Introduction
ADAM10 (a disintegrin and metalloprotease 10) is a
member of the ADAMs family, which contain two important structures,
a matrix metalloprotease (MMP) domain and disintegrin domain (DI).
The MMP domain is important for cleavage-dependent activation of
proteins, including various cell signaling molecules, such as Notch
receptors and ligands (1),
cadherins (epithelial (E)-cadherin and neural (N)-cadherin)
(2,3), epidermal growth factor (EGF)
(4) and the adhesion molecule L1
(5). ADAM10 may also act as an
α-secretase to cleave amyloid precursor protein and decrease
amyloid β protein production (6).
ADAM10 may also be important for the development of the nervous
system, where it regulates proliferation, migration,
differentiation and survival of various cells, including axonal
growth and myelination (7). It is
clear that ADAM10 is important for neural development, however, it
may also be important in certain nervous system diseases, including
Alzheimer's disease and inflammatory responses (7,8), the
spatial and temporal expression patterns of ADAM10 during
vertebrate brain development remain to be fully elucidated.
Previous studies have determined that the expression of ADAM10 mRNA
was restricted to specific brain regions, including developing
blood vessels, neuroepithelial regions and differentiating gray
matter (9,10); however, the exact cellular
localization of ADAM10 in the adult brains remains unclear.
The present study constructed an ADAM10
complementary RNA (cRNA) probe to investigate the expression
pattern of the Adam10 gene in the central nervous system
(CNS) of adult mice by in situ hybridization (ISH).
Immunohistochemical staining was used to identify the types of ISH
staining-positive cells with neuron or astrocyte-specific
antibodies. The results demonstrated that the expression of the
ADAM10 gene was predominantly in the neurons of the cerebral
cortex, hippocampus, thalamus and cerebellar granular cells in the
CNS of adult mice.
Materials and methods
Animals
A total of 10 healthy C57/BL6 mice (age, 10 weeks; 5
female, 5 male; weight, 250-300 g), were provided by the Animal
Experiment Center of Zhejiang University (Hangzhou, China). They
were randomly divided into a control group (a total of six mice,
three mice for mixed cultures of neurons and glial cells, three for
tissue sections) and experimental group (four mice, two mice were
used for mixed culturing of neuron and glial cells, and two for
tissue sections). The mice were housed at an ambient temperature of
37C with 50% humidity under a 12-h light/dark cycle. All mice had
access to food and water ad libitum. After 2 weeks, all the
mice were sacrificed via cervical dislocation, and the tissues were
immediately used for the experiments or stored at -80°C. The
current study was approved by the Animal Advisory Committee at
Zhejiang University. All the animals were treated in accordance
with national and institutional guidelines on the care of animals
in research.
In situ hybridization
To investigate the expression pattern of the
Adam10 gene in the CNS of adult mice, an ADAM10
complementary RNA probe was constructed. Cell type-specific
antibody immunohistochemical staining was applied to identify the
types of the ISH staining-positive cells.
Generation of Adam10 cRNA probes
The method for generation of the ADAM10 cRNA probe
was described previously with a few modifications (9,10).
More detailed protocols are described below.
RNA extraction and cDNA cloning
Total RNA from adult C57/BL6 mice brains was
prepared using the TRIzol reagent, according to the manufacturer's
protocol (cat. no. 15596-026, Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and a glass homogenizer, this was incubated
subsequently with DNase I for 1 h at 37°C to remove any
contaminating DNA. RNA was reverse transcribed to cDNA using a
M-MLV RTase cDNA Synthesis kit (cat. no. D6130, Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's protocol. The
polymerase chain reaction (PCR) amplification of ADAM10 cDNA was
performed using the KOD-Plus kit (Toyobo Co., Ltd., Osaka, Japan)
and the following primers, sense, 5′GGTGAAACGCATAAGAATC-3′ and
antisense, 5′CACTGAACTGCTTGCTCC-3′ under conditions as follows:
Denaturation at 94°C for 4 min; 35 cycles of denaturation at 94°C
for 30 sec, annealing at 56°C for 40 sec, and extension at 68°C for
90 sec; and extension at 68°C for 5 min. The amplified PCR
fragments were analyzed on agarose gels. The identity of the PCR
products was confirmed with restriction analysis. For restriction
reactions, the electrophoresed PCR products were purified from
agarose gels using QIAquick Gel Extraction kit (Qiagen GmbH,
Hilden, Germany) and cloned into pGEM-T Easy plasmid (Promega
Corporation, Madison, WI, USA) following the manufacturer's
protocol.
Probe synthesis
For probe synthesis, digoxigenin-labeled antisense
and sense cRNA probes were transcribed in vitro from the
purified pGEM-T Easy plasmids according to the manufacturer's
protocol (Promega Corporation). Sense cRNA probes served as
negative controls for in situ hybridization. Probes were
purified with a mini Quick Spin RNA Column (cat. no. 11814427001,
Roche Diagnostics GmbH) and were then precipitated by sodium
acetate. Incorporation of label and correct probe size was
identified by RNA formaldehyde denaturing gel electrophoresis and
blotting.
