Introduction
Lung cancer is a highly prevalent type of cancer
(1), and is a leading cause of
cancer-associated mortality worldwide (2). In 2012, ~11.9% of cancer cases
diagnosed were attributed to lung cancer in Europe (3). In China, lung cancer accounts for
18.5% of cancer cases, and has a mortality rate of 23.1%, ranking
it the most prevalent and most lethal among all cancer types
(4). Lung cancer is classified as
follows: Small cell lung cancer (SCLC) and non-SCLC (NSCLC)
(5). Adenocarcinoma, squamous cell
carcinoma and large cell cancer are the three main subtypes of
NSCLC (6). Following surgical
treatment, the majority of cases of NSCLC will relapse within 5
years; therefore, it is of great importance to fully elucidate the
mechanism underlying the progression of NSCLC.
Microarray screening has been used to identify
differentially expressed genes (DEGs) in cancer samples, in order
to better understand the mechanisms underlying cancer development
(7). Navab et al (8) identified a prognostic gene-expression
signature that contained a subset of 11 genes, which were validated
in numerous independent NSCLC gene expression databases. In
addition, since lung adenocarcinoma and squamous cell carcinoma
exhibit different histology, gene expression levels, particularly
the levels of relevant markers such as cytokeratin 5/6, differ
between them (9). Therefore,
specific genes and microRNAs may be used to distinguish between
these two types of cancer (10),
and the genetic signatures of these cancer types may differ
(11). However, the differences in
expression between lung adenocarcinoma and squamous cell carcinoma
have yet to be fully elucidated.
The present study used gene expression files
downloaded from the Gene Expression Omnibus (GEO) database,
compared the DEGs detected between lung adenocarcinoma and squamous
cell carcinoma samples, and conducted function and pathway
enrichment analyses. In addition, a protein-protein interaction
(PPI) network of the DEGs was constructed. Genes that exhibited
higher degrees in the networks were selected as the key genes in
the two lung cancer groups.
Materials and methods
Microarray data
The GSE6044 gene expression profile was downloaded
from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The profile
included data from 10 lung adenocarcinoma samples, 10 lung squamous
cell carcinoma samples, and five matched normal lung tissue
samples. The microarray data were based on the GPL201 [HG-Focus]
platform (Affymetrix Human HG-Focus Target Array; Affymetrix, Inc.,
Santa Clara, CA, USA).
Data processing
The raw CEL data were rectified and standardized by
Robust Multichip Average using the Affy package in R language
(bioconductor.org/packages/release/bioc/html/affy.html),
in order to obtain the corresponding expression data of the probes.
Subsequently, redundant probes that did not correspond with an
Entrez Gene ID were deleted, whereas the median value was used for
probes that corresponded with several Entrez Gene IDs.
DEGs screening
Student's t-test was used to screen for DEGs in the
two types of lung cancer samples. The P-values were further
adjusted using the false discovery rate (FDR) approach, according
to the Benjamini-Hochberg (BH) method (12). Genes with a FDR <0.1 and fold
change (FC) >1.5 or <0.67 were considered DEGs between the
cancer and normal samples.
Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment
In order to identify the pathways associated with
the two cancer groups, tools in the Database for Annotation,
Visualization and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov) (13,14)
were used to screen the pathways enriched for in the DEGs from the
cancer samples using the Expression Analysis Systematic Explorer
score (a modified Fisher's exact t-test) with BH multiple testing
correction (12). A KEGG pathway
with a BH-corrected P<0.05 was considered to be significantly
enriched.
Global network construction of
cancer-associated genes
A total of 14 cancer-associated pathways were
downloaded from the KEGG database (www.genome.jp/kegg), including colorectal cancer,
pancreatic cancer, glioma, thyroid cancer, acute myeloid leukemia,
chronic myeloid leukemia, basal cell carcinoma, melanoma, renal
cell carcinoma, bladder cancer, prostate cancer, endometrial
cancer, SCLC and NSCLC. Annotated genes from the 14 pathways were
used to construct the global network according to their interaction
in pathways by the Cytoscape software (www.cytoscape.org). The expression (upregulated or
downregulated) of these genes were further annotated by mapping to
the present gene analysis results in two types of lung cancer
samples in comparison with normal samples. Subsequently, the
enriched DEGs were screened to construct the PPI network in which
the interaction relationships between proteins were downloaded from
the Human Protein Reference Database (HPRD; www.hprd.org) database (15).
