| Target cDNA | Primer sequence
(5′-3′) | Product size
(bp) | GenBank ID |
|---|
| CD73 |
F:CTGCCTCTGTCCTCTTTCTT | | |
| R:
TATTTTCTGACCACCCACTCA | 311 | NM_002526 |
| CD44 | F:
GGCTCACTCAAGCTCTTTAACT | | |
| R:
TAGACCTCCCTTATTTCTATCGT | 283 | NM_000610 |
| CD90 | F:
GGGTTGGAGAAGGAGGTAAAG | | |
| R:
CGCAGAAGTCCCTGAGAAG | 345 | NM_006288 |
| Alpl |
F:CACTCACTCCAAGACCACC | | |
| R:
GTTTTGTGTTCCCTTTTAACCA | 216 | NM_000478 |
| Runx2 |
F:GCTTTACCATTCTTAGGTTTCTG | | |
| R:
TACAATACAAATAGTCCCTTTGAA | 199 | NM_001024630 |
| CollagenI |
F:CCCTGGAAAGAATGGAGATGAT | | |
| R:
ACTGAAACCTCTGTGTCCCTTCA | 138 | NM_000088 |
| Col10a1 | F:
TTTCAAAATTCGACTAGAAGTGG | | |
| R:
CTTGAAAGAATGGTTGAGAACAG | 217 | NM_000493 |
| Acan | F:
CAGCTGAATGTATTGGATGAGAA | | |
| R:
GGGGAGGGGAGAAGGTTG | 286 | NM_013227 |
| PPARγ2 | F:
AGGAGCAGAGCAAAGAGGT | | |
| R:
TTGGTCGTTCAAGTCAAGAT | 131 | NM_015869 |
| LPL | F:
TACACCAAACTGGTGGGACA | | |
| R:
TGGATCGAGGCCAGTAATTC | 174 | NM_000237 |
| Nestin | F:
CAGGAGAAACAGGGCCTACA | | |
| R:
TGGGAGCAAAGATCCAAGAC | 242 | NM_006617 |
| TUJ1 | F:
CCTTCATCGGGAACAGCACG | | |
| R:
ACTCCTCCTCGTCGTCTTCGTA | 233 | NM_006086 |
Reverse-transcription quantitative (RT-q)PCR
analysis was performed with SYBR Green PCR Master Mix (Applied
Biosystems, Thermo Fisher Scientific, Inc.) on an ABI 7500 Fast Dx
Real-Time PCR Instrument (Thermo Fisher Scientific, Inc.). Primers
used were identical to those used for real-time PCR (Table I). Reaction conditions were as
follows: Initial denaturation for 2 min at 50°C and 10 min at 95°C,
followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Three
independent experiments were performed in triplicate to confirm the
reproducibility of the results. GAPDH was used as internal control.
Relative expression levels were calculated using the
2−ΔΔCq method and values were presented as the mean
expression level ± standard deviation (SD) (24).
Osteogenic differentiation and Alizarin
Red S staining
For osteogenic differentiation, cells were seeded in
a six-well plate at a density of 2×104
cells/cm2 and cultured in maintenance medium. When the
cell monolayer reached 70% confluence, the medium was replaced with
osteogenic differentiation medium [maintenance medium, 100 nmol/l
dexamesone, 10 mM β-glycerol phosphate, 50 µM L-ascorbic
acid 2-phosphate and 50 nM vitamin D3 (all from Sigma-Aldrich)],
followed by incubation for two weeks with the culture medium
replaced twice a week. If the Alizarin Red S staining was not
satisfactory, the cultures were cultivated for another two weeks.
The control cells were kept in normal maintenance medium for the
duration.
Alizarin Red S staining was performed once calcified
nodules were observed at two or four weeks of induction. The medium
was removed and the cells were fixed with 4% paraformaldehyde (m/v;
Sigma-Aldrich). The plates were rinsed three times with 1X PBS and
stained with 40 mM Alizarin Red S (pH 4.2; Sigma-Aldrich) for 5–10
min at room temperature. Excess dye was removed in case of
over-staining by washing three times with 1X PBS. Cells were
observed and images were captured using an inverted fluorescence
microscope (Nikon Ti-E; Nikon Corporation, Tokyo, Japan).
Adipogenic differentiation and Oil Red O
staining
For adipogenic differentiation, cells were seeded
into six-well plates. When the cells reached 70% confluence, the
medium was changed to adipogenic differentiation medium
[maintenance medium supplemented with 1×106 mol/l
dexamesone, 10 µg/ml insulin, 0.5 mmol/l
3-isobutyl-1-methylxanthine and 60 µmol/l indomethacin (all
from Sigma-Aldrich)]. The medium was replaced twice a week for two
weeks.
Oil Red O staining was performed after adipogenic
induction. The medium was removed and the cells were fixed with 4%
(m/v) paraformaldehyde. Oil Red O stock solution was prepared by
dissolving 0.5 g Oil Red O (Sigma-Aldrich) in isopropanol, which
was diluted with deionized water at a 3:2 ratio, followed by
filtration using filter paper. Cells were stained for 10 min at
room temperature and excess dye was subsequently removed with 75%
ethanol. Stained cells were observed under an inverted fluorescence
microscope (Nikon Ti-E).