Combined in situ hybridization and
immunohistochemistry on cryosections
In situ hybridization was performed as
described previously with minor modifications (11,12).
Briefly, cryostat sections (thickness, 15 µm) were fixed
with 4% formaldehyde in phosphate-buffered saline (PBS), pretreated
with proteinase K and acetic anhydride and hybridized overnight at
65°C with cRNA probes at ~1-2 µg/ml in hybridization
solution (50% formamide, 5X saline sodium citrate, 1X Denhardt's
solution, 100 µg/ml herring sperm DNA (KPL, Inc.,
Gaithersburg, MD, USA), 300 µg/ml yeast tRNA and 5 mM EDTA).
Subsequently, the sections were washed and the uncombined cRNA was
removed by RNase. The sections were incubated with alkaline
phosphatase-coupled sheep anti-digoxigenin antibody (1:5,000; Roche
Diagnostics GmbH; cat. no. 11093274910) for 1 h at room
temperature. To visualize the labeled mRNA, a solution of
4-nitroblue tetrazolium chloride (NBT; Boehringer Ingelheim, Ltd.,
Ingelheim, Germany) and 5-bromo-4-chloro-3-indoyl-phosphate (BCIP;
Boehringer Ingelheim, Ltd.) was added.
Immunohistochemistry
Following in situ hybridization, the sections
were processed for immunohistochemistry following the
immunostaining protocol described by Tiveron et al (12), with a few modifications. Briefly,
following the blocking step with 10% bovine serum albumin (Beyotime
Institute of Biotechnology, Haimen, China) for 1 h at room
temperature, post-hybridized slides were incubated with anti-RNA
binding protein, fox-1 homolog 3 (NeuN) antibody (diluted 1:100 in