Results
DEGs selection
A total of 8,348 genes were obtained from the
GSE6044 gene expression profile. Under the screening criteria of
FDR<0.1 and FC >1.5 or P<0.67, 95 upregulated DEGs and 241
downregulated DEGs (up vs. down, 1:3) were detected in the lung
adenocarcinoma samples, whereas 204 upregulated DEGs and 285
downregulated DEGs (up vs. down, 1:1.5) were detected in the lung
squamous cell carcinoma samples, as compared with the normal lung
tissue samples. The majority of DEGs were downregulated in both
lung cancer groups.
Significantly enriched pathways in the
DEGs from the two lung cancer groups
In order to determine the differences between the
two types of lung cancer at the level of related pathways, DAVID
was used to conduct pathway enrichment. The DEGs in the lung
squamous cell carcinoma group were significantly enriched in the
following three pathways: Hsa04110, cell cycle (P=3.93E-06),
involving genes YWAQ, MCM3, CHEK2, GADD45 G, RAD21 and PCNA;
hsa03030, DNA replication (P=8.50E-06), involving genes MCM3, FEN1,
RFC4, POLA1, PCNA and MCM2; and hsa03430, mismatch repair
(P=0.01309), involving genes RFC4, MSH6, RFC2 and MSH3.
The DEGs may be involved in the lung adenocarcinoma
group by participating in metabolism pathways, including Hsa00010,
glycolysis/gluconeogenesis (ADH1B, ADH1C, ALDH3A2, LDHA, AKR1A1 and
FBP1) and hsa00980, metabolism of xenobiotics by cytochrome P450
(ADH1B, GSTA3, CYP2F1 and ADH6). However, no significant pathways
were enriched when using the cut-off value of BH-corrected
P<0.05.
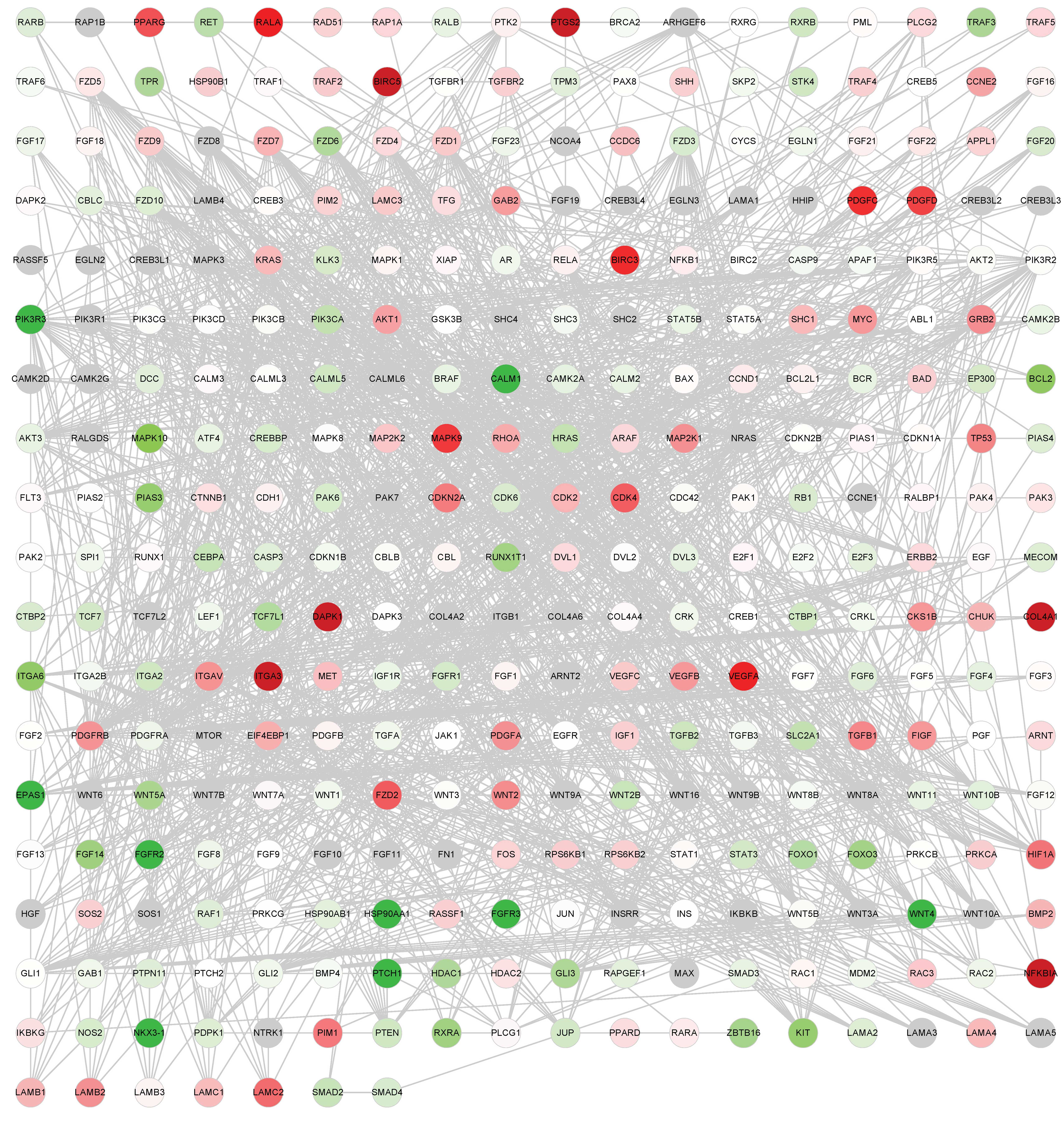
Global network of cancer-associated
genes
Annotated genes from the 14 cancer-associated
pathways downloaded from KEGG were used to construct the global
network. There were 341 genes and 1,569 edges (interactions between
genes) in the network, as presented in Fig. 1A and B.
