Introduction
Glioblastoma multiforme (GBM), which accounts for
~40% of all primary brain tumors, is one of the most malignant
tumors in humans, with patients having a median overall survival
time of 12–15 months (1). Previous
studies have demonstrated that tumor cell populations in GBM are
heterogeneous in terms of morphology and differentiation status
(2). A highly tumorigenic and
self-renewing subpopulation of cells, which display stem-like
behavior, is the glioblastoma stem cells (GSCs) (2,3).
Notably, the property of 'stemness' in GSCs is believed to be key
for tumor formation, differentiation, proliferation and resistance
to chemo- and radiotherapy, possibly explaining the high frequency
of treatment failure and tumor relapse observed in glioblastoma
(4,5). The differentiation of GSCs may lead
to the inhibition of their self-renewing ability and tumorigenic
potential, as well as increasing their sensitivity to treatment.
The promotion of differentiation is considered to be a potential
strategy to eradicate GSCs, and therefore it is important to
understand the detailed molecular mechanisms involved in GSC
differentiation (6,7).
Advances in next-generation deep sequencing
technologies have identified a large number of non-coding RNAs
(ncRNAs) termed long non-coding RNAs (lncRNAs). These molecules are
>200 nucleotides in length and are a class of single-stranded
RNAs, which lack protein-coding ability (8,9).
Rather than being irrelevant transcriptional noise, studies have
revealed that lncRNAs possess critical regulatory roles in numerous
biological processes, including immune responses, cellular
metabolism, stem cell differentiation and tumorigenesis (10,11).
Furthermore, certain studies have demonstrated that lncRNA
signatures correlate with glioma malignancy grade, histological
differentiation and prognosis (12,13).
The diverse mechanisms underlying the regulatory roles of lncRNAs
include interactions with mRNAs, functioning as miRNA sponges and
acting as tethers, guides, decoys and scaffolds for proteins
(14,15).
Numerous studies of GSC differentiation have
revealed certain underlying molecular mechanisms involved in this
process. Certain transcription factors (TFs), protein-coding genes
and ncRNAs have been proposed to be involved in the maintenance of
GSC stemness, and the regulation of these factors may facilitate
GSC differentiation (16–18). However, the role of lncRNAs in the
GSC differentiation process remains to be elucidated.
In the present study, using patient-derived GSC
lines and their differentiation-induced GSC (DGSC) counterparts, a
high-throughput microarray analysis was performed to identify the
profile changes of lncRNAs and mRNAs. Using bioinformatics
analysis, aberrant lncRNA and mRNA expression was integrated and
potential regulatory roles of lncRNAs in GSC differentiation were
proposed.
Materials and methods
GSC culture and differentiation
GSCs (G0, G1, G2, G3, G4 and G5) were isolated from
neurosurgical samples from six patients with GBM who were
hospitalized in the Department of Neurosurgery of Beijing Tiantan
Hospital (Beijing, China). Informed consent was obtained from the
patients and approval was given from the Ethics Committee of
Beijing Tiantan Hospital (KY2014-021-02; Beijing, China). Tumor
tissue was sectioned into 1-mm3 fragments using scissors
and washed with Dulbecco's modified Eagle's medium/nutrient mixture
F12 (DMEM/F12; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) three times. The fragments were then digested with 0.02%
trypsin for 20 min and dissociated into single cells. The cells
were resuspended and maintained in GSC-propagating medium composed
of 2 mM GlutaMAX (Invitrogen; Thermo Fisher Scientific, Inc.), 20
ng/ml recombinant human epidermal growth factor (R&D Systems,
Inc., Minneapolis, MN, USA), 20 ng/ml basic fibroblast growth
factor (Invitrogen; Thermo Fisher Scientific, Inc.), N2 supplement
(Invitrogen; Thermo Fisher Scientific, Inc.) and B27 supplement
(Invitrogen; Thermo Fisher Scientific, Inc.) in DMEM/F12. The above
procedure was completed within 1 h of the surgical removal of
tissue. GSCs (2×104/mouse; 6 mice) were subcutaneously
injected into the left hind flank of 6-week-old female non-obese
diabetic/severe combined immunodeficiency (NOD/SCID) mice
(VitalStar Biotechnology Co., Ltd., Beijing, China). The mice were
sacrificed by decapitation 40 days later, and tumors were harvested
and stained with hematoxylin and eosin. The animal use was approved
by the Institutional Animal Care and Use Committee of Capital
Medical University. All animal procedures were performed in
accordance with the 1996 Guide for the Care and Use of Laboratory
Animals (19). The rats were
housed in an air-conditioned room with a constant temperature of
22°C and a 12 h light/12 h dark cycle. The humidity in the housing
room was 50–60%. The rats had free access to food and water. GSCs
were sustained in GSC-propagating medium and it took ~18 h for them
to undergo symmetric divisions. To induce GSC differentiation,
following 5 passages which took about half a month in total
following the resuscitation in GSC-propagating medium, GSCs were
resuspended and cultured in DMEM/F12 medium containing 10% fetal
bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) without
N2 and B27 supplements for 4 days, as previously described
(16,20).