0.1 M PBS; EMD Millipore, Billerica, MA, USA; cat. no. MAB377) and
anti-glial fibrillary acidic protein (GFAP) antibody (diluted 1:200
in 0.1 M PBS; EMD Millipore; cat. no. AB5804) overnight at 4°C.
Subsequent to three washes for 10 min with 0.01 M PBS, the slides
were incubated with horseradish peroxidase (HRP)-conjugated rabbit
anti-mouse (1:1,000; Gene Tech, Shanghai, China; cat. no. GP016129)
and pig anti-rabbit (1:1,000; Gene Tech, Shanghai, China; cat. no.
GP021729) secondary antibodies for 1 h at room temperature and then
washed twice for 10 min in 0.01 M PBS. Color development was
performed by using the DAB kit (OriGene Technologies, Inc.,
Beijing, China) according to the manufacturer's protocol. ADAM10
mRNA abundance in each anatomical region was determined from
optical density measurements. The measurement was performed by use
of the Image-Pro Plus 6.0 (Media Cybernetics, Inc., Rockville, MD,
USA). The mean activity in the tissue fields was calculated. For
each field, at least five sections were measured and the mean value
was used. Density values for each parameter are presented according
to their respective percentile distributions: +++, >50% above
background, strong; ++, >25% above background, moderate; +,
<25% above the background, low but positive signal.
Image analysis
All sections were viewed and photographed under a
light microscope (Olympus BX40; Olympus Corporation, Hamburg,
Germany) equipped with a digital camera (Olympus DP70; Olympus