Two upregulated DEGs and seven downregulated DEGs in
the adenocarcinoma group, and eight upregulated DEGs and five
downregulated DEGs in the squamous cell carcinoma group were
selected in the global network under the FDR<0.1 and FC >1.5
or <0.67 criteria. These DEGs were used to construct the PPI
network based on the HRPD database; the network consisted of 21
DEGs and 517 relationships with two common genes, NKX3-1 and
MAPK10) in Fig. 2A and B. The
degrees (number of interacting partners) of the genes in the PPI
networks were calculated (Table
I), and the top five genes were identified, as follows:
HSP90AA1, BCL2, CDK2, KIT and HDAC2, which had degrees of 81, 64,
63, 50 and 46, respectively (Table
II). HSP90AA1 exhibited the highest degree, and was the most
downregulated gene detected in the adenocarcinoma group.
Furthermore, the expression differences of these 19 DEGs were
compared between the two types of lung cancer (Table II). The results demonstrated that
some genes exhibited similar expression trends between the groups,
such as NKX3-1 and MAPK10, whereas DVL3 was upregulated in squamous
cell carcinoma samples; however, its expression was not
significantly altered in adenocarcinoma samples. The expression of
BCL2 was significantly downregulated in adenocarcinoma samples, but
not changed in squamous cell carcinoma samples.
 | Table IDegrees of the top 10 genes in the
protein-protein interaction network. |
Table I
Degrees of the top 10 genes in the
protein-protein interaction network.
| Symbol | Degree |
|---|
| HSP90AA1 | 81 |
| BCL2 | 64 |
| CDK2 | 63 |
| KIT | 50 |
| HDAC2 | 46 |
| CDK4 | 36 |
| FGFR3 | 30 |
| TGFBR2 | 28 |
| MAP2K1 | 27 |
| RAD51 | 23 |
 | Table IIComparison of 19 differentially
expressed genes between the two types of lung cancer. |
Table II
Comparison of 19 differentially
expressed genes between the two types of lung cancer.