Chondrogenic differentiation and Alcian
Blue staining
Cells at 70% confluence were induced in chondrogenic
differentiation medium [maintenance medium supplemented with 100
ng/ml transforming growth factor (TGF)-β3, 50 µg/ml ascorbic
acid-2-phosphate, and 10 µg/ml insulin (all from
Sigma-Aldrich)]. Medium was replaced every three or four days over
three weeks.
For Alcian Blue staining, the culture medium was
removed and cells were washed once with 1X PBS. Cells were then
fixed with 4% paraformaldehyde for 20 min, incubated in 1% Alcian
Blue staining solution (pH 2.5; Sigma-Aldrich) for 20 min at room
temperature, washed three times with double distilled
H2O and observed under an inverted fluorescence
microscope (Nikon Ti-E).
Neurogenic differentiation and
immunofluorescence analysis
For neurogenic differentiation, SHEDs at passages
3–7 were seeded into a six-well plate at 2×104
cells/cm2. The induction process comprised two stages
lasting a total of two weeks. At the first stage which lasted four
days, the induction medium contained neurobasal medium containing
0.5% B27 supplement (both from Invitrogen), a cocktail of 200 ng/ml
Sonic hedgehog, 100 ng/ml fibroblast growth factor 8 (both from
ImmunoTools GmbH, Friesoythe, Germany) and 50 ng/ml basic
fibroblast growth factor (Sigma-Aldrich). At the second stage, the
induction medium contained neurobasal media supplemented with 0.5%
B27 and 10 ng/ml brain derived neurotrophic factor (ImmunoTools
GmbH) for the next 10 days. The induction medium was replaced every
three to four days.
For immunofluorescence analysis, SHEDs were fixed
with 4% paraformaldehyde for 1 h at day 14 after induction, washed
with 1X PBS and blocked in 1% normal goat serum (Sigma-Aldrich) for
1 h. The cells were then incubated with rabbit anti-human glial
fibrillary acidic protein polyclonal antibody (GFAP; dilution,
1:500; cat. no. Z0334; Dako, Glostrup, Denmark) or mouse anti-human
neuron specific class III β-tubulin monoclonal antibody (TUJ1;
dilution, 1:500 dilution; cat. no. T8660; Sigma-Aldrich) at 4°C
overnight. After washing three times with PBS, the cells were
incubated with the following secondary antibodies for 2 h at room
temperature in the dark: Cy 3-conjugated AffiniPure donkey
anti-rabbit IgG polyclonal antibody (dilution, 1:300; cat. no.