Corporation). The measurements were performed using Image-Pro Plus
6.0 software. Photomicrographs were adjusted in contrast and
brightness using Adobe Photoshop (Adobe Systems, Inc., San Jose,
CA, USA) for optimal display of the staining patterns in the
figures.
Results
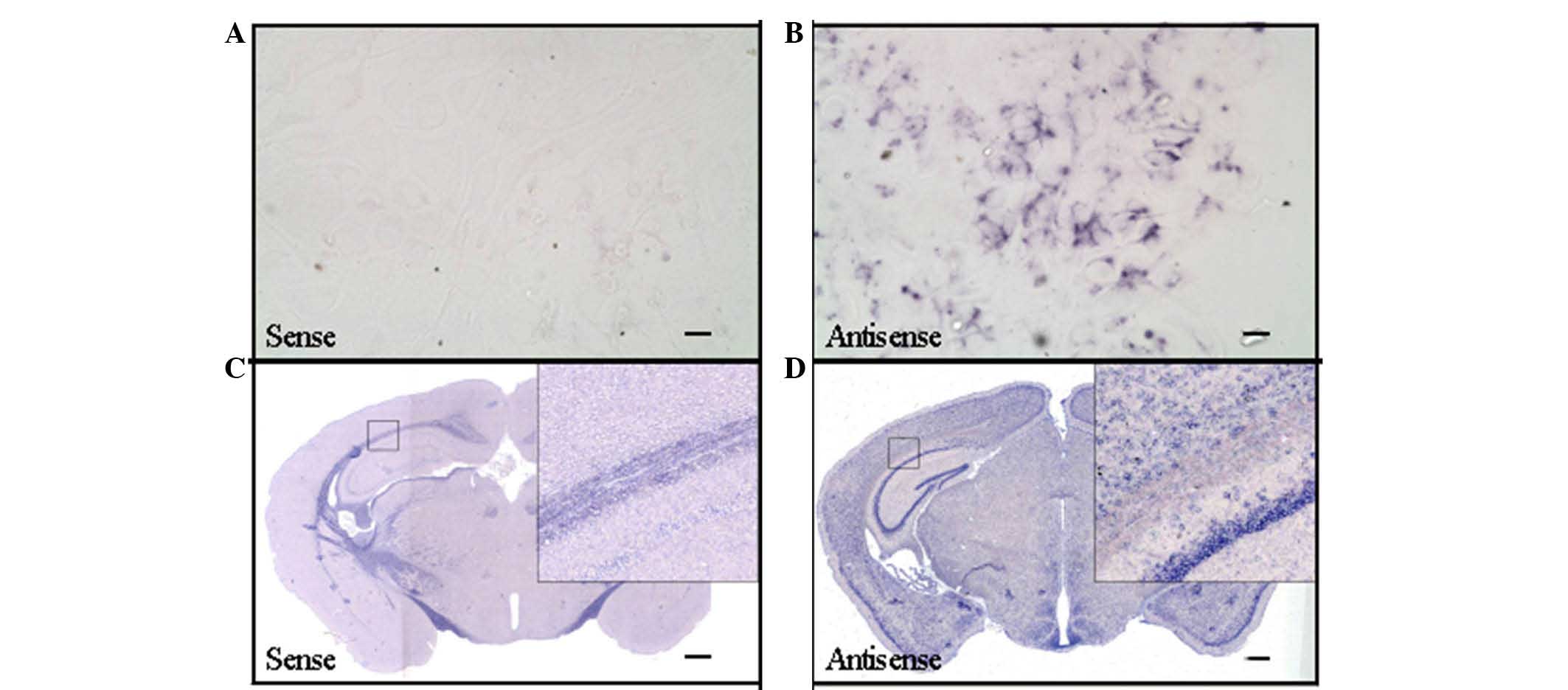
The effectiveness of the ADAM10 cRNA
probe was indicated by detection of immunoreactivity with the
antisense probe
To detect the effectiveness of the ADAM10 cRNA probe
for in situ hybridization, T7 RNA transcriptase and SP6 RNA
transcriptase were used in in vitro transcription to obtain
the antisense and sense cRNA probes, respectively. Sense cRNA
probes served as a negative control for in situ
hybridization. The in situ hybridization results presented
in Fig. 1 indicated that
hybridization signal may be detected only in the antisense ADAM10
cRNA probe slices using mixed cultures of neurons and glial cells
and C57BL6 mouse brain slices (Fig. 1B
and D). The immunoreactivity was not be detected by use of the
sense ADAM10 cRNA probe (Fig. 1A and
C).
ADAM10 mRNA expression is distributed in
selected regions of the adult mouse brain
In situ hybridization determined that ADAM10
mRNA distribution is present in selected regions of the adult mouse
brain (Table I). ADAM10 mRNA was
markedly expressed throughout the telencephalon, including the
parietal and piriform cortex, the hippocampus (CA1-CA3 and the
dentate gyrus; Fig. 2). Within the
diencephalon, the hybridization signal was moderate, particularly