| Symbol | Gene ID | Adenocarcinoma
| Squamous cell
carcinoma
|
|---|
| Dir | FC | BH-P | Dir | FC | BH-P |
|---|
| FGFR3 | 2261 | −1 | 0.120 | 0.006 | −1 | 0.400 | 0.368 |
| EPAS1 | 2034 | −1 | 0.503 | 0.195 | −1 | 0.290 | 0.040 |
| KIT | 3815 | −1 | 0.591 | 0.565 | −1 | 0.310 | 0.012 |
| NKX3-1 | 4824 | −1 | 0.338 | 0.020 | −1 | 0.348 | 0.029 |
| TGFBR2 | 7048 | 1 | 1.210 | 0.769 | −1 | 0.435 | 0.093 |
| WNT4 | 54361 | −1 | 0.452 | 0.017 | −1 | 0.591 | 0.116 |
| HSP90AA1 | 3320 | −1 | 0.479 | 0.027 | −1 | 0.771 | 0.352 |
| MAPK10 | 5602 | −1 | 0.510 | 0.032 | −1 | 0.577 | 0.064 |
| BCL2 | 596 | −1 | 0.546 | 0.056 | −1 | 0.877 | 0.908 |
| HDAC2 | 3066 | 1 | 1.147 | 0.806 | 1 | 1.751 | 0.037 |
| CDK2 | 1017 | 1 | 1.310 | 0.153 | 1 | 1.809 | 0.056 |
| RAD51 | 5888 | 1 | 1.205 | 0.657 | 1 | 1.812 | 0.048 |
| MAP2K1 | 5604 | 1 | 1.451 | 0.288 | 1 | 1.840 | 0.060 |
| ITGA3 | 3675 | 1 | 2.120 | 0.049 | 1 | 1.008 | 0.981 |
| DVL3 | 1857 | −1 | 0.942 | 0.912 | 1 | 2.279 | 0.066 |
| CDK4 | 1019 | 1 | 1.644 | 0.236 | 1 | 2.799 | 0.004 |
| CKS1B | 1163 | 1 | 1.427 | 0.754 | 1 | 3.061 | 0.040 |
| DAPK1 | 1612 | 1 | 3.083 | 0.095 | 1 | 1.148 | 0.749 |
| FZD7 | 8324 | 1 | 1.325 | 0.792 | 1 | 4.668 | 0.068 |
Discussion
In the present study, DEGs in lung adenocarcinoma
and lung squamous cell carcinoma samples were screened by comparing
gene expression between the cancer samples and normal lung tissue
samples. The majority of DEGs were revealed to be downregulated.
Compared with lung adenocarcinoma, there were more DEGs in the lung
squamous cell carcinoma group. The disheveled homolog DVL3 was
upregulated in squamous cell carcinoma and was downregulated in
lung adenocarcinoma. Disheveled family proteins have been reported
to be associated with the development of cancer, as the cytoplasmic
mediators to activate the WNT/β-catenin signaling pathway (16). In addition, positive DVL3
expression has previously been detected in NSCLC samples (17). DVL3 has also been identified as a
candidate driver for genomic amplification of chromosome 3q26-29 in
lung squamous cell carcinoma of the lung. Knocking down of DVL3
protein can lead to cell growth inhibition (18) Therefore, DVL3 may be considered a
reliable biomarker to distinguish between these two types of lung
cancer. Furthermore, BCL2 was downregulated in lung adenocarcinoma;
however, it was not detected as a DEG in lung squamous cell
carcinoma. It has previously been reported that the BCL2 protein
acts as an inhibitor of apoptosis, and its family members, Bcl-2
and Bcl-xL, are overexpressed in numerous types of tumor cell,
including NSCLC (19). Targeted
inhibition of BCL2 overcomes the resistance of NSCLC cell lines to
chemotherapy (20). However,
recent meta-analysis studies suggest BCL2-negative expression is
associated with poor prognosis (21,22),
indicating its cancer inhibition roles, which was also confirmed by
analysis of lung adenocarcinoma samples in the present study.
A PPI network analysis demonstrated that the top
five genes were HSP90AA1, BCL2, CDK2, KIT and HDAC2, indicating
these genes may be important for lung cancer. These results were
consistent with those of previous studies although their roles
require further elucidation. The HSP90AA1 gene (generally referred
to as HSP90) is located on chromosome 14q32.2 (23). HSP90 is an emerging focus of cancer
therapy due to its ability to simultaneously inhibit several
signaling pathways (24). Sequist
et al (25) reported that a
potent inhibitor of HSP90, IPI-504, exerts clinical activity in
patients with NSCLC, particularly among patients with anaplastic
lymphoma kinase gene rearrangements. However, HSP90AA1 was observed
to be downregulated in the current study, indicating its underlying
tumor suppressor effect, which was also demonstrated by Li et
al (26) who identified the
expression of HSP90AA1 was lower in nasopharyngeal carcinoma
radioresistant cells than that in sensitive cells.