Immunofluorescence
GSCs grown as neurospheres were collected by
centrifugation at 150 × g for three min at room temperature, then
the cells were transferred to microscope slides, while the DGSCs
were grown and stained in 24-well plates. The cells were fixed with
4% formaldehyde, permeabilized with 0.1% Triton X-100 for 10 min
and then blocked with 10% normal goat serum (Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) for 5 min. The cells were
then incubated at 4°C overnight with the following primary
antibodies: Mouse anti-cluster of differentiation (CD) 133
(dilution 1:500; catalog no., MAB4399; EMD Millipore, Billerica,
MA, USA); rabbit anti-nestin (dilution 1:250; catalog no., ab82375;
Abcam, Cambridge, UK); rabbit anti-glial fibrillary acidic protein
(GFAP; dilution 1:500; catalog no., ab33922; Abcam); rabbit
anti-βIII tubulin (Tuj1; dilution 1:500; catalog no., ab18207;
Abcam); mouse anti-O4 (dilution 1:200; catalog no., MAB345; EMD
Millipore). Following washing with phosphate-buffered saline 3
times, the cells were incubated with fluorescein isothiocyanate
(FITC)-conjugated anti-mouse (dilution 1:1,000, catalog no.,
F-11,021; Thermo Fisher Scientific, Inc.) and anti-rabbit (dilution
1:2,000; catalog no., 65-6111; Thermo Fisher Scientific, Inc.)
secondary antibodies and Texas Red (TR) -conjugated anti-mouse
(dilution 1:2,000; catalog no., T-6390; Thermo Fisher Scientific,
Inc.) and anti-rabbit (dilution 1:1,000 catalog no., T-2767; Thermo
Fisher Scientific, Inc.) secondary antibodies. The cell nuclei were
counterstained with DAPI (Sigma-Aldrich, St. Louis, MO, USA).
Images were captured using a DMI 4,000 Leica fluorescent microscope
and processed using the Leica Application Suite version 4.2 Imaging
System software (Leica Microsystems GmbH, Wetzlar, Germany).
RNA extraction and microarray
analysis
Total RNA was extracted from GSCs and DGSCs using
TRIzol® reagent (Sigma-Aldrich) according to the
manufacturer's instructions. RNA purity and concentration was
assessed using a NanoDrop ND-1,000 spectrophotometer (Thermo Fisher
Scientific, Inc.). RNA integrity was determined by formaldehyde
denaturing gel electrophoresis. Microarray hybridization was
performed by CapitalBio Corporation (Beijing, China). Briefly, each
RNA sample was amplified and transcribed into double-stranded
complementary DNA (cDNA). The labeled cDNA was then hybridized to
the lncRNA+mRNA Human Gene Expression Microarray version 4.0,
4×180K chip. Data normalization, quality control and the
calculation of differences in gene expression were performed with
the GeneSpring GX version 11.5.1 software package (Agilent
Technologies, Inc., Santa Clara, CA, USA). lncRNAs and mRNAs were
defined as differentially expressed (DE) if the fold-change values
were >2.0, or if P<0.05. The microarray data was uploaded to
the Gene Expression Omnibus (GEO) database (GSE68343; www.ncbi.nlm.nih.gov/geo/). In addition,
hierarchical clustering with average linkage was used for
calculating the distinguishable lncRNA and mRNA expression patterns
with Cluster version 3.0 software (bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv).