in the septal nucleus (Fig. 2A),
the thalamus and hypothalamus (dorsomedial hypothalamic nucleus;
Fig. 2B and C) and surrounding
areas of the third ventricle (Fig. 2C
and D). In the striatum, a weaker hybridization signal for
ADAM10 was also detected (Fig. 2A and
B).
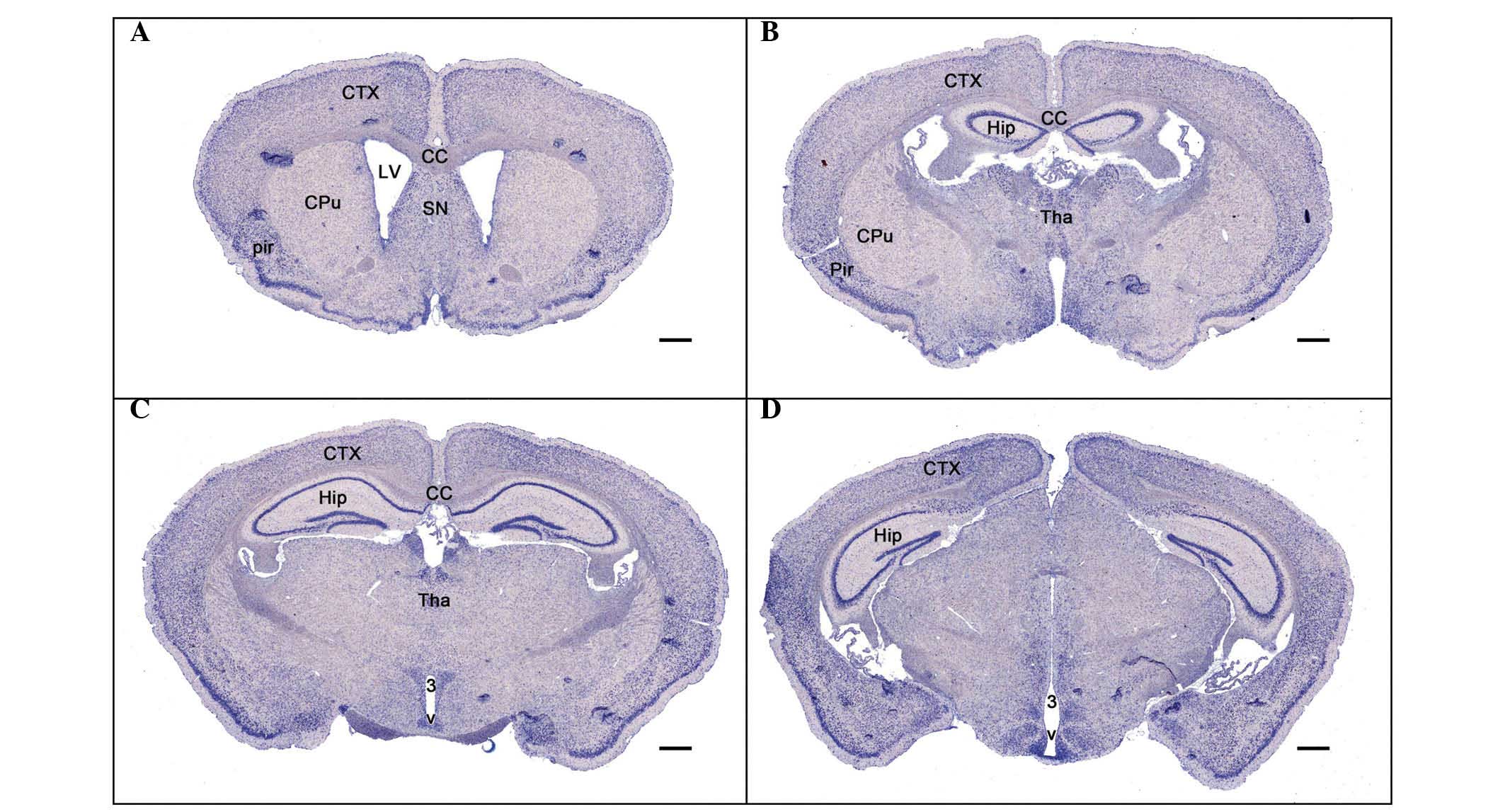 | Figure 2Distribution of Adam10 mRNA expression
in the adult mouse brain. (A–D) Adam10 cRNA probe in situ
hybridization results in cerebral coronal sections from (A) rostral
to (D) cauda1. Scale bar, 500 µm. CTX, cortex; Pir, piriform
cortex; CPu, caudate putamen; LV, lateral ventricle; SN, septal
nucleus; CC, corpus callosum; Hip, hippocampus; Tha, thalamus; 3 V,
the third ventricle; Adam10, a disintegrin and metallopeptidase
domain 10. |
 | Table IDistribution of Adam10 mRNA expression
in the adult mouse brain using in situ hybridization. |
Table I
Distribution of Adam10 mRNA expression
in the adult mouse brain using in situ hybridization.
| Brain region | Adam10 mRNA
abundance |
|---|
| Piriform cortex | +++ |
| Frontoparietal
cortex | +++ |
| Septal nucleus | ++ |
| Hippocampus | |
| CA1-CA3 | +++ |
| Dentate gyrus | +++ |
| Thalamus | ++ |
| Hypothalamus | ++ |
| Striatum | + |
| Cerebellum | |
| Molecular layer | ++ |
| Purkinje cell
layer | +++ |
| Internal granular
cell layer | +++ |
| White matter | ++ |
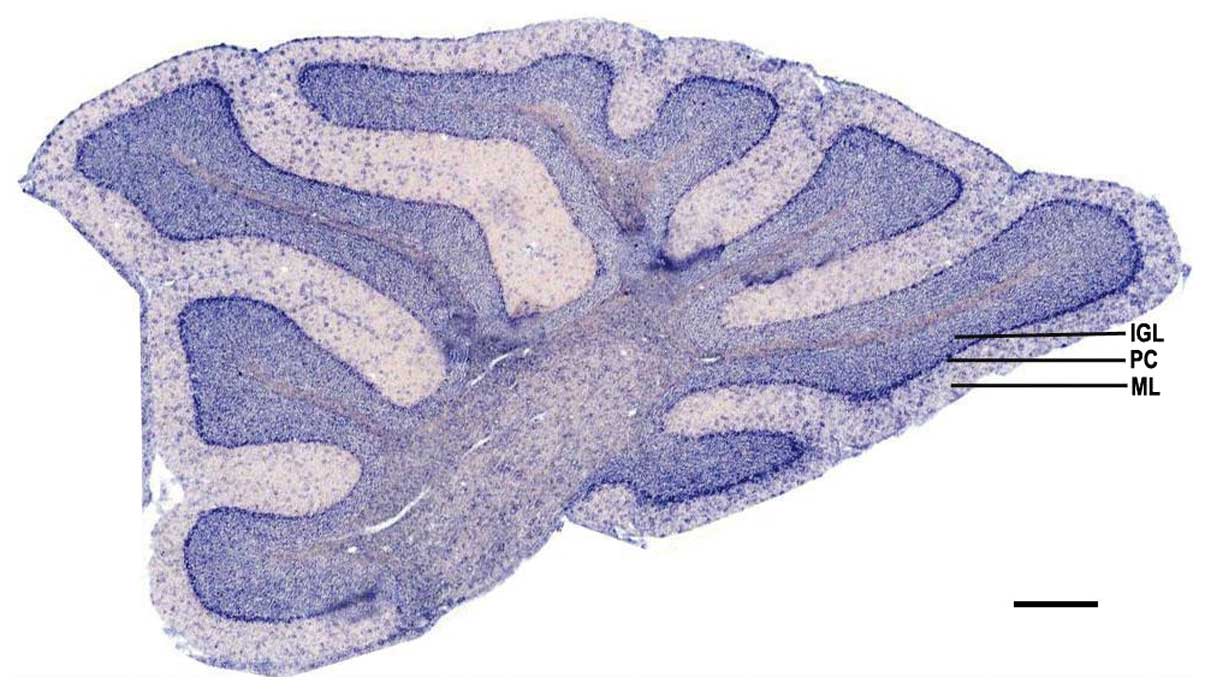
ADAM10 mRNA expression is distributed in
selected regions of the cerebellum of adult mice
To detect the expression of ADAM10 mRNA in the