711-165-152; Jackson Immuno Research Labs, West Grove, PA, USA) or
Alexa Fluor 488 AffiniPure Goat Anti-Mouse IgG polyclonal antibody
(dilution, 1:300; cat. no. 115-545-146; Jackson Immuno Research
Labs) Nuclei were stained with Hoechst 33258 (1:10,000 dilution;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA). After washing,
the cells were examined under an inverted fluorescence microscope
(Nikon Ti-E).
Statistical analysis
Statistical analyses were performed using SPSS
version 11.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism
version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Values are
expressed as the mean ± SD and images are representative of three
independent experiments. Student's t-test was used for statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference between values.
Results
Isolation and cultivation of dental pulp
stem cells obtained from deciduous teeth
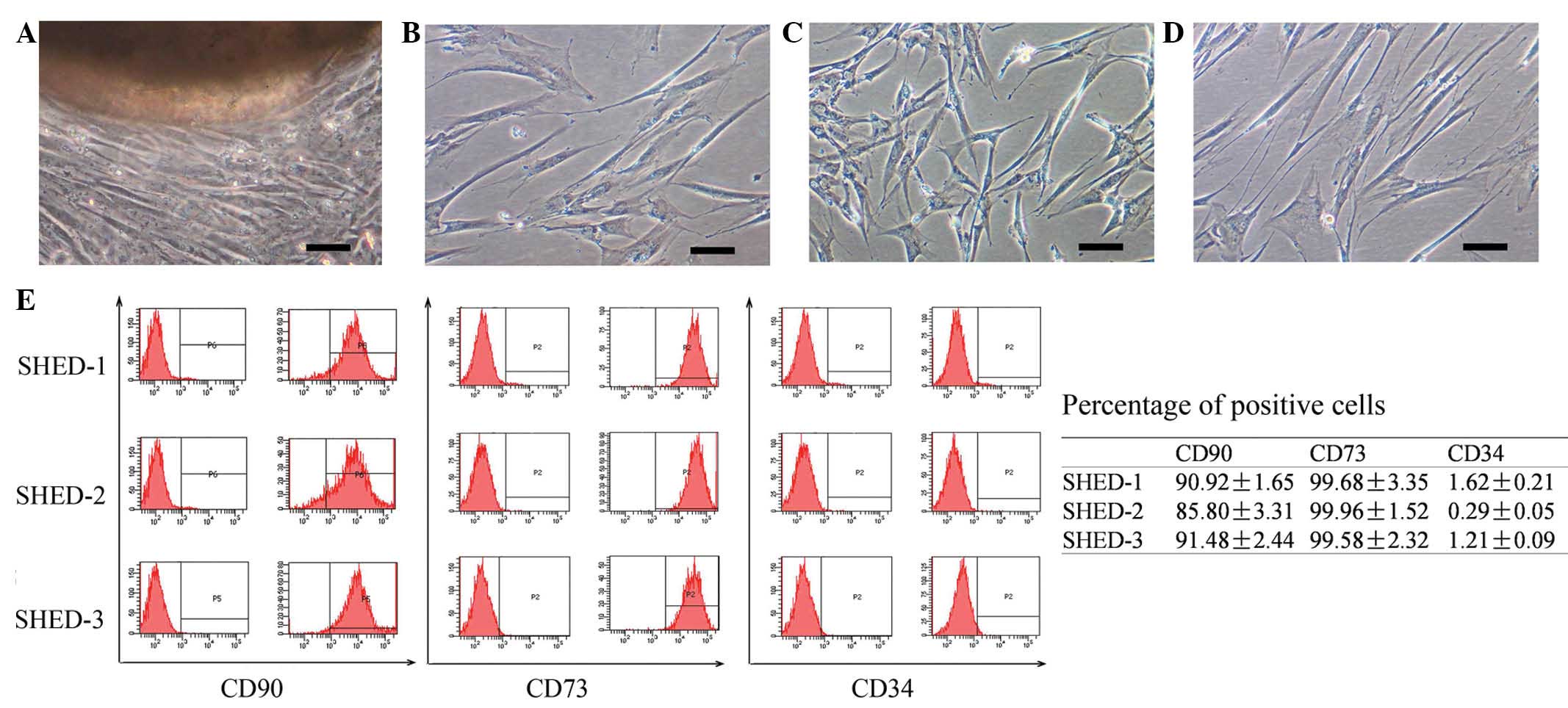
SHEDs were observed to grow out from the obtained
dental pulp tissue clumps within 2–3 weeks of culture (Fig. 1A). Most cells had a typical
fibroblast-like appearance. A small percentage of cells were
polygonal, spindle shaped or oval shaped. Each cell had 1–2
projections (Fig. 1B–D). After
passage, the cells were attached to the bottom of the culture flask
within 2 h and reached 90% confluence within ~1 week. The SHEDs
were cultured for 20 passages without any observed decreases of the
cell growth rate.
Characterization of SHEDs
The isolated SHEDs were analyzed by flow cytometry
to determine the expression of cell surface markers. The analysis
indicated that the SHEDs highly expressed CD73 (89.4±3.6%) and CD90
(99.7±0.2%), while not expressing the hematopoietic stem cell
marker CD34 (1.0±0.6%) (Fig.
1E).
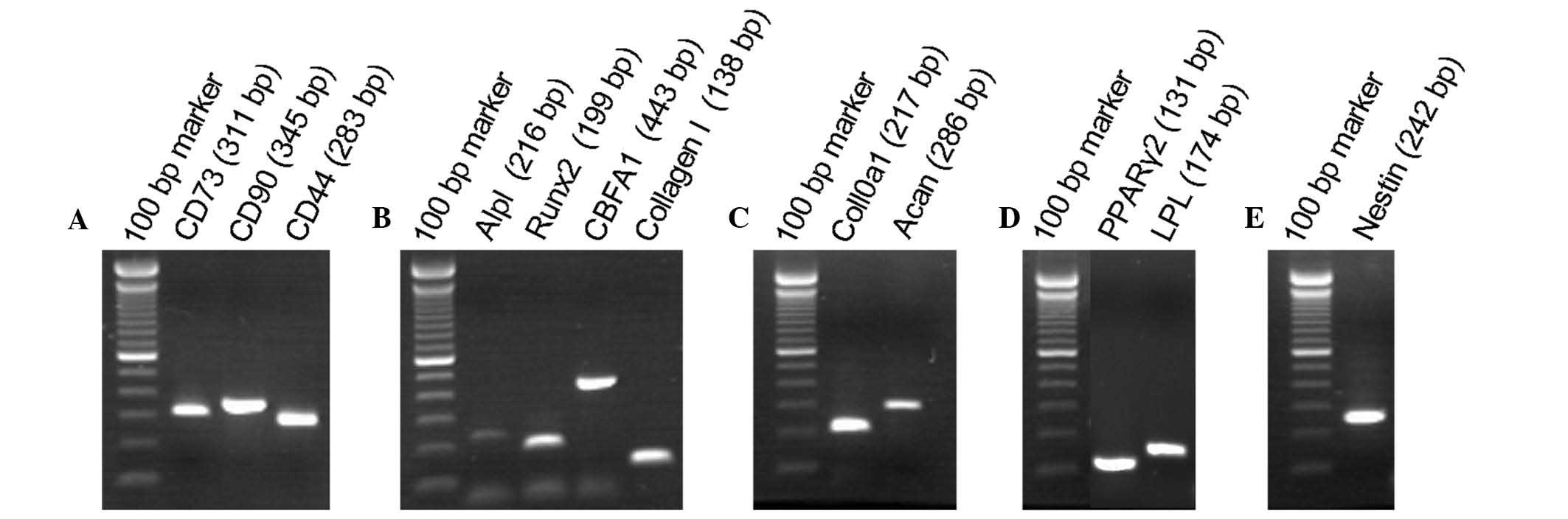
The expression of marker genes in the SHEDs was also
determined by reverse transcription-PCR. It was revealed that the
SHEDs not only expressed the mesenchymal stem cell marker genes as
CD73, CD90 and CD44 (Fig. 2A), but
also expressed genes characteristic for osteoblasts, adipocytes,
chondroblasts and neurocytes. Specifically, the SHEDs expressed
Alpl, Runx2, CBFA1 and CollagenI as osteoblastic markers (Fig. 2B). Col10a1 and Acan as chondroblast
markers (Fig. 2C), the adipocyte
marker genes PPARγ2 and LPL (Fig.