Co-expression network construction and
lncRNA target prediction
The lncRNA-mRNA co-expression network was
constructed based on associations between DE lncRNAs and DE mRNAs
during GSC differentiation. Using the open source bioinformatics
software Cytoscape (Institute of Systems Biology, Seattle, WA,
USA), a network of the selected lncRNA-mRNA pairs was drawn
(Pearson correlation coefficient >0.99 or <−0.99; and
P<0.05). Subsequently, the lncRNA-mRNA co-expression network was
analyzed using a cis- or trans-regulatory prediction
program. A target gene was defined as cis if the mRNA
transcribed was within a 10 kilo-base (kb) window upstream or
downstream of the lncRNA genomic location, using gene annotations
from the University of California, Santa Cruz (Santa Cruz, CA, USA;
genome.ucsc.edu/). As lncRNAs may act as
competing endogenous RNAs, which regulate the mRNA transcripts by
competing for shared microRNAs, the trans- predictions were
primarily executed by searching the pairs of lncRNAs and mRNAs that
had similar sequences in the 3′ untranslated region using the
BLAST-Like Alignment Tool (default parameter settings) software
(genome.ucsc.edu/cgi-bin/hgBlat).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation
Total RNA was reverse transcribed to cDNA using the
PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan). Using a
7500 Real Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with SYBR® Premix Ex Taq™ II (Tli
RNaseH Plus; Takara Bio, Inc.), RT-qPCR was performed according to
the manufacturer's instructions. The specific primers for each gene
are presented in Table I. All
experiments were performed in triplicate. Gene expression was
normalized to GAPDH in each sample.
 | Table IReverse transcription-quantitative
polymerase chain reaction primers. |
Table I
Reverse transcription-quantitative
polymerase chain reaction primers.
| Name | Primer sequence
|
|---|
| Forward | Reverse |
|---|
|
ENSG00000235427.1 |
5′-AAAACCACTGAGACACGGAGGC-3′ |
5′-CCAGGGACAGGCAGACATCA-3′ |
| TAX1BP3 |
5′-CCTACATCCCGGGCCAGC-3′ | 5′-
CACCTCCAATGCTGAAACCCA-3′ |
| CAV1 |
5′-CGACCCTAAACACCTCAACGA-3′ | 5′-
GGCAGACAGCAAGCGGTAAAA-3′ |
| RPTOR |
5′-GTGGTGGACTGGGAGCAGGAGA-3′ |
5′-TGAGCGGTGGGAATCACAGGA-3′ |
|
ENSG00000261924.1 |
5′-AGGAATGACATGAACACGAGGGAA-3′ |
5′-CCAGGGCGATATGTGGAGCAA-3′ |
| P2RX5-TAX1BP3 |
5′-CGAGGCGAAGCGTGGAA-3′ |
5′-TGGTGTAAGGGAGAAGCAGAGG-3′ |
| CD133 |
5′-TACCAAGGACAAGGCGTTCACAGA-3′ |
5′-GTGCAAGCTCTTCAAGGTGCTG-3′ |
| Nestin |
5′-CGTTGGAACAGAGGTTGGAG-3′ |
5′-TAAGAAAGGCTGGCACAGGT-3′ |
| Tuj1 |
5′-GTACGAAGACGACGAGGAGG-3′ |
5′-GCCTGGAGCTGCAATAAGAC-3′ |
| GFAP |
5′-GTCCATGTGGAGCTTGACG-3′ |
5′-GCAGGTCAAGGACTGCAACT-3′ |
| SOX2 |
5′-ATGCACAACTCGGAGATCAG-3′ |
5′-TATAATCCGGGTGCTCCTTC-3′ |
| GAPDH |
5′-CTATAAATTGAGCCCGCAGCC-3′ |
5′-GCGCCCAATACGACCAAATC-3′ |
Analysis of TF binding sites and lncRNA
conservation
Genomic annotations of human lncRNA genes and
transcripts were downloaded from the GENCODE website (www.gencodegenes.org/). Peak lists of chromatin
immunoprecipitation sequencing (ChIP-Seq) datasets performed on 4
TFs [POU domain, class 3, transcription factor (POU3F), sex
determining region Y-box 2 (SOX2), spalt-like transcription factor
2 (SALL2) and oligodendrocyte lineage transcription factor (OLIG)
2] were available from the deep sequencing of ChIP-Seq libraries in
GSCs (21). Subsequently, the
peaks of the TFs were overlapped with the regulatory regions of the
lncRNA genes (5 kb upstream from the transcription start site) and
the gene body of the lncRNA using the InterSectBed tool from
Bedtools (bedtools.readthedocs.io/en/latest/). If the regulatory
regions or the gene body of an lncRNA gene had at least one TF
peak, the lncRNA gene was determined as having the potential to be
regulated by the TF. Conservation analysis was conducted using
phastCons data (compgen.cshl.edu/phast/) as described previously
(22). If the average phastCons
score was >0.1, the lncRNA was defined as conserved.