cerebellum, cerebellar sagittal sections were used for in
situ hybridization. The hybridization results indicated that
the positive signals were predominantly distributed in the internal
granular cell layer and purkinje cell layer. There were also
scattered positive cells distributed in the molecular layer
(Fig. 3).
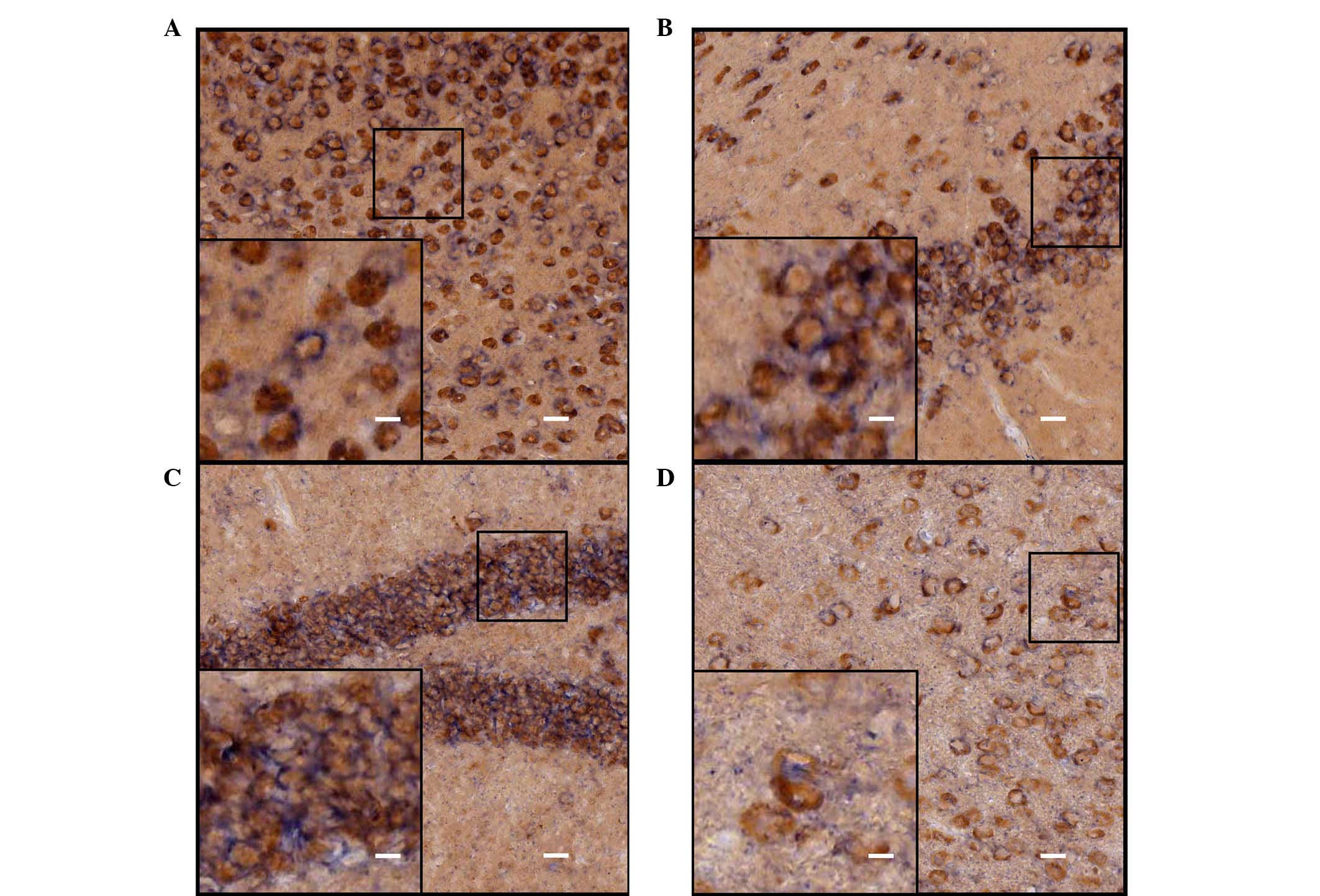
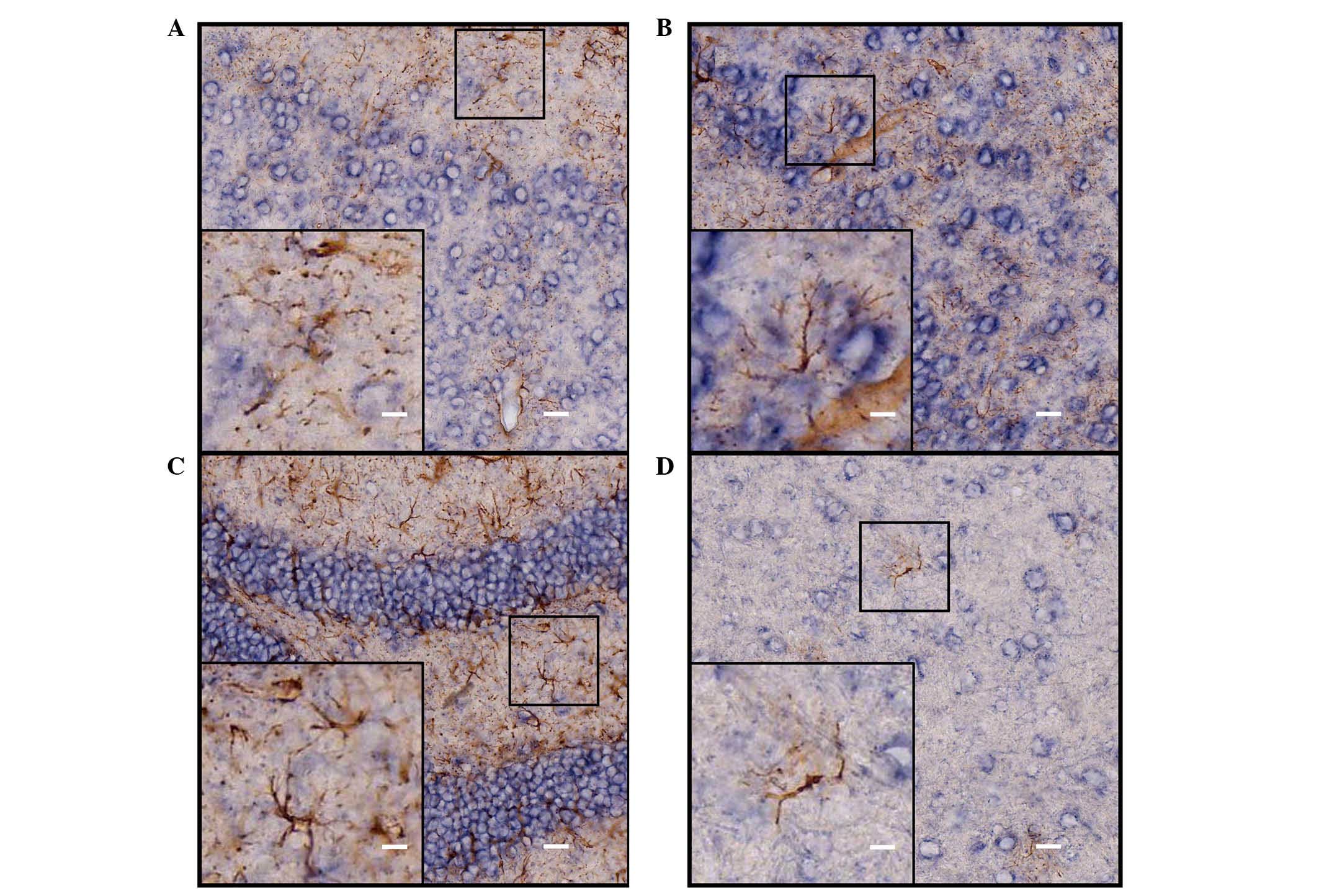
Identification of the ISH-positive cells
in the cerebrum of adult mice
Neurons and astrocytes are the basic components of
the CNS, and each performs important functions. In order to further
identify cell locations of ADAM10 in neurons or astrocytes,
immunohistochemical staining with different cell-specific
antibodies (astroglial marker, GFAP and the neuronal marker, NeuN)
were used to determine the types of the ISH-positive cells. The
hybridization signal was developed by alkaline phosphatase and
NBT/BCIP staining, and the positive cells were a blue-purple color
following staining. HRP-labeled chromogenic reagents were used for
immunohistochemistry, and the positive cells were stained brown.
Results from the double staining indicated that the majority of the
ADAM10 ISH-positive cells coexpressed NeuN (Fig. 4), and that a number of cells were
not NeuN immunoreactive (Fig. 4D);
however, these cells were also negative for GFAP staining (Fig. 5) in various selected regions of
adult mouse brain, including the parietal cortex, piriform cortex,
hippocampal dentate gyrus and thalamus.