CDK2 is a member of the cyclin-dependent kinases,
and inhibition of CDK2 has been reported to induce anaphase
catastrophe and lead to apoptosis in NSCLC (27). As expected, CDK2 was also
upregulated in the lung cancer samples in the present study,
particularly in squamous cell carcinoma. Furthermore, Abrams et
al (28) evaluated the
activity of the indolinone kinase inhibitor SU11248 against the
receptor tyrosine kinase KIT in SCLC; the results suggested that
SU11248 may have clinical potential for the treatment of SCLC via
direct KIT-mediated antitumor activity. Nevertheless, KIT was
downregulated in squamous cell carcinoma in the present study,
which may be attributed to the lower malignancy in NSCLC compared
with SCLC. Histone deacetylases (HDACs) have been identified as
therapeutic targets due to their regulatory function on DNA
structure and organization (29).
HDAC inhibitors (HDACIs) represent a novel class of anticancer
agents. The HDACIs LBH589 (29),
trichostatin A (30) and
suberoylanilide hydroxamic acid (31) exert profound anti-growth activity
against lung cancer cells. In addition, the other DEGs identified
in the present study may be considered potential biomarkers for
lung cancer therapy.
In conclusion, the present study identifies various
crucial genes, including HSP90AA1, BCL2, CDK2, KIT and HDAC2, that
display different expression trends in lung adenocarcinoma and lung
squamous cell carcinoma via DEG analysis compared with normal lung
tissue and global network construction. These genes may be
potential biomarkers for differentiating these two types of lung
cancer in clinic.
References
|
1
|
Spruit MA, Janssen PP, Willemsen SC,
Hochstenbag MM and Wouters EF: Exercise capacity before and after
an 8-week multidisciplinary inpatient rehabilitation program in
lung cancer patients: A pilot study. Lung Cancer. 52:257–260. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Inamura K, Fujiwara T, Hoshida Y, Isagawa
T, Jones MH, Virtanen C, Shimane M, Satoh Y, Okumura S, Nakagawa K,
et al: Two subclasses of lung squamous cell carcinoma with
different gene expression profiles and prognosis identified by
hierarchical clustering and non-negative matrix factorization.
Oncogene. 24:7105–7113. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang S, Dai M, Ren JS, Chen YH and Guo
LW: Estimates and prediction on incidence, mortality and prevalence
of lung cancer in China in 2008. Zhonghua Liu Xing Bing Xue Za Zhi.
33:391–394. 2012.In Chinese. PubMed/NCBI
|
|
5
|
Li S, Huang S and Peng SB: Overexpression
of G protein-coupled receptors in cancer cells: Involvement in
tumor progression. Int J Oncol. 27:1329–1339. 2005.PubMed/NCBI
|
|
6
|
Raponi M, Zhang Y, Yu J, Chen G, Lee G,
Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y and Beer DG:
Gene expression signatures for predicting prognosis of squamous
cell and adenocarcinomas of the lung. Cancer Res. 66:7466–7472.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar
|
|
8
|
Navab R, Strumpf D, Bandarchi B, Zhu CQ,
Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L,
Barczyk M, et al: Prognostic gene-expression signature of
carcinoma-associated fibroblasts in non-small cell lung cancer.
Proc Natl Acad Sci USA. 108:7160–7165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsuta K, Tanabe Y, Yoshida A, Takahashi F,
Maeshima AM, Asamura H and Tsuda H: Utility of 10
immunohistochemical markers including novel markers (desmocollin-3,
glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis
of squamous cell carcinoma from adenocarcinoma of the Lung. J
Thorac Oncol. 6:1190–1199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hamamoto J, Soejima K, Yoda S, Naoki K,
Nakayama S, Satomi R, Terai H, Ikemura S, Sato T, Yasuda H, et al:
Identification of microRNAs differentially expressed between lung
squamous cell carcinoma and lung adenocarcinoma. Mol Med Rep.
8:456–462. 2013.PubMed/NCBI
|
|
11
|
Liu J, Yang XY and Shi WJ: Identifying
differentially expressed genes and pathways in two types of
non-small cell lung cancer: Adenocarcinoma and squamous cell
carcinoma. Genet Mol Res. 13:95–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haynes W: Benjamini-Hochberg method.
Encyclopedia of Systems Biology. Dubitzky W, Wolkenhauer O, Cho KH
and Yokota H: Springer; New York, NY: pp. 782013, View Article : Google Scholar
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peri S, Navarro JD, Amanchy R, Kristiansen
TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B,
Gandhi TK, Gronborg M, et al: Development of human protein
reference database as an initial platform for approaching systems
biology in humans. Genome Res. 13:2363–2371. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Semënov MV and Snyder M: Human dishevelled
genes constitute a DHR-containing multigene family. Genomics.