Statistical analysis
Statistical analyses were performed using SPSS
software, version 13.0 (SPSS, Inc., Chicago, IL, USA). Values are
presented as the mean ± standard deviation, and Student's
t-test was used for comparing sample sets. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characterization of GSCs isolated from
surgical GBM samples and determination of GSC differentiation
GSCs were cultured in GSC-propagating media, where
they formed characteristic renewable neurospheres and were able to
proliferate indefinitely (Fig.
1Aa). Immunofluorescence revealed that GSCs exhibited an
increased proportion of cells positive for CD133 (Fig. 1Ab), and increased expression of
nestin, a neural progenitor cell marker (Fig. 1Ac), compared with DGSCs (Fig. 1Ad and e). The tumorigenesis of GSCs
was confirmed by tumor formation in NOD/SCID mice following
inoculation with 2×104 GSCs (data not shown).
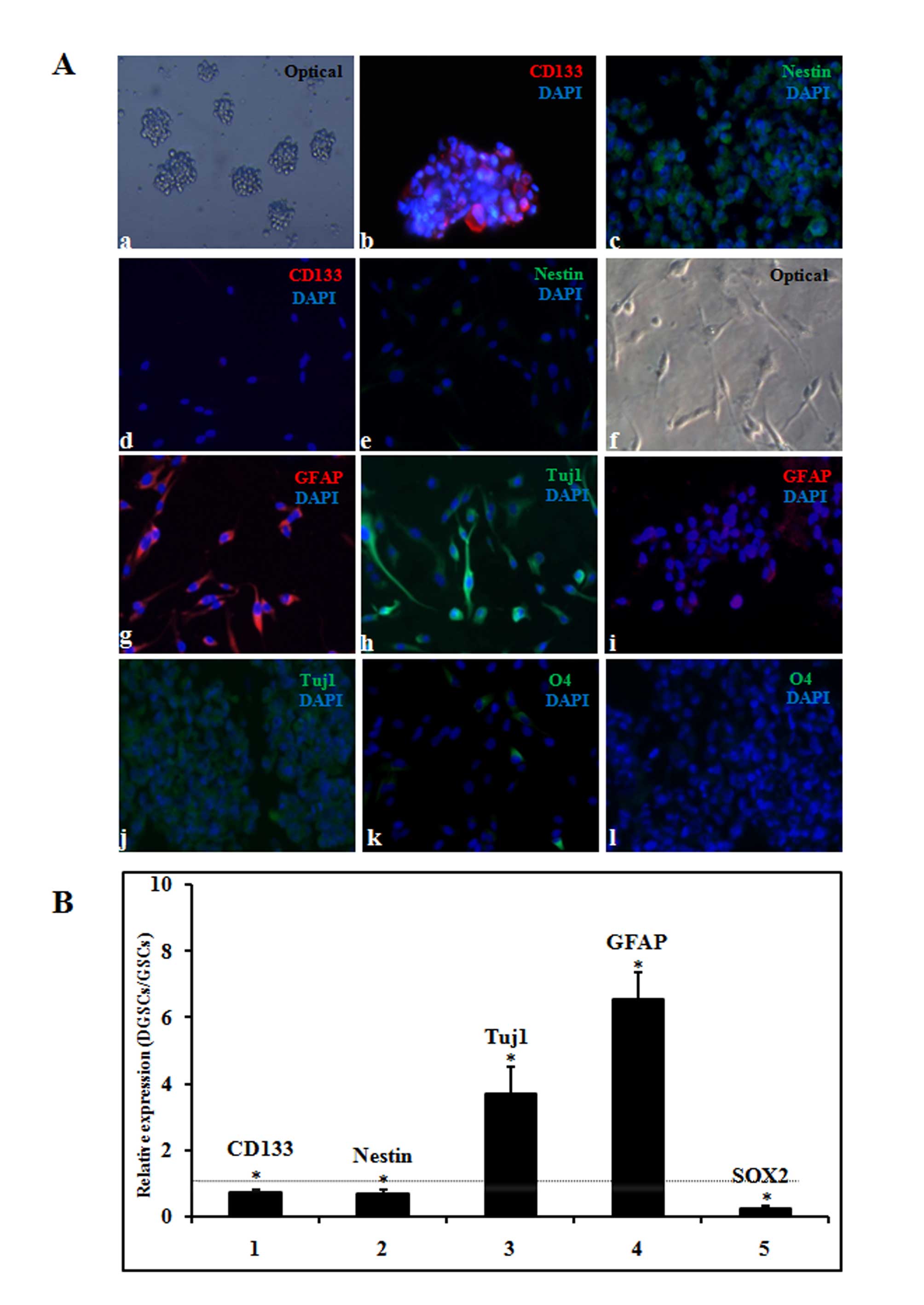 | Figure 1Determination of GSC differentiation.
(A) Differentiation was determined by immunofluorescence. (a)
Morphology of the tumor spheres in stem cell media (magnification,
×100). Expression of the stem cell markers (b) CD133 and (c) nestin
was determined by immunostaining (magnification, ×200). Expression
of (d) CD133 and (e) nestin indicated that GSCs were
differentiated. (f) Serum-induced differentiation of GSCs was
observed using an optical microscope (magnification, ×200).
Expression of (g) GFAP and (h) Tuj1 indicated that GSCs were
differentiated. Expression of the differentiation markers (i) GFAP
(j) Tuj1, and (k and l) O4 was determined by immunostaining
(magnification, ×200). DAPI staining was used to indicate the
nuclei of GSCs. (B) Differentiation was confirmed by reverse
transcription-quantitative polymerase chain reaction analysis of
the following genes: CD133, nestin, Tuj1, GFAP and SOX2. The
relative expression represents the relative fold change in DGSC
values to GSC values. Data are presented as the mean ± standard
error. *P<0.05 for DGSC expression vs. GSC
expression. The dotted line indicates a relative fold change of 1.
GSC, glioblastoma stem cell; DGSC, differentiated glioblastoma stem
cell; CD133, cluster of differentiation 133; Tuj1, βIII tubulin;
GFAP, glial fibrillary acidic protein; SOX2, sex determining region
Y-box 2. |
The differentiation of GSCs was induced in DMEM/F12
medium containing 10% FBS for 4 days. Following serum exposure, the
GSCs acquired glial- and neurite-like cell features, with
protrusions and adherence to the flask, observed using an optical
microscope (Fig. 1Af). In
addition, immunofluorescent staining analysis confirmed astrocytic-
and neural-cell differentiation, indicated by the positive staining
with anti-GFAP and anti-Tuj1 in the majority of DGSCs (Fig. 1Ag and h), but not in GSCs (Fig. 1i and j). Furthermore, it was
observed that a small proportion of the cells differentiated into
the oligodendrocytic lineage, as demonstrated by positive staining
for O4 sulfatides in DGSCs (Fig.
1Ak), but not in GSCs (Fig.
1Al). The results of the present study are consistent with a
previous study (16), and indicate
that GSCs may be efficiently induced into astrocytic and neural
lineages, and partly induced into oligodendrocytic lineages.
RT-qPCR detected similar trends in CD133 (P=0.02), nestin (P=0.03),
GFAP (P<0.001) and Tuj1 (P<0.001) expression in the GSC
differentiation process. In addition, SOX2 (P<0.001), which
functions in the maintenance of GSC stemness, was markedly
decreased in DGSCs compared with the expression observed in GSCs
(Fig. 1B).
lncRNAs and mRNAs are differentially
expressed during GSC differentiation
Microarray analysis was performed to investigate the
expression of lncRNAs and mRNAs during GSC differentiation. A total
of 1,545 lncRNAs were differentially expressed greater than 2-fold
(P<0.05) during the GSC differentiation process, of which 650
lncRNAs were upregulated and 895 were downregulated. In addition,
2,729 mRNAs were differentially expressed greater than 2-fold
(P<0.05) between GSCs and DGSCs, of which 2,179 were upregulated
and 550 were downregulated. Subsequently, Cluster 3.0 software was
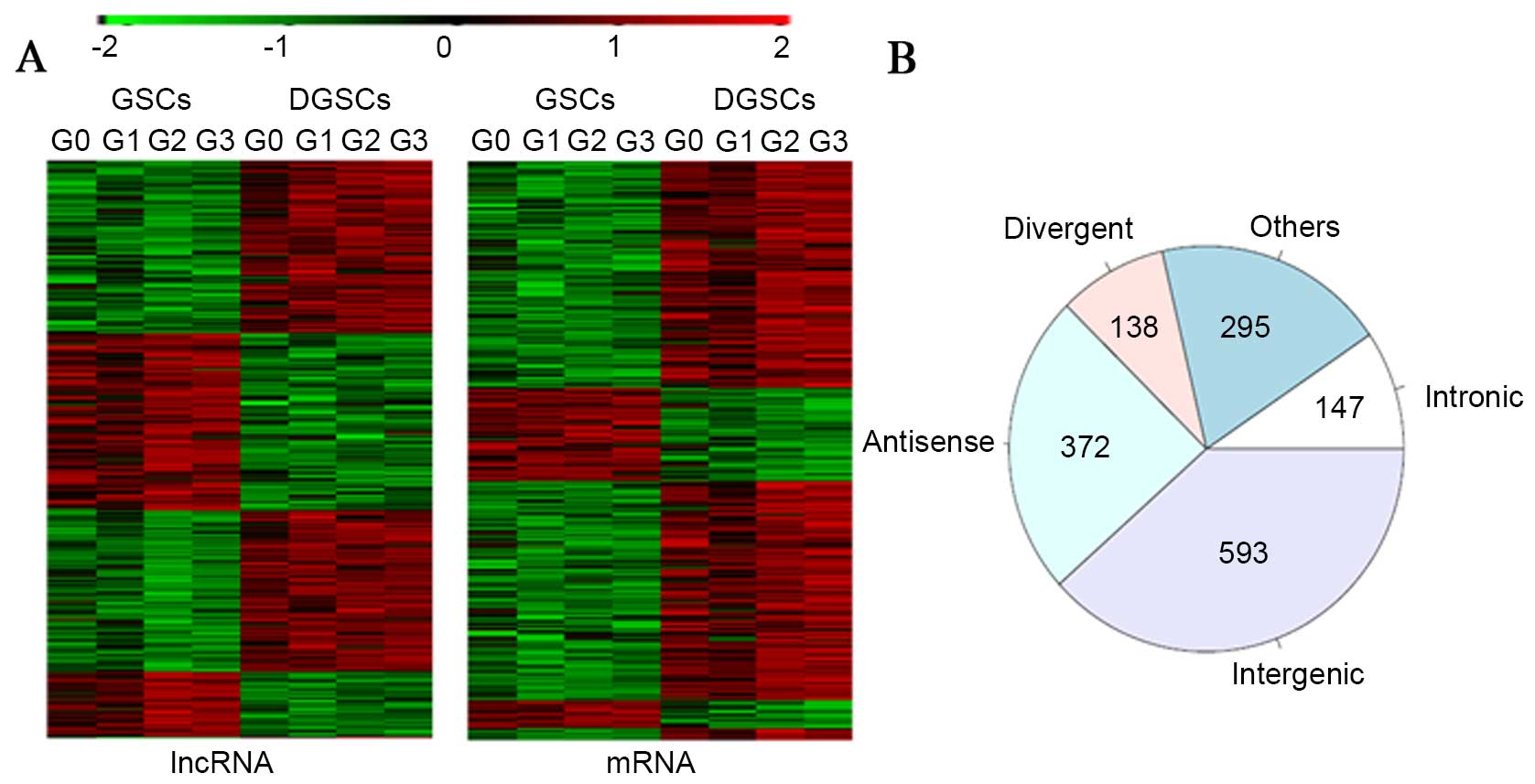
used for the clustering of the lncRNA and mRNA expression data. As
presented in Fig. 2A, the GSCs
exhibited similar expression patterns of the lncRNAs and mRNAs, but
were distinct from the DGSCs. The results of the present study
indicated that the expression patterns of lncRNAs and mRNAs may be
associated with stemness, and that lncRNAs and mRNAs may have
potential roles in regulating GSC differentiation.
To aid the interpretation of the functionality of
the lncRNAs, the DE lncRNAs were classified into five categories:
Antisense, intergenic, intronic, divergent and others, based on
their locations relative to nearby protein-coding genes (23). The anatomy of the lncRNA loci
implied that the lncRNAs have potential functions in regulating
their neighboring protein-coding genes. According to this