Discussion
In the present study, an ADAM10 cRNA probe for in
situ hybridization was constructed to investigate the
expression pattern of the ADAM10 gene in the CNS of adult mice.
Immunohistochemical staining was used to identify the type of cell
that was ISH staining-positive. The results demonstrated that
expression of the ADAM10 gene was restricted to neurons of the
cerebral cortex, hippocampus, thalamus and cerebellar granular
cells in the CNS of adult mice.
ADAM10, termed Kuzbanian in Drosophila
(13) is essential to embryonic
development and control of neurogenesis and axon extension in the
CNS (14–16). ADAM10 has been confirmed as a
candidate α-secretase responsible for cleaving various proteins,
including APP, heparin-binding EGF, EGF receptor, E-cadherin,
N-cadherin, protocadherin C3 and vascular endothelial-cadherin
(2–4,17–20).
ADAM10-deficient mice developed only to embryonic day (E)9.5 with
multiple defects in the CNS, somites and the cardiovascular system
(1). A previous study determined
that conditional knock-out of ADAM10 in neural progenitor cells
(NPCs), NPC-derived neurons and glial cells in mice, leads to
perinatal mortality with a disrupted neocortex and a markedly
reduced ganglionic eminence (21).
ADAM10 gene knockout results in abnormalities of the cardiovascular
system and CNS, suggesting that the ADAM10 gene is important for
the development of the CNS. However, its specific roles and precise
underlying molecular mechanisms remain to be further
elucidated.
Previous studies have demonstrated that the
expression of ADAM10 in the brain of adult mice was restricted to
specific areas. Within the telencephalon and diencephalon, the
expression of ADAM10 mRNA was more widespread. In the
mesencephalon, ADAM10 mRNAs was expressed in the inferior
colliculus. The highest expression was detected in the cerebral
cortex (9). These results are
similar to the findings of the present study, that ADAM10 mRNA was
distributed in specific regions of adult mouse brain, including
notable expression in the parietal and piriform cortex, the
hippocampus and the cerebral cortex. Moderate expression was also
observed in the septal nucleus, the thalamus, hypothalamus and
surrounding areas of the third ventricle. The present study also
determined that a weaker hybridization signal for ADAM10 may be
detected in the striatum. In a previous study, Lin et al
(10) indicated that ADAM10 was
predominantly expressed by developing blood vessels, restricted
neuroepithelial regions and in differentiating gray matter. In
addition, ADAM10 was observed to be expressed by oligodendrocytes
at later embryonic stages in numerous fiber tracts (10). The present study aimed to detect
the ADAM10 expression in neurons and astrocytes in the CNS of adult
mice; thus, it was not determined whether blood vessels expressed
ADAM10. However, based on previous studies that identified strong
ADAM10 expression in blood vessels persisting at E10 and E12, prior
to decreasing at later stages and is no longer detectable in brain
at E19 (10), the present study
hypothesizes that the mature blood vessels would not express ADAM10
in the CNS of adult mice. In addition, the ADAM10 expression in
oligodendrocytes was not investigated in the current study, as
there is increasing evidence suggesting that oligodendrocytes
express ADAM10 in the developing and adult brain (10,22).
In conclusion, the present study is consistent with
previous results by other groups (9,10)
that demonstrate ADAM10 is expressed in a number of restricted
regions of the brain of adult mice. To the best of our knowledge,
this is the first study to demonstrate that the ADAM10 is expressed
by neurons in the brain of adult mice. These results may provide
the basis for future investigations at the experimental level using
Cre/Loxp conditional knockout technology (23).
Acknowledgments
The present study was supported by the National
Youth Fund (grant no. 81400931) and the Natural Foundation of
Beijing (grant no. 7153715). The authors would like to thank
Professor Duan Shumin (Medical College of Zhejiang University,
Zhejiang, China) for providing technical support.
Abbreviations:
|
Par
|
parietal cortex
|
|
Pir
|
piriform cortex
|
|
Hip
|
hippocampus
|
|
Tha
|
thalamus
|
|
DG
|
dentate gyrus
|
|
IGL
|
granule cell layer
|
|
ML
|
molecular layer
|
|
PC
|
purkinje cell layer
|
|
GFAP
|
glial fibrillary acidic protein
|
References
|
1
|
Hartmann D, de Strooper B, Serneels L,
Craessaerts K, Herreman A, Annaert W, Umans L, Lübke T, Lena Illert
A, von Figura K and Saftig P: The disintegrin/metalloprotease ADAM
10 is essential for Notch signalling but not for alpha-secretase
activity in fibroblasts. Hum Mol Genet. 11:2615–2624. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reiss K, Maretzky T, Ludwig A, Tousseyn T,
de Strooper B, Hartmann D and Saftig P: ADAM10 cleavage of
N-cadherin and regulation of cell-cell adhesion and beta-catenin
nuclear signalling. EMBO J. 24:742–752. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maretzky T, Reiss K, Ludwig A, Buchholz J,
Scholz F, Proksch E, de Strooper B, Hartmann D and Saftig P: ADAM10
mediates E-cadherin shedding and regulates epithelial cell-cell
adhesion, migration, and beta-catenin translocation. Proc Natl Acad
Sci USA. 102:9182–9187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yan Y, Shirakabe K and Werb Z: The
metalloprotease Kuzbanian (ADAM10) mediates the transactivation of
EGF receptor by G protein-coupled receptors. J Cell Biol.