2D) as well as the neuronal stem cell marker Nestin (Fig. 2E). The diversity of markers
expressed confirmed the multi-lineage differentiation ability of
the SHEDs.
Osteogenic differentiation of SHEDs
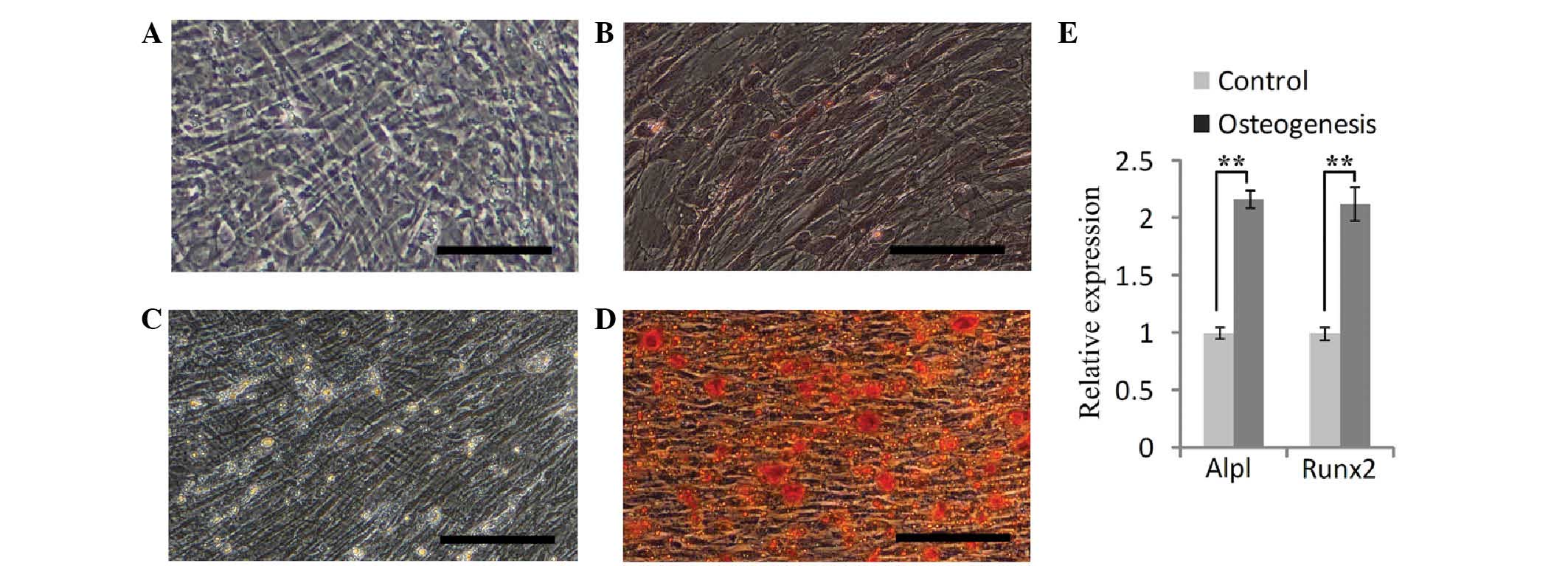
To test the osteogenic differentiation ability of
the SHEDs, the cells were cultured in osteogenic induction medium
for two weeks. Alizarin Red S staining showed that the SHEDs formed
a small number of calcified nodules (Fig. 3A and B). After the osteogenic
induction process was performed for a further two weeks, a large
number of calcified nodules was observed (Fig. 3C and D). RT-qPCR analysis showed
that after four weeks of induction, the expression of Alpl and
Runx2 was increased by 2.17- and 2.13-fold of that in uninduced
cells (Fig. 3E). These results
indicated that SHEDs possess an osteogenetic capacity.
Adipogenic differentiation of SHEDs
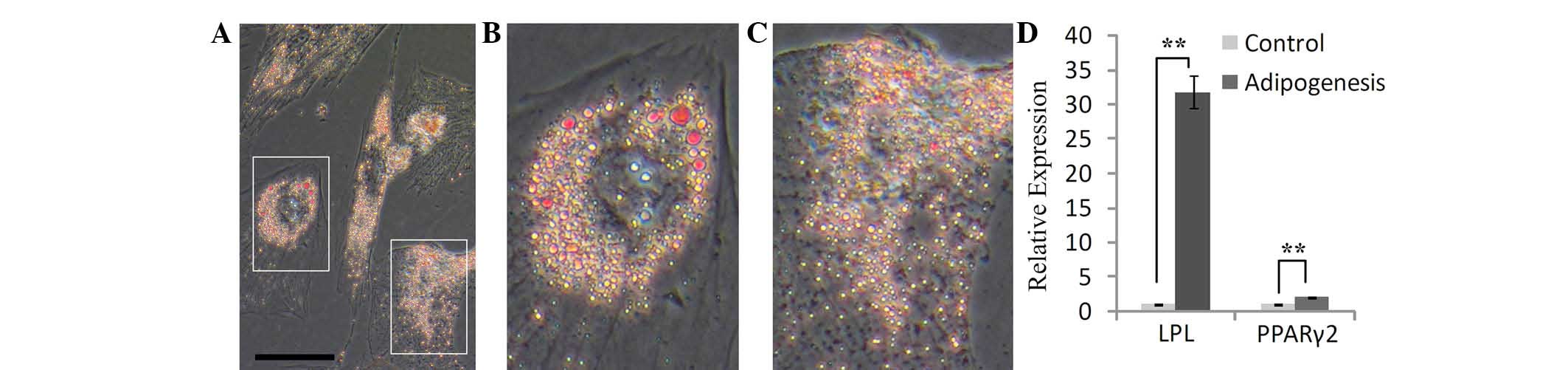
After SHEDs were cultured in adipogenic medium, the
growth of the cells was decreased. The morphology of certain cells
changed from spindle-like to polygonal shapes within seven days,
and cell hypertrophy was observed. Oil Red O staining revealed the
presence of numerous lipid vacuoles after two weeks of induction
(Fig. 4A–C). In addition, RT-qPCR
showed that the expression of PPARγ2 and LPL was obviously
increased in induced SHEDs compared to that in control cells; in
particular, LPL increased to 31.7-fold of that of the control cells
(Fig. 4D). This result indicated
that SHEDs are able to differentiate into adipose cells under the
induction conditions applied.
Chondrogenic differentiation of
SHEDs
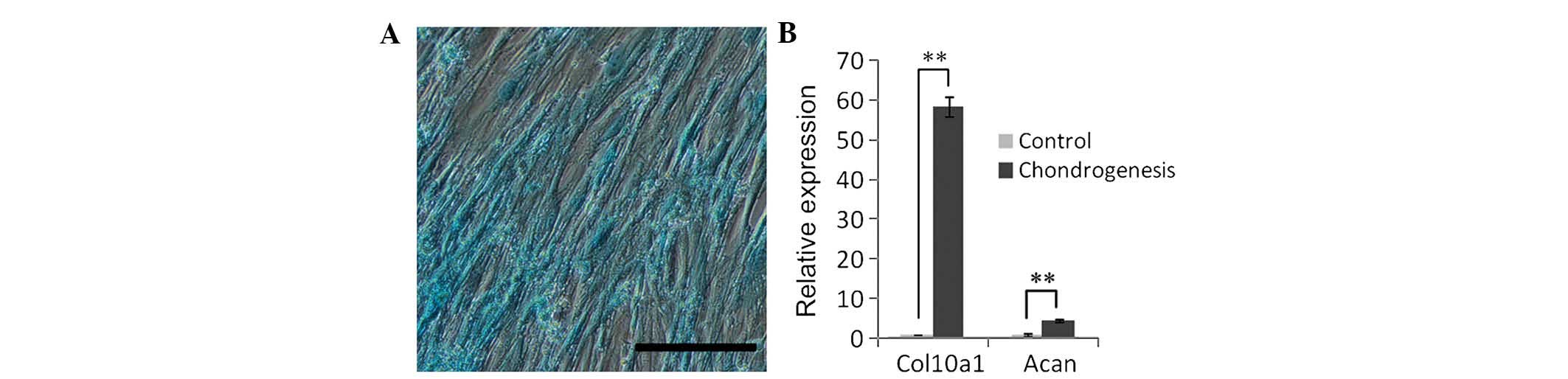
To assess their chondrogenic differentiation
capacity, SHEDs were cultured in induction medium containing
TGF-β3. Alcain blue was used to detect the extracellular matrix
proteoglycan, and positive staining was observed after chondrogenic
induction for three weeks (Fig.
5A). Furthermore, RT-qPCR was used to analyze the expression of
Acan and Col10a1, revealing an upregulation by 4.7- and 58.43-fold,
respectively, compared with that in the control group (Fig. 5B). These results indicated that
SHEDs were able to differentiate into chondroblast cells.
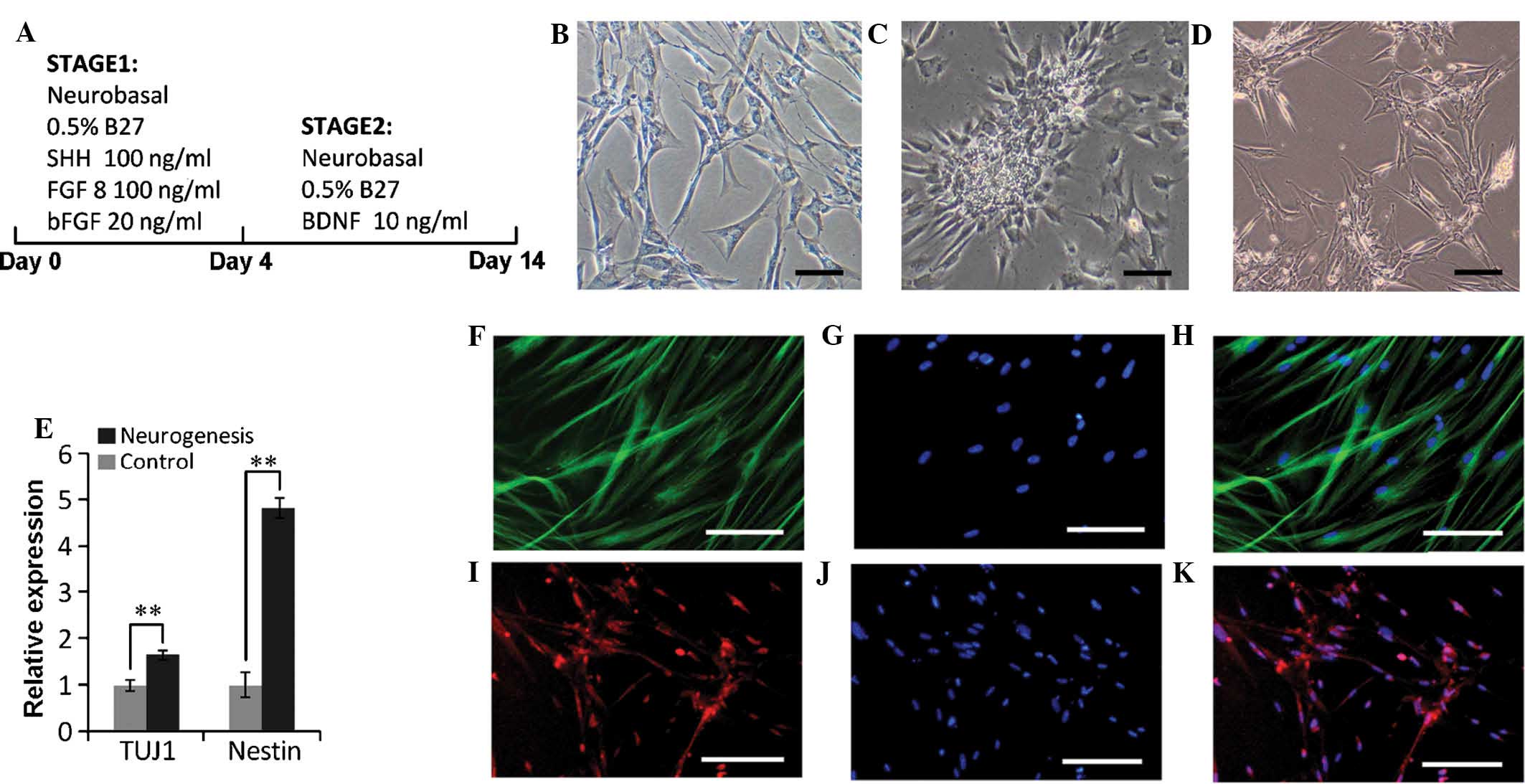
Neuronal differentiation of SHEDs
Developmental studies showed that DPSCs originate
from neuroectodermal tissue. Previous studies have indicated that
DPSCs contain a subset of progenitor cells that can differentiate
towards mature neurons under the appropriate conditions (25,26).
A two-stage protocol was used to induce the SHEDs to differentiate
into neuronal cells (Fig. 6A).
After the first stage, the cells formed rosette-shaped clusters and
after the second stage, a percentage of cells exhibited a
neuron-like appearance (Fig.
6B–D). RT-qPCR indicated that TUJ1 and Nestin were obviously
upregulated by 1.67- and 4.84-fold, respectively, of that of
undifferentiated control cells (Fig.
6E). In addition, immunofluorescence staining was performed for
the neuron-specific markers TUJ1 and GFAP, revealing a high
expression of TUJ1 (Fig. 6F–H) and
GFAP (Fig. 6I–K) in these
neuron-shaped cells. All of these results showed that SHEDs were be
able to differentiate into neuron-like cells.
Discussion
Human DPSCs represent a novel type of stem cells
featuring high proliferative potential, capacity for self-renewal
and multi-lineage differentiation (5,9,23).
Among them, SHEDs were first isolated by Miura et al
(6) in 2003 from dental pulp of
exfoliated deciduous teeth. The proliferation rate and
colony-forming ability of SHEDs was higher than that of stem cells
from permanent teeth. Furthermore, it has been shown that in SHEDs,
the expression of pluripotent markers, including OCT4, SOX2, NANOG
and REX1, was higher (>2.0 fold) compared with that in stem
cells from permanent teeth (19).
Owing to their higher proliferation rate and higher expression
levels of pluripotent markers, SHEDs are considered to be a more
immature form of stem cells than those obtained from permanent
teeth.
There are two methods to isolate SHEDs from pulp
tissue: Enzymatic dissociation of pulp tissue and outgrowth from
tissue explants (27). Although
enzyme digestion is considered to be the most common method used to
acquire dental pulp stem cells (28–30),
it has been reported that the outgrowth method can also be used to
acquire multipotent stem cells (31,32).
In the present study, the outgrowth method was used to isolate the
cells from pulp tissue, as only a small amount of pulp tissue is
available from deciduous teeth and the method was easy and
convenient.
The results of the present study showed that SHEDs
expressed various markers of bone, adipose, cartilage and neural
cells, probably due to the heterogeneous populations of stem cells.
Pulp is composed of different cell types, including odontoblasts,
vessels, nerves, firoblasts and multiple stem cells (33,34).
In the present study, a specific sub-type of SHEDs was examined.
For use as a source for cell therapy, the heterogenous SHEDs are
preferred, as they are enabled to cope with the various
environmental cues after cell transplantation. However, in other
cases, pre-selected cells [sorted using magnetic-activated cell
sorting (MACS)] were shown to be more effective than heterogeneous
stem cells. For example, human
c-kit+/CD34+/CD45− DPSCs have been
demonstrated to be a promising sub-population for bone-tissue
engineering (35,36).
The present study found that SHEDs shared multiple
characteristics with mesenchymal stem cells. First, SHEDs were
demonstrated to have a marked capacity to proliferate. The SHEDs
were passaged once per week until passage 20, with their growth
rate remaining constant over this duration. Furthermore, SHEDs
expressed mesenchymal stem cell markers. Flow cytometry results
demonstrated that >90% of SHEDs expressed CD73 and CD90, and
RT-qPCR illustrated that SHEDs also expressed CD44. This was
consistent with the findings of a previous study (37). Finally, SHEDs were found to have
potential for multi-lineage differentiation, including osteogenic,
adipogenic, chrongenic and neurogenic differentiation (6,12–15,38).
The differentiation potential of stem cells is
important when considering their potential to regenerate specific
tissues, including bone, cartilage or adipose tissue. The present
study demonstrated that SHEDs were able to differentiate into cells
that form large lipid droplets, calcium salts, cartilage or
neural-like tissue with the up-regulation of the corresponding
marker genes. However, as not all of the SHEDs had multiple
differentiation ability, pre-selection of a cell sub-population by
MACS for engineering of different tissue types such as bone,
cartilage, nerve and vessels may be a better choice.
The present study reported on the isolation, culture
and characterization of SHEDs. An improved outgrowth from tissue
explant method was developed to isolate the SHEDs. These cells
expressed stem cell markers such as CD44, CD73 and CD90. In
response to appropriate stimuli, the cells were able to
differentiate into bone, adipose, cartilage and neural cells, as
evidenced by the expression of the respective tissue-specific
markers. The present study therefore further paved the road for the
utilization of SHEDs for tissue engineering.
Acknowledgments
This work was supported by the research grants from
Research Scholar Fund of Liaocheng People's Hospital of Shandong
province (no. 2011LCYYF001) and the Special Fund for Post Doctoral
Innovation Projects of Shandong Province (no. 201303025).
References
|
1
|
Caplan AI: Mesenchymal stem cells. J
Orthop Res. 9:641–650. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dimarino AM, Caplan AI and Bonfield TL:
Mesenchymal stem cells in tissue repair. Front Immunol. 4:2012013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiang Y, Jahagirdar BN, Reinhardt RL,
Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund
T, Blackstad M, et al: Pluripotency of mesenchymal stem cells
derived from adult marrow. Nature. 418:41–49. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jin HJ, Bae YK, Kim M, Kwon SJ, Jeon HB,
Choi SJ, Kim SW, Yang YS, Oh W and Chang JW: Comparative analysis
of human mesenchymal stem cells from bone marrow, adipose tissue,
and umbilical cord blood as sources of cell therapy. Int J Mol Sci.
14:17986–18001. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:13625–13630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miura M, Gronthos S, Zhao M, Lu B, Fisher
LW, Robey PG and Shi S: SHED: Stem cells from human exfoliated
deciduous teeth. Proc Natl Acad Sci USA. 100:5807–5812. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morsczeck C, Götz W, Schierholz J,
Zeilhofer F, Kühn U, Möhl C, Sippel C and Hoffmann KH: Isolation of
precursor cells (PCs) from human dental follicle of wisdom teeth.
Matrix Biol. 24:155–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seo BM, Miura M, Gronthos S, Bartold PM,
Batouli S, Brahim J, Young M, Robey PG, Wang CY and Shi S:
Investigation of multipotent postnatal stem cells from human
periodontal ligament. Lancet. 364:149–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Walboomers XF, Shi S, Fan M and
Jansen JA: Multilineage differentiation potential of stem cells
derived from human dental pulp after cryopreservation. Tissue Eng.
12:2813–2823. 2006. View Article : Google Scholar
|
|
10
|
Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang
S, Shi S and Huang GT: Characterization of the apical papilla and
its residing stem cells from human immature permanent teeth: A
pilot study. J Endod. 34:166–171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morsczeck C, Völlner F, Saugspier M,
Brandl C, Reichert TE, Driemel O and Schmalz G: Comparison of human
dental follicle cells (DFCs) and stem cells from human exfoliated
deciduous teeth (SHED) after neural differentiation in vitro. Clin
Oral Investig. 14:433–440. 2010. View Article : Google Scholar
|
|
12
|
Ma L, Makino Y, Yamaza H, Akiyama K,
Hoshino Y, Song G, Kukita T, Nonaka K, Shi S and Yamaza T:
Cryopreserved dental pulp tissues of exfoliated deciduous teeth is
a feasible stem cell resource for regenerative medicine. PLoS One.
7:e517772012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nourbakhsh N, Soleimani M, Taghipour Z,
Karbalaie K, Mousavi SB, Talebi A, Nadali F, Tanhaei S, Kiyani GA,
Nematollahi M, et al: Induced in vitro differentiation of
neural-like cells from human exfoliated deciduous teeth-derived
stem cells. Int J Dev Biol. 55:189–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eslaminejad MB, Vahabi S, Shariati M and
Nazarian H: In vitro growth and characterization of stem cells from
human dental pulp of deciduous versus permanent teeth. J Dent
(Tehran). 7:185–195. 2010.
|
|
15
|
Koyama N, Okubo Y, Nakao K and Bessho K:
Evaluation of pluripotency in human dental pulp cells. J Oral
Maxillofac Surg. 67:501–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lizier NF, Kerkis A, Gomes CM, Hebling J,
Oliveira CF, Caplan AI and Kerkis I: Scaling-up of dental pulp stem
cells isolated from multiple niches. PLoS One. 7:e398852012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Casagrande L, Demarco FF, Zhang Z, Araujo
FB, Shi S and Nör JE: Dentin-derived BMP-2 and odontoblast
differentiation. J Dent Res. 89:603–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishkitiev N, Yaegaki K, Calenic B,
Nakahara T, Ishikawa H, Mitiev V and Haapasalo M: Deciduous and
permanent dental pulp mesenchymal cells acquire hepatic morphologic
and functional features in vitro. J Endod. 36:469–474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kerkis I, Kerkis A, Dozortsev D,
Stukart-Parsons GC, Gomes Massironi SM, Pereira LV, Caplan AI and
Cerruti HF: Isolation and characterization of a population of
immature dental pulp stem cells expressing OCT-4 and other
embryonic stem cell markers. Cells Tissues Organs. 184:105–116.