42:302–310. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei Q, Zhao Y, Yang ZQ, Dong QZ, Dong XJ,
Han Y, Zhao C and Wang EH: Dishevelled family proteins are
expressed in non-small cell lung cancer and function differentially
on tumor progression. Lung Cancer. 62:181–192. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Qian J, Hoeksema MD, Zou Y,
Espinosa AV, Rahman SM, Zhang B and Massion PP: Integrative
genomics analysis identifies candidate drivers at 3q26-29 amplicon
in squamous cell carcinoma of the lung. Clin Cancer Res.
19:5580–5590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Q, Yang S and Kang MQ: Influence of
survivin and Bcl-2 expression on the biological behavior of
non-small cell lung cancer. Mol Med Rep. 5:1409–1414.
2012.PubMed/NCBI
|
|
20
|
Han ZX, Wang HM, Jiang G, Du XP, Gao XY
and Pei DS: Overcoming paclitaxel resistance in lung cancer cells
via dual inhibition of stathmin and Bcl-2. Cancer Biother
Radiopharm. 28:398–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Wang S, Wang L, Wang R, Chen S,
Pan B, Sun Y and Chen H: Prognostic value of Bcl-2 expression in
patients with non-small-cell lung cancer: A meta-analysis and
systemic review. Onco Targets Ther. 8:3361–3369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao XD, He YY, Gao J, Zhao C, Zhang LL,
Tian JY and Chen HL: High expression of Bcl-2 protein predicts
favorable outcome in non-small cell lung cancer: Evidence from a
systematic review and meta-analysis. Asian Pac J Cancer Prev.
15:8861–8869. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gallegos Ruiz MI, Floor K, Roepman P,
Rodriguez JA, Meijer GA, Mooi WJ, Jassem E, Niklinski J, Muley T,
van Zandwijk N, et al: Integration of gene dosage and gene
expression in non-small cell lung cancer, identification of HSP90
as potential target. PLoS One. 3:e00017222008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Workman P, Burrows F, Neckers L and Rosen
N: Drugging the cancer chaperone HSP90: Combinatorial therapeutic
exploitation of oncogene addiction and tumor stress. Ann NY Acad
Sci. 1113:202–216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sequist LV, Gettinger S, Senzer NN,
Martins RG, Jänne PA, Lilenbaum R, Gray JE, Iafrate AJ, Katayama R,
Hafeez N, et al: Activity of IPI-504, a novel heat-shock protein 90
inhibitor, in patients with molecularly defined non-small-cell lung
cancer. J Clin Oncol. 28:4953–4960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y
and Liu Y: Genome-wide analyses of radioresistance-associated miRNA
expression profile in nasopharyngeal carcinoma using next
generation deep sequencing. PLoS One. 8:e844862013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu S, Danilov AV, Godek K, Orr B, Tafe LJ,
Rodriguez-Canales J, Behrens C, Mino B, Moran CA, Memoli VA, et al:
CDK2 inhibition causes anaphase catastrophe in lung cancer through
the centrosomal protein CP110. Cancer Res. 75:2029–2038. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abrams TJ, Lee LB, Murray LJ, Pryer NK and
Cherrington JM: SU11248 inhibits KIT and platelet-derived growth
factor receptor beta in preclinical models of human small cell lung
cancer. Mol Cancer Ther. 2:471–478. 2003.PubMed/NCBI
|
|
29
|
Geng L, Cuneo KC, Fu A, Tu T, Atadja PW
and Hallahan DE: Histone deacetylase (HDAC) inhibitor LBH589
increases duration of gamma-H2AX foci and confines HDAC4 to the
cytoplasm in irradiated non-small cell lung cancer. Cancer Res.
66:11298–11304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Platta CS, Greenblatt DY, Kunnimalaiyaan M
and Chen H: The HDAC inhibitor trichostatin A inhibits growth of
small cell lung cancer cells. J Surg Res. 142:219–226. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Komatsu N, Kawamata N, Takeuchi S, Yin D,
Chien W, Miller CW and Koeffler HP: SAHA, a HDAC inhibitor, has
profound anti-growth activity against non-small cell lung cancer
cells. Oncol Rep. 15:187–191. 2006.
|