158:221–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gutwein P, Mechtersheimer S, Riedle S,
Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M and Altevogt DP:
ADAM10-mediated cleavage of L1 adhesion molecule at the cell
surface and in released membrane vesicles. FASEB J. 17:292–294.
2003.
|
|
6
|
Lammich S, Kojro E, Postina R, Gilbert S,
Pfeiffer R, Jasionowski M, Haass C and Fahrenholz F: Constitutive
and regulated alpha-secretase cleavage of Alzheimer's amyloid
precursor protein by a disintegrin metalloprotease. Proc Natl Acad
Sci USA. 96:3922–3927. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang P, Baker KA and Hagg T: The ADAMs
family: Coordinators of nervous system development, plasticity and
repair. Prog Neurobiol. 79:73–94. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pruessmeyer J and Ludwig A: The good, the
bad and the ugly substrates for ADAM10 and ADAM17 in brain
pathology, inflammation and cancer. Semin Cell Dev Biol.
20:164–174. 2009. View Article : Google Scholar
|
|
9
|
Kärkkäinen I, Rybnikova E, Pelto-Huikko M
and Huovila AP: Metalloprotease-disintegrin (ADAM) genes are widely
and differentially expressed in the adult CNS. Mol Cell Neurosci.
15:547–560. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin J, Luo J and Redies C: Different
expression of five members of the ADAM family in the developing
chicken brain. Neuroscience. 157:360–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaeren-Wiemers N and Gerfin-Moser A: A
single protocol to detect transcripts of various types and
expression levels in neural tissue and cultured cells: In situ
hybridization using digoxigenin-labelled cRNA probes.
Histochemistry. 100:431–440. 1993. View Article : Google Scholar
|
|
12
|
Tiveron MC, Hirsch MR and Brunet JF: The
expression pattern of the transcription factor Phox2 delineates
synaptic pathways of the autonomic nervous system. J Neurosci.
16:7649–7660. 1996.PubMed/NCBI
|
|
13
|
Rooke J, Pan D, Xu T and Rubin GM: KUZ, a
conserved metalloprotease-disintegrin protein with two roles in
Drosophila neurogenesis. Science. 273:1227–1231. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fambrough D, Pan D, Rubin GM and Goodman
CS: The cell surface metalloprotease/disintegrin Kuzbanian is
required for axonal extension in Drosophila. Proc Natl Acad Sci
USA. 93:13233–13238. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan D and Rubin GM: Kuzbanian controls
proteolytic processing of Notch and mediates lateral inhibition
during Drosophila and vertebrate neurogenesis. Cell. 90:271–280.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen YY, Hehr CL, Atkinson-Leadbeater K,
Hocking JC and McFarlane S: Targeting of retinal axons requires the
metalloproteinase ADAM10. J Neurosci. 27:8448–8456. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sahin U, Weskamp G, Kelly K, Zhou HM,
Higashiyama S, Peschon J, Hartmann D, Saftig P and Blobel CP:
Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six
EGFR ligands. J Cell Biol. 164:769–779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schulz B, Pruessmeyer J, Maretzky T,
Ludwig A, Blobel CP, Saftig P and Reiss K: ADAM10 regulates
endothelial permeability and T-cell transmigration by proteolysis
of vascular endothelial cadherin. Circ Res. 102:1192–1201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maretzky T, Scholz F, Köten B, Proksch E,
Saftig P and Reiss K: ADAM10-mediated E-cadherin release is
regulated by proin-flammatory cytokines and modulates keratinocyte
cohesion in eczematous dermatitis. J Invest Dermatol.
128:1737–1746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reiss K, Maretzky T, Haas IG, Schulte M,
Ludwig A, Frank M and Saftig P: Regulated ADAM10-dependent
ectodomain shedding of gamma-protocadherin C3 modulates cell-cell
adhesion. J Biol Chem. 281:21735–21744. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jorissen E, Prox J, Bernreuther C, Weber
S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A,
Tesseur I, et al: The disintegrin/metalloproteinase ADAM10 is
essential for the establishment of the brain cortex. J Neurosci.
30:4833–4844. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jangouk P, Dehmel T, Meyer Zu Hörste G,
Ludwig A, Lehmann HC and Kieseier BC: Involvement of ADAM10 in
axonal outgrowth and myelination of the peripheral nerve. Glia.
57:1765–1774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsien JZ, Chen DF, Gerber D, Tom C, Mercer
EH, Anderson DJ, Mayford M, Kandel ER and Tonegawa S: Subregion-
and cell type-restricted gene knockout in mouse brain. Cell.
87:1317–1326. 1996. View Article : Google Scholar : PubMed/NCBI
|