2006. View Article : Google Scholar
|
|
20
|
Ferro F, Spelat R, D'Aurizio F, Puppato E,
Pandolfi M, Beltrami AP, Cesselli D, Falini G, Beltrami CA and
Curcio F: Dental pulp stem cells differentiation reveals new
insights in Oct4A dynamics. PLoS One. 7:e417742012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sonoyama W, Liu Y, Fang D, Yamaza T, Seo
BM, Zhang C, Liu H, Gronthos S, Wang CY, Wang S, et al: Mesenchymal
stem cell-mediated functional tooth regeneration in swine. PLoS
One. 1:e792006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakamura S, Yamada Y, Katagiri W, Sugito
T, Ito K and Ueda M: Stem cell proliferation pathways comparison
between human exfoliated deciduous teeth and dental pulp stem cells
by gene expression profile from promising dental pulp. J Endod.
35:1536–1542. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang GT, Gronthos S and Shi S:
Mesenchymal stem cells derived from dental tissues vs. those from
other sources: Their biology and role in regenerative medicine. J
Dent Res. 88:792–806. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Govindasamy V, Abdullah AN, Ronald VS,
Musa S, Ab Aziz ZA, Zain RB, Totey S, Bhonde RR and Abu Kasim NH:
Inherent differential propensity of dental pulp stem cells derived
from human deciduous and permanent teeth. J Endod. 36:1504–1515.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hilkens P, Gervois P, Fanton Y,
Vanormelingen J, Martens W, Struys T, Politis C, Lambrichts I and
Bronckaers A: Effect of isolation methodology on stem cell
properties and multilineage differentiation potential of human
dental pulp stem cells. Cell Tissue Res. 353:65–78. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arora V, Arora P and Munshi AK: Banking
stem cells from human exfoliated deciduous teeth (SHED): Saving for
the future. J Clin Pediatr Dent. 33:289–294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gay IC, Chen S and MacDougall M: Isolation
and characterization of multipotent human periodontal ligament stem
cells. Orthod Craniofac Res. 10:149–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nishino Y, Yamada Y, Ebisawa K, Nakamura
S, Okabe K, Umemura E, Hara K and Ueda M: Stem cells from human
exfoliated deciduous teeth (SHED) enhance wound healing and the
possibility of novel cell therapy. Cytotherapy. 13:598–605. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bakopoulou A, Leyhausen G, Volk J,
Tsiftsoglou A, Garefis P, Koidis P and Geurtsen W: Assessment of
the impact of two different isolation methods on the
osteo/odontogenic differentiation potential of human dental stem
cells derived from deciduous teeth. Calcif Tissue Int. 88:130–141.
2011. View Article : Google Scholar
|
|
32
|
Spath L, Rotilio V, Alessandrini M,
Gambara G, De Angelis L, Mancini M, Mitsiadis TA, Vivarelli E, Naro
F, Filippini A, et al: Explant-derived human dental pulp stem cells
enhance differentiation and proliferation potentials. J Cell Mol
Med. 14:1635–1644. 2010. View Article : Google Scholar
|
|
33
|
Avery JK: Structural elements of the young
normal human pulp. Oral Surg Oral Med Oral Pathol. 32:113–125.
1971. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi S and Gronthos S: Perivascular niche
of postnatal mesenchymal stem cells in human bone marrow and dental
pulp. J Bone Miner Res. 18:696–704. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Laino G, Carinci F, Graziano A, d'Aquino
R, Lanza V, De Rosa A, Gombos F, Caruso F, Guida L, Rullo R, et al:
In vitro bone production using stem cells derived from human dental
pulp. J Craniofac Surg. 17:511–515. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Laino G, d'Aquino R, Graziano A, Lanza V,
Carinci F, Naro F, Pirozzi G and Papaccio G: A new population of
human adult dental pulp stem cells: A useful source of living
autologous fibrous bone tissue (LAB). J Bone Miner Res.
20:1394–1402. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vakhrushev IV, Antonov EN, Popova AV,
Konstantinova EV, Karalkin PA, Kholodenko IV, Lupatov AY, Popov VK,
Bagratashvili VN and Yarygin KN: Design of tissue engineering
implants for bone tissue regeneration of the basis of new
generation polylactoglycolide scaffolds and multipotent mesenchymal
stem cells from human exfoliated deciduous teeth (SHED cells). Bull
Exp Biol Med. 153:143–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taghipour Z, Karbalaie K, Kiani A, Niapour
A, Bahramian H, Nasr-Esfahani MH and Baharvand H: Transplantation
of undifferentiated and induced human exfoliated deciduous
teeth-derived stem cells promote functional recovery of rat spinal
cord contusion injury model. Stem Cells Dev. 21:1794–1802. 2012.
View Article : Google Scholar
|