Introduction
A number of studies have revealed that the
concentration of high mobility group box 1 protein (HMGB1), an
inflammatory factor, is markedly increased in the bone fracture
microenvironment (1–3). HMGB1 was also shown to promote the
osteogenic differentiation of mesenchymal stem cells (MSCs)
(2). Previous studies have
demonstrated the directional migration of MSCs towards the bone
fracture site, and have shown that the osteogenic differentiation
ability of MSCs has an essential role in the wound-healing process
(4–6). In addition, MSCs synthesize various
cytokines, including stem cell factor, thrombopoietin and
interleukin-6 (7,8), which exert an important influence on
the behavior and activity of peripheral cells, and on the MSCs
themselves (9,10).
Ras-associated protein-1 (Rap1), a member of the Ras
superfamily of small GTPases, has the function of reversing the
oncogenic potential of Ras, inducing a change in cell morphology
activated by mutant K-ras, and transmitting oncogenic signals
(11,12). Rap1, similarly to other GTPases,
demonstrates binary switches by cycling between inactive GDP-bound
and active GTP-bound conformations, and regulates multiple cellular
signaling pathways after receiving a cellular stimulus (12–17).
In addition, the migrational ability of MSCs was reported to
increase via activation of the Rap1 signaling pathway (18). Furthermore, HMGB1 has been
confirmed to activate the RAS/mitogen-activated protein kinase
(MAPK) signaling pathway of MSCs (19). Given that Rap1 is a member of the
Ras family, an hypothesis was developed for the present study that
HMGB1 may also potentiate MSC migration via Rap1 activation, and a
series of experiments have been performed to verify this
hypothesis.
In the present study, a scratch assay has been used
to determine the effect of HMGB1 on the mobility of MSCs. To
further study the molecular mechanism of MSC activation via HMGB1,
a Quantibody® array was applied to detect the cytokines
synthesized by MSCs with and without treatment with HMGB1. The
identification of differentiated cytokines under these two
conditions was used to reveal the effect of HMGB1 on the MSCs.
These results may provide a basis for developing novel approaches
in bone-fracture-healing therapy.
Materials and methods
Reagents
Recombinant human HMGB1 protein (Sigma-Aldrich, St.
Louis, MO, USA) was commercially purchased. A concentration of 25
ng/ml protein was used in the experiments detailed below. The Rap1
inhibitor (Cell Signaling Technology, Inc., Danvers, MA, USA),
which is a transferase inhibitor, was used at a concentration of 5
µM. The reagents listed above were handled and used
according to the manufacturer's protocols.
Isolation and culture expansion of human
bone marrow MSCs
MSCs (Cyagen Biosciences Inc., Guangzhou, China)
were commercially purchased. Adherent cells were trypsinized using
0.25% trypsin (Cyagen Biosciences Inc.) for ~30 sec and passaged
after the cell confluence had reached ~80%, and the cells at
passages 3 to 5 were used in the experiments detailed below.
Typically, these cells exhibited the capacity of differentiation
into osteoblasts, adipoblasts and chondrocytes under specific
inductive conditions.
Transwell migration assay
Cell migration was performed with Transwell chambers
(pore size, 8-µm diameter; Corning Costar, Inc., Corning,
NY, USA). Complete™ medium (Cyagen Biosciences Inc.) containing
0.1% fetal calf serum (FCS; Cyagen Biosciences Inc.) was added into
the wells of a 24-well plate, and subsequently, serum-starved MSCs
(1×105) suspended in a volume of 100 µl Complete™
medium containing 0.1% FCS were added into the upper chamber. Prior
to the addition of HMGB1, the transwell plate (with MSCs in the
upper chamber and medium containing 0.1% FCS only in the lower
chamber), was first incubated at 37°C for 1 h. Following the
addition of HMGB1, the plate was subsequently incubated at 37°C for
3 h, followed by membrane fixation with 4% paraformaldehyde
(Beyotime Institute of Biotechnology, Haimen, China) and staining
with 0.1% crystal violet (Beyotime Institute of Biotechnology). The
membrane was subsequently washed, and the cells on the underside of
the membrane were observed under a light microscope (Leica DMI/LM;
Leica Microsystems GmbH, Wetzlar, Germany). Numbers of cells were
counted in five to ten random fields for each membrane.
Cell scratch assay
Cell migration was determined using a scratch assay.
The cells were cultivated to 90% confluence on 12-well plates. The
groups were as follows: Control, without HMGB1; treatment, MSCs
cultured with 25 ng/ml HMGB1; supernatant, MSCs pretreated with 25
ng/ml HMGB1 and following 48 h, the supernatant of the MSCs was
extracted to culture a fresh batch of MSCs. Subsequently, cell
scrapers (Corning Costar Inc.) were used to scratch the confluent
cells. The extent of cellular growth was observed at 0 and 48 h.
All the experiments were repeated in triplicate.
Western blot analysis
Cells were harvested and lysed in
radioimmunoprecipitation assay buffer containing proteinase
inhibitors (Cyagen Biosciences Inc.). Following measurement of the
protein concentration using a BCA kit (Beyotime Institute of
Biotechnology), protein samples were separated using 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and
the proteins were transferred onto a polyvinylidene difluoride
(PVDF) membrane by western blotting. The membranes were blocked in
5% skimmed milk for 1 h, and incubated with the following
antibodies: Rabbit anti-human GTP-Rap1 (Active Rap1 Detection kit,
cat. no. CST8818; dilution, 1:500) from Cell Signaling Technology,
Inc., rabbit anti-human Rap1 (cat. no. ab47234; Abcam, Cambridge,
MA, USA; dilution, 1:500) and rabbit anti-human β-actin (cat. no.
sc-130301; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
separately at 4°C overnight. After an incubation with goat
anti-rabbit peroxidase-linked secondary antibody (cat. no. 31210;
Thermo Fisher Scientific, Inc., dilution, 1:5,000) at 25°C for 3 h,
immunoreactive proteins were visualized using an enhanced
chemiluminescence (ECL) reagent (Thermo Fisher Scientific, Inc.).
Relative quantification of bands in the western blot was performed
using ImageJ software [National Institutes of Health (NIH),
Bethesda, MA, USA).
Antibody arrays
Soluble proteins in the medium of the stromal cell
lines were measured using the Human Cytokine Array G1000
(AAH-CYT-G1000; RayBiotech, Inc., Norcross, GA, USA), according to
the manufacturer's protocol. These arrays are able to detect up to
120 proteins. Stromal cells were plated 3 days prior to the
experiment in Dulbecco's modified Eagle's medium (DMEM; Cyagen
Biosciences Inc.) containing 10% FBS, and were 75–90% confluent
when the cell lysis solution was collected and filtered. Medium
containing 10% FBS was also hybridized to the arrays, and used
subsequently for normalization. Ten technical and biological
replicates were performed, and these demonstrated a very high
correlation (correlation coefficient >0.9; data not shown).
Hybridization was performed overnight at 4°C. All slides were
scanned using a GenePix® 4000B Microarray Scanner (Axon
Instruments, Inc., Union City, CA, USA) and analysed using the
software GenePix® Pro 6.0. The F532 median - 2B532 score
was used, and averaged across triplicates on each array. The
results were subsequently normalized using internal controls, and
the values for cytokines in clear medium containing 10% FBS were
subtracted.
Statistical analysis
Statistical significance was performed using the
two-tailed Student's t-test, assuming equal variances. The
chi-squared test was used to compare rates. P<0.05, P<0.01
and P<0.001 were taken to indicate statistically significant
values.
Results
Effects of HMGB1 on the increasing
migrational ability of MSCs
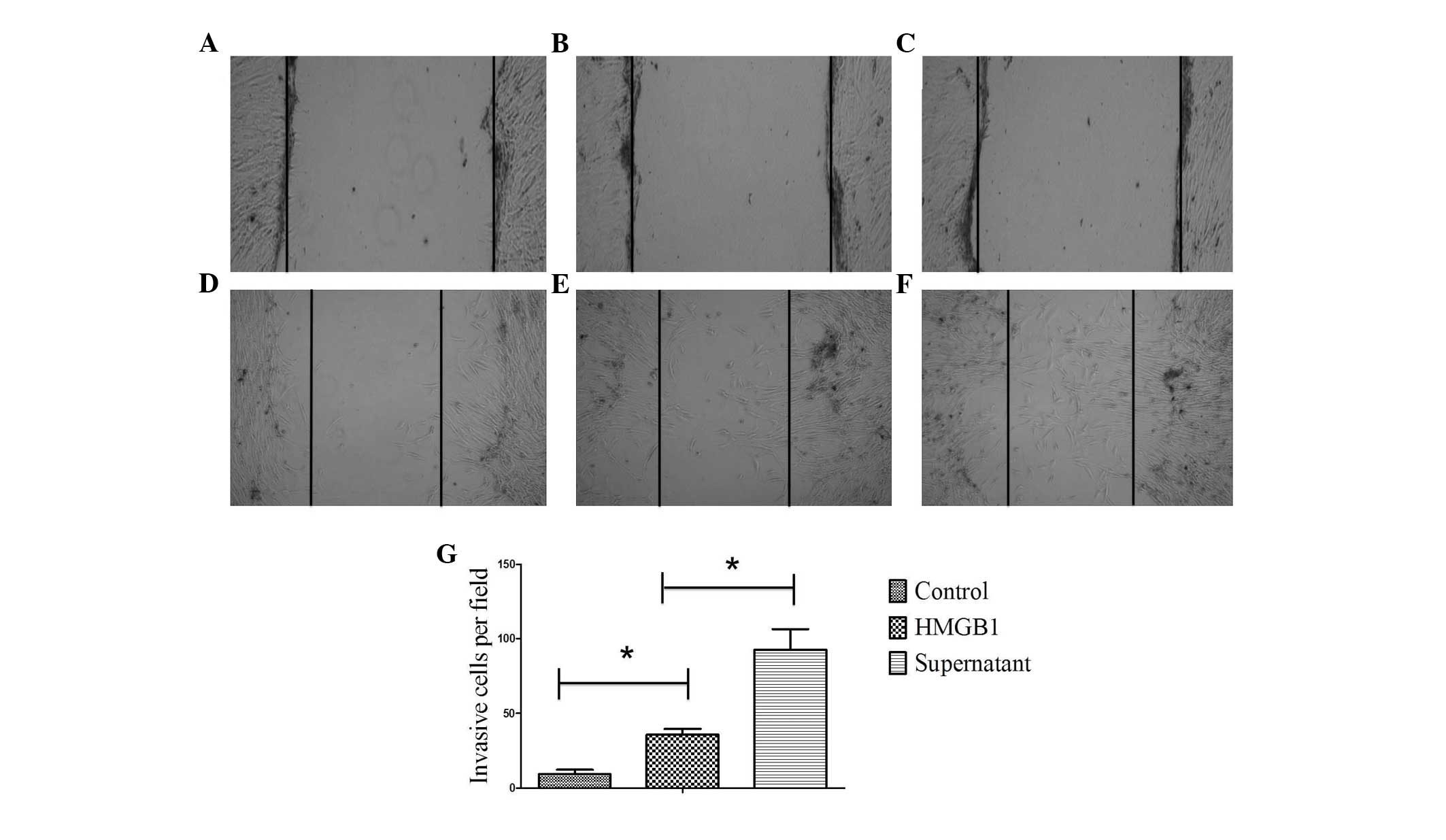
To investigate the effect of HMGB1 on the migration
of MSCs, a scratch assay was performed (Fig. 1). The MSCs of all groups moved
towards the blank area to a certain extent after 48 h, indicating a
certain level of migrational ability. Compared with the control
group, MSCs cultured with 25 ng/ml HMGB1 significantly surpassed
the baseline (P<0.05), indicating that the migrational ability
of MSCs was increased on stimulation with HMGB1 (Fig. 1B and C). In another experiment,
MSCs were pretreated with 25 ng/ml HMGB1. After 48 h, the
supernatant of the MSCs was extracted to culture a new batch of
MSCs (Fig. 1D), and a scratch
assay was performed. A greater number of the MSCs from the second
treatment crossed the baseline compared with that observed in the
group of MSCs directly cultured with HMGB1 (P<0.05). This
confirmed that HMGB1 could significantly increase the migrational
ability of MSCs (Fig. 1E).
Effects of HMGB1 on cellular chemokine
synthesis by MSCs
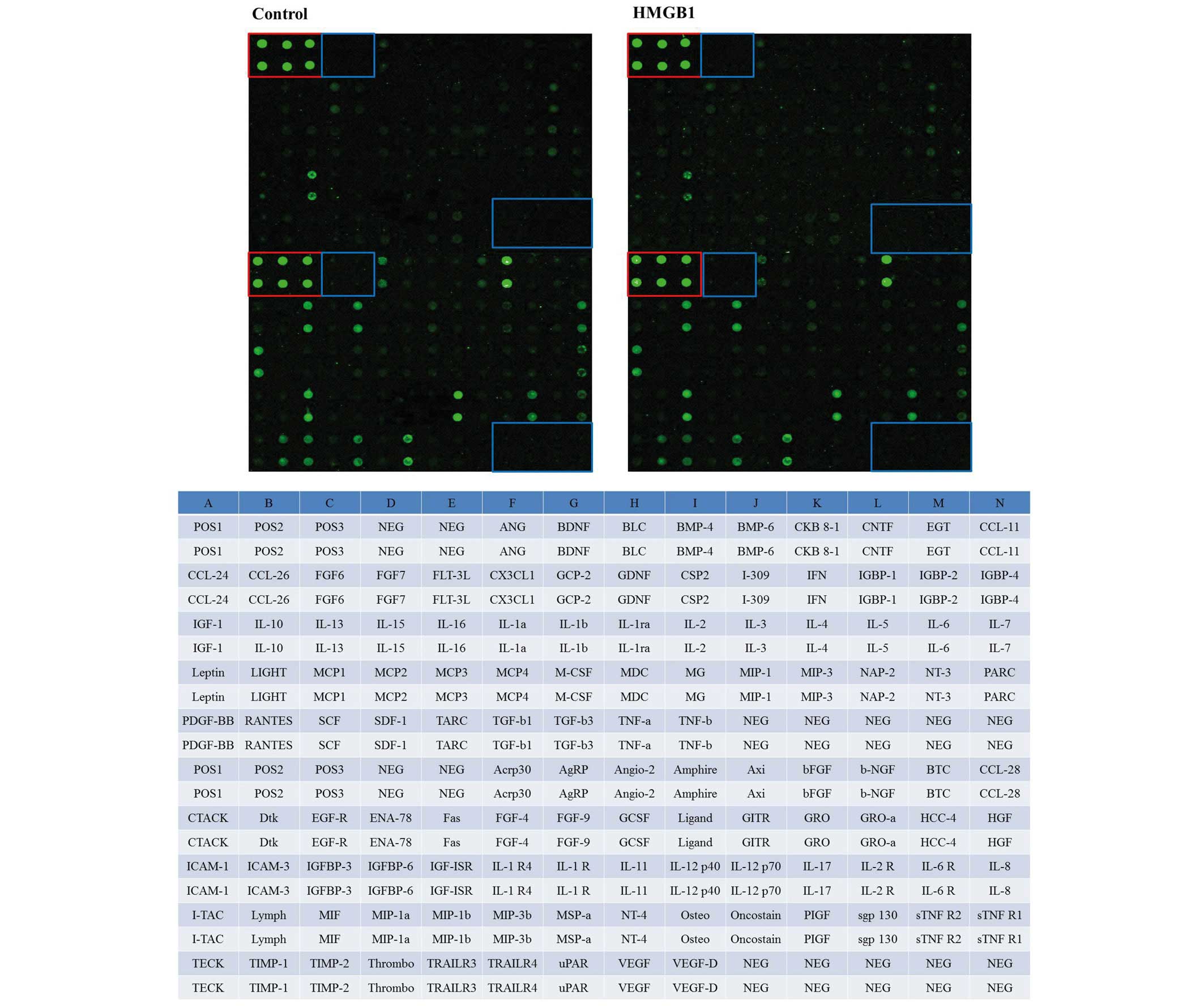
To determine the influence of HMGB1 stimulation on
human MSCs at the protein level, a Quantibody® array was
performed. The treatment group was stimulated with 25 ng/ml HMGB1
and compared with non-stimulated cultures (Fig. 2).
Underlying our search criteria of a differential
synthesis (>2.0-fold change or <0.5-fold change), the
bioinformatics data analysis identified that seven cytokines were
differentially synthesized between the two groups. Increased
cytokine synthesis (relative level, >1.5-fold change) of bone
morphogenetic protein-4 (BMP-4), neurotrophin-3 (NT-3), LIGHT [or
tumor necrosis factor superfamily member 14 (TNFSF14)], monocyte
chemoattractant protein 4 (MCP-4), Dtk and macrophage inflammatory
protein-1β (MIP-1β) following induction with HMGB1 was measured
(Fig. 3). A reduced level of
cytokine synthesis (relative level, <0.5-fold change) was
observed for β-nerve growth factor (β-NGF; Fig. 3A and B).
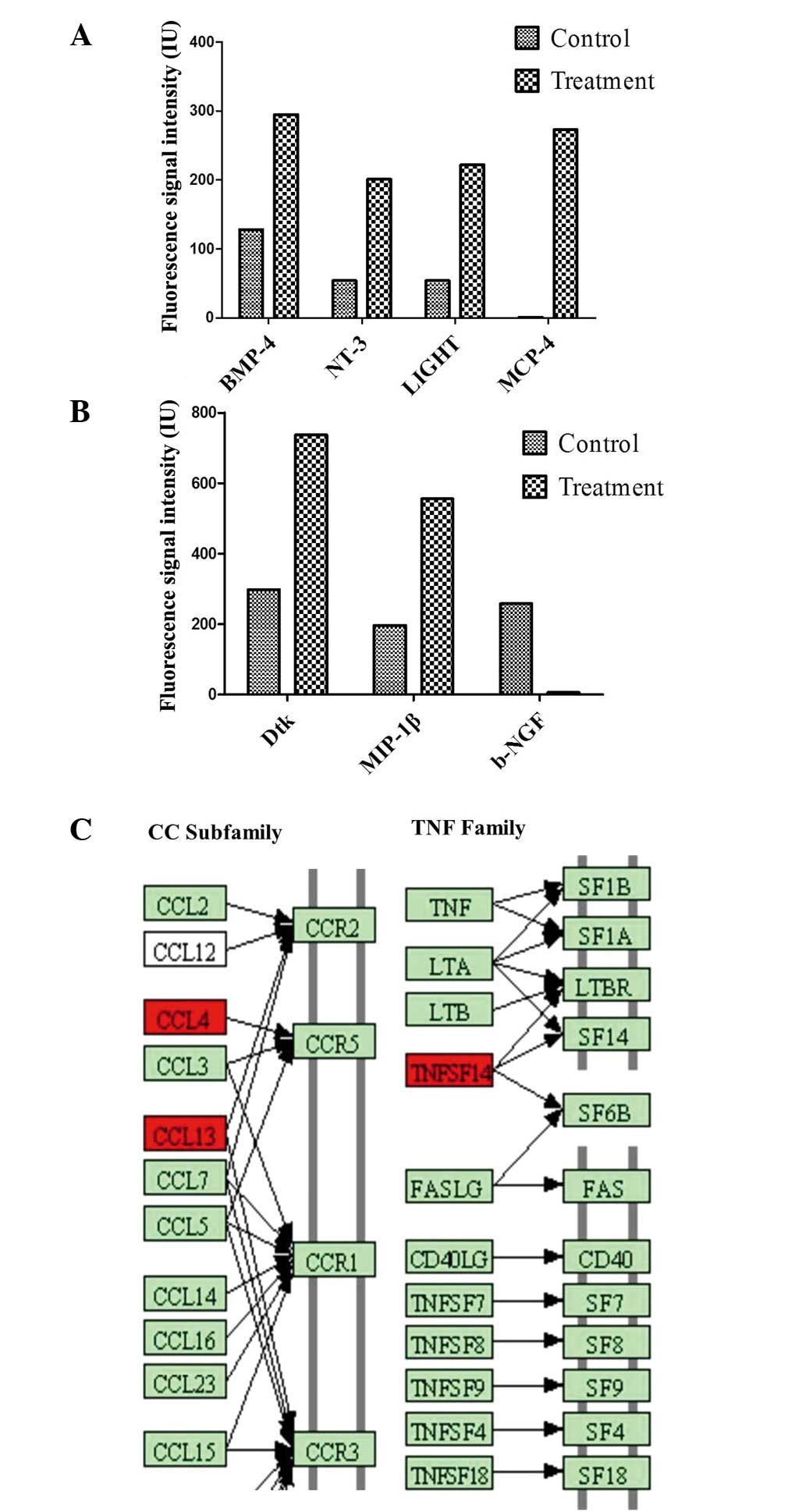 | Figure 3(A and B) Seven cytokines were
differentially synthesized (>2.0-fold change or <0.5-fold
change), based on the results of Quantibody® array. (C)
Cytokine-cytokine receptor interaction between the synthesized
cytokines. CCL4, CCL13 and TNFSF14 of the differentially secreted
cytokines (>2.5-fold induction), involved in cellular movement,
cellular development and cell death, are presented in their
cytokine-cytokine receptor interaction pathway (CCR2, CCR5, CCR3,
LTB R, SF14 and SF6B) according to the cytokine-cytokine receptor
interaction pathway (0460hsa) in the Kyoto Encyclopedia of Genes
and Genomes database (red indicates ʻincreasedʼ). BMP-4, bone
morphogenetic protein-4; NT-3, neurotrophin-3; MCP-4, monocyte
chemoattractant protein 4; MIP-1β, macrophage inflammatory
protein-1β; CCLx, chemokine ligand x; CCRx, chemokine receptor x;
TNFSF14, tumor necrosis factor superfamily member 14; LTA/B(R),
lymphotoxin-α/β (receptor). |
Hierarchical clustering of those seven cytokines
with normalized cytokine synthesis values disclosed two distinct
groups: The HMGB1-treated group and the non-stimulated control
group. Six differentially secreted cytokines were visualized as
being induced, and one cytokine as being repressed. Notably, among
the seven cytokines, three of them were identified as being
involved in the nuclear factor-κB (NF-κB) signaling pathway,
namely, MCP-4, MIP-1β and LIGHT. Subsequently, two cytokines were
identified as being involved in the neurotrophic factor-mediated
Trk receptor signaling pathway: NT-3 and β-NGF. Seven
differentially secreted cytokines were also revealed to be
associated with the response to an external stimulus (7), cell migration (6), localization of the cell (6), regulation of programmed cell death
(5) and the immune system process
(5), according to Gene Ontology
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID). As expected, numerous differentially synthesized
cytokines were associated with more than one biological
process.
Underlying our search criteria of a differential
secretion, bioinformatics data analysis resulted in the selection
of three cytokines (relative level, >2.5-fold control). These
were visualized as being induced between the two groups. Increased
levels of cytokine secretion were measured for chemokine ligand 4
(CCL4), CCL13 and TNFSF14 following induction with HMGB1. The
detected differentially secreted cytokines serve roles in other
molecular and cellular functions, including cellular growth and
proliferation, cell death, cell morphology and cellular
development. On the basis of the three differentially secreted
cytokines, the cytokine-cytokine receptor interaction pathway
(0460hsa in the Kyoto Encyclopedia of Genes and Genomes database)
was the dominant pathway that was influenced by HMGB1. Thus, the
secretion of CCL5 and CCL26 was induced (Fig. 3C).
HMGB1 induces MSCs to activate the Rap1
signaling pathway and enhance MSC migration
The present study identified that six cytokines, of
the differentially synthesized cytokines, are involved in cell
migration based on the results of the Quantibody® array.
Subsequently, among the six cytokines, β-NGF was identified as
being involved in the Rap1 signaling pathway. Therefore, the Rap1
signaling pathway was proposed to be significant to the current
study. However, a limitation of the study is that the array was not
repeated three times. Thus, to verify the hypothesis that the Rap1
signaling pathway of MSCs is also activated by HMGB1 stimulation,
western blot analysis was performed. Compared with the expression
of GTP-Rap1 in the control group, that in MSCs increased
significantly following treatment with 25 ng/ml HMGB1. By contrast,
the expression of GTP-Rap1 declined significantly following
treatment with the Rap1 inhibitor. These results indicated that
HMGB1 was able to activate the Rap1 signaling pathway in MSCs.
However, the Rap1 inhibitor demonstrated marked inhibition of the
Rap1 signaling pathway activated by HMGB1 (Fig. 4A).
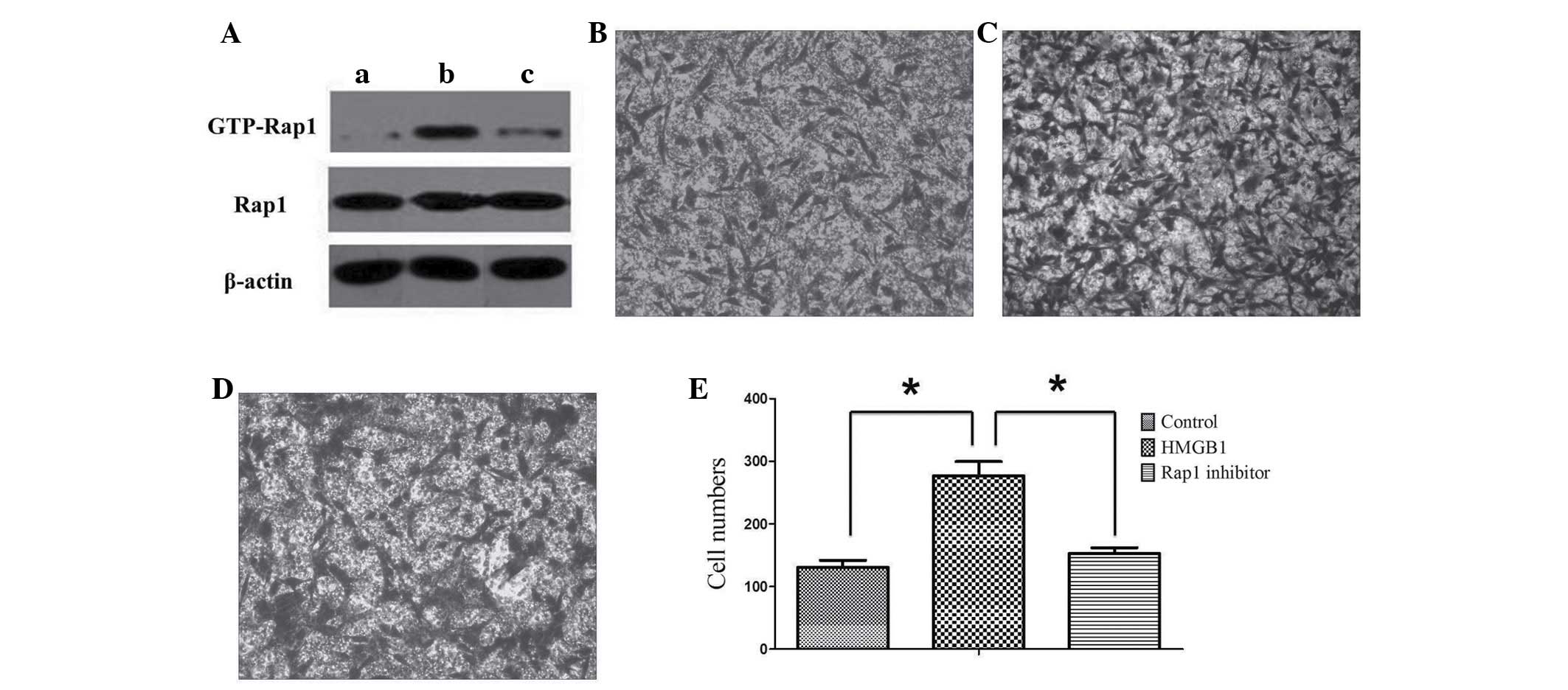 | Figure 4Rap1 activation is required for MSC
migration. (A) Western blot analysis of the Rap1 signaling activity
upon treatment with HMGB1 (25 ng/ml) and/or Rap1 inhibitor (note,
β-actin was run during the same experiment, but on different parts
of the gel). MSCs were maintained (A,a) in the basal medium alone,
(A,b) with 25 ng/ml HMGB1 or (A,c) with 25 ng/ml HMGB1 and Rap1
inhibitor. (B–E) A transwell migration assay was performed. MSCs
were maintained (B) in the basal medium alone, (C) with 25 ng/ml
HMGB1 or (D) with 25 ng/ml HMGB1 and Rap1 inhibitor. (E) The
migratory cells were counted in the HMGB1, inhibitor or control
groups, respectively. Results were obtained from three independent
experiments and the data are expressed as the mean ± standard
deviation (n=3 in each experiment). *P<0.05. HMGB1,
high mobility group box 1 protein. |
To investigate whether activation of the Rap1
signaling pathway could increase the mobility of MSCs, a transwell
migration assay was performed. The number of MSCs migrating towards
the other side of the membrane increased markedly following
treatment with HMGB1. However, following treatment with the Rap1
inhibitor, the number of migrating cells was markedly reduced
compared with that in the group that was not treated with the
inhibitor. This result suggested that activation and inhibition of
the Rap1 signaling pathway significantly enhanced and suppressed
the migration of MSCs, respectively (Fig. 4B–E).
Discussion
In the first scratch assay, it was observed that the
mobility of MSCs treated with an appropriate concentration of HMGB1
was greater compared with that of the control group, suggesting
that HMGB1 enhanced the migration of MSCs. In another experiment,
the supernatant of MSCs induced with HMGB1 was used to culture a
separate batch of MSCs. The migration of MSCs treated with the
supernatant was enhanced compared with that of the MSCs of the
control group and the group directly treated with an identical
concentration of HMGB1. Despite the crude experimental design,
these results suggested that certain cytokines in the supernatant
were also able to enhance the migration of MSCs. Therefore, it was
hypothesized that, following HMGB1 stimulation, MSCs synthesize
certain cytokines to enhance their mobility further. However, this
hypothesis requires further verification, since the present
experiment did not control for all the potentially confounding
variables.
To confirm whether the synthesis of specific
cytokines by MSCs was enhanced following HMGB1 stimulation, and to
identify the cytokines responsible for enhancing the mobility of
MSCs, a Quantibody® array was performed, which revealed
that the synthesis of CCL4, CCL13 and TNFSF14 increased markedly.
TNFSF belongs to the family of type II transmembrane proteins,
which contains 19 members, with approximately 150 homologous amino
acids in the extracellular C-terminal domain. TNFSF is formed by
ten β-strands, folded into a helical conformation, which forms a
binding site for its corresponding receptors. The majority of the
members of this family form trimers with their corresponding
receptors, and manifest their biological activities in the trimeric
form (20). Following the binding
of TNFSFs with their ligands, certain members activate the MAPK
signaling pathway to stimulate cellular activities (21,22),
whereas others recruit the death-induced signaling complex. These
molecules subsequently activate caspases, and thereby induce cell
apoptosis (23,24). The CCL subfamily members contains
over 20 members, with two neighboring cysteine residues in the
N-terminal domain. These predominantly activate monocytes and
certain of the T cell subfamilies (25). CCL4, also termed MIP-1β, activates
natural killer cells and multiple immune cells (26,27).
CCL13, also termed MCP-4, is encoded by a gene located in human
chromosome 17 within a large cluster of other CC chemokines. After
binding with its receptors, it is involved in the body's allergic
reaction (28–31). Most importantly, CCL4 and CCL13
have been revealed to enhance the migration of MSCs (32,33).
Therefore, it was hypothesized that HMGB1 may enhance MSC migration
by promoting the synthesis of CCL4 and CCL13, which would increase
the mobility of MSCs.
To confirm our hypothesis about the Rap1 signaling
pathway, the expression of relevant proteins of MSCs that are
induced by HMGB1 was investigated using western blot analysis. The
results indicated that the expression of GTP-Rap1 in MSCs increased
markedly following HMGB1 stimulation. Since the level of GTP-bound
Rap1 represents the activation of Rap1, it was deduced that HMGB1
activated the Rap1 signaling pathway of MSCs. Furthermore, the Rap1
inhibitor was used to block the Rap1 signaling pathway in MSCs. By
means of the transwell migration assay, the mobility of MSCs was
found to decrease substantially following treatment with the Rap1
inhibitor. This result suggested that HMGB1 activates the Rap1
signaling pathway, and enhances the migration of MSCs.
In the present study, a preliminary analysis of the
Quantibody® array results was performed, and these
results require further verification. Nevertheless, several of the
results are intriguing, and merit further study. For example,
several differentially synthesized cytokines were identified that
are involved in two signaling pathways: The NF-κB signaling pathway
and the neurotrophin signaling pathway. However, whether these two
signaling pathways are activated by HMGB1 stimulation requires
verification. Such a confirmation would help to elucidate the
mechanism underlying the effect of HMGB1 on MSCs.
In conclusion, activation of the Rap1 signaling
pathway has been shown to enhance the migration of MSCs. However,
downstream signaling following activation of the Rap1 signaling
pathway requires further investigation. It has been reported that
Rap1 can activate β1 and β2 integrins in T cells, which could
improve cell mobility (18,34).
However, whether Rap1 in MSCs may also improve the synthesis of
integrins, and whether the integrins have a direct effect on the
migration of MSCs, remains to be elucidated. These questions will
form the basis of our next research endeavours.
Acknowledgments
This work was supported by grants from the National
Natural Science Foundation of China (grant nos. 81271973 and
81201397), the Zhejiang Provincial Natural Science Foundation of
China (no. Y2090283) and Zhejiang Medical and Health Science and
Technology Plan Project (no. 2011ZDA011).
References
|
1
|
Naglova H and Bucova M: HMGB1 and its
physiological and pathological roles. Bratisl Lek Listy.
113:163–171. 2012.PubMed/NCBI
|
|
2
|
Meng E, Guo Z, Wang H, Jin J, Wang J, Wang
H, Wu C and Wang L: High mobility group box 1 protein inhibits the
proliferation of human mesenchymal stem cells and promotes their
migration and differentiation along osteoblastic pathway. Stem
Cells Dev. 17:805–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Charoonpatrapong K, Shah R, Robling AG,
Alvarez M, Clapp DW, Chen S, Kopp RP, Pavalko FM, Yu J and Bidwell
JP: HMGB1 expression and release by bone cells. J Cell Physiol.
207:480–490. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Granero-Moltó F, Weis JA, Miga MI, Landis
B, Myers TJ, O'Rear L, Longobardi L, Jansen ED, Mortlock DP and
Spagnoli A: Regenerative effects of transplanted mesenchymal stem
cells in fracture healing. Stem Cells. 27:1887–1898. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Glass GE, Chan JK, Freidin A, Feldmann M,
Horwood NJ and Nanchahal J: TNF-alpha promotes fracture repair by
augmenting the recruitment and differentiation of muscle-derived
stromal cells. Proc Natl Acad Sci USA. 108:1585–1590. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marsell R and Einhorn TA: The biology of
fracture healing. Injury. 42:551–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sidney LE, Kirkham GR and Buttery LD:
Comparison of osteogenic differentiation of embryonic stem cells
and primary osteoblasts revealed by responses to IL-1β, TNF-α and
IFN-γ. Stem Cells Dev. 23:605–617. 2014. View Article : Google Scholar
|
|
8
|
Tso GH, Law HK, Tu W, Chan GC and Lau YL:
Phagocytosis of apoptotic cells modulates mesenchymal stem cells
osteogenic differentiation to enhance IL-17 and RANKL expression on
CD4+ T cells. Stem Cells. 28:939–954. 2010.PubMed/NCBI
|
|
9
|
Guo J, Jie W, Shen Z, Li M, Lan Y, Kong Y,
Guo S, Li T and Zheng S: SCF increases cardiac stem cell migration
through PI3K/AKT and MMP-2/-9 signaling. Int J Mol Med. 34:112–118.
2014.PubMed/NCBI
|
|
10
|
Mingari MC, Moretta A and Moretta L:
Regulation of KIR expression in human T cells: A safety mechanism
that may impair protective T-cell responses. Immunol Today.
19:153–157. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raaijmakers JH and Bos JL: Specificity in
Ras and Rap signaling. J Biol Chem. 284:10995–10999. 2009.
View Article : Google Scholar :
|
|
12
|
Katagiri K, Maeda A, Shimonaka M and
Kinashi T: RAPL, a Rap1-binding molecule that mediates Rap1-induced
adhesion through spatial regulation of LFA-1. Nat Immunol.
4:741–748. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rebstein PJ, Cardelli J, Weeks G and
Spiegelman GB: Mutational analysis of the role of Rap1 in
regulating cytoskeletal function in Dictyostelium. Exp Cell Res.
231:276–283. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bos JL, Franke B, M'Rabet L, Reedquist K
and Zwartkruis F: In search of a function for the Ras-like GTPase
Rap1. FEBS Lett. 410:59–62. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McLeod SJ and Gold MR: Activation and
function of the Rap1 GTPase in B lymphocytes. Int Rev Immunol.
20:763–789. 2001. View Article : Google Scholar
|
|
16
|
Katagiri K, Ohnishi N, Kabashima K, Iyoda
T, Takeda N, Shinkai Y, Inaba K and Kinashi T: Crucial functions of
the Rap1 effector molecule RAPL in lymphocyte and dendritic cell
trafficking. Nat Immunol. 5:1045–1051. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Creasy CL and Chernoff J: Cloning and
characterization of a member of the MST subfamily of Ste20-like
kinases. Gene. 167:303–306. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen CP, Huang JP, Chu TY, Aplin JD, Chen
CY and Wu YH: Human placental multipotent mesenchymal stromal cells
modulate trophoblast migration via Rap1 activation. Placenta.
34:913–923. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng S, Zhou G, Luk KD, Cheung KM, Li Z,
Lam WM, Zhou Z and Lu WW: Strontium promotes osteogenic
differentiation of mesenchymal stem cells through the Ras/MAPK
signaling pathway. Cell Physiol Biochem. 23:165–174. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mauri DN, Ebner R, Montgomery RI, Kochel
KD, Cheung TC, Yu GL, Ruben S, Murphy M, Eisenberg RJ, Cohen GH, et
al: LIGHT, a new member of the TNF superfamily and lymphotoxin
alpha are ligands for herpesvirus entry mediator. Immunity.
8:21–30. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu TL, Chang YC, Chen SJ, Liu YJ, Chiu
AW, Chio CC, Chen L and Hsieh SL: Modulation of dendritic cell
differentiation and maturation by decoy receptor 3. J Immunol.
168:4846–4853. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugiyama T, Suzuki H and Takahashi T:
Light-induced rapid Ca2+ response and MAPK
phosphorylation in the cells heterologously expressing human OPN5.
Sci Rep. 4:53522014.
|
|
23
|
Yu KY, Kwon B, Ni J, Zhai Y, Ebner R and
Kwon BS: A newly identified member of tumor necrosis factor
receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J
Biol Chem. 274:13733–13736. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuai J, Nickbarg E, Wooters J, Qiu Y, Wang
J and Lin LL: Endogenous association of TRAF2, TRAF3, cIAP1 and
Smac with lymphotoxin beta receptor reveals a novel mechanism of
apoptosis. J Biol Chem. 278:14363–14369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Irving SG, Zipfel PF, Balke J, McBride OW,
Morton CC, Burd PR, Siebenlist U and Kelly K: Two inflammatory
mediator cytokine genes are closely linked and variably amplified
on chromosome 17q. Nucleic Acids Res. 18:3261–3270. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bystry RS, Aluvihare V, Welch KA,
Kallikourdis M and Betz AG: B cells and professional APCs recruit
regulatory T cells via CCL4. Nat Immunol. 2:1126–1132. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cocchi F, DeVico AL, Garzino-Demo A, Arya
SK, Gallo RC and Lusso P: Identification of RANTES, MIP-1 alpha and
MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T
cells. Science. 270:1811–1815. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garcia-Zepeda EA, Combadiere C, Rothenberg
ME, Sarafi MN, Lavigne F, Hamid Q, Murphy PM and Luster AD: Human
monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine
with activities on monocytes, eosinophils and basophils induced in
allergic and nonallergic inflammation that signals through the CC
chemokine receptors (CCR)-2 and -3. J Immunol. 157:5613–5626.
1996.PubMed/NCBI
|
|
29
|
Naruse K, Ueno M, Satoh T, Nomiyama H, Tei
H, Takeda M, Ledbetter DH, Coillie EV, Opdenakker G, Gunge N, et
al: A YAC contig of the human CC chemokine genes clustered on
chromosome 17q11.2. Genomics. 34:236–240. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blanpain C, Migeotte I, Lee B, Vakili J,
Doranz BJ, Govaerts C, Vassart G, Doms RW and Parmentier M: CCR5
binds multiple CC-chemokines: MCP-3 acts as a natural antagonist.
Blood. 94:1899–1905. 1999.PubMed/NCBI
|
|
31
|
Lamkhioued B, Garcia-Zepeda EA, Abi-Younes
S, Nakamura H, Jedrzkiewicz S, Wagner L, Renzi PM, Allakhverdi Z,
Lilly C, Hamid Q and Luster AD: Monocyte chemoattractant protein
(MCP)-4 expression in the airways of patients with asthma.
Induction in epithelial cells and mononuclear cells by
proinflammatory cytokines. Am J Respir Crit Care Med. 162:723–732.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lejmi E, Perriraz N, Clément S, Morel P,
Baertschiger R, Christofilopoulos P, Meier R, Bosco D, Bühler LH
and Gonelle-Gispert C: Inflammatory chemokines MIP-1δ and MIP-3α
are involved in the migration of multipotent mesenchymal stromal
cells induced by hepatoma cells. Stem Cells Dev. 24:1223–1235.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang F, Wang C, Wang H, Lu M, Li Y, Feng
H, Lin J, Yuan Z and Wang X: Ox-LDL promotes migration and adhesion
of bone marrow-derived mesenchymal stem cells via regulation of
MCP-1 expression. Mediators Inflamm. 2013:6910232013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Burbach BJ, Medeiros RB, Mueller KL and
Shimizu Y: T-cell receptor signaling to integrins. Immunol Rev.
218:65–81. 2007. View Article : Google Scholar : PubMed/NCBI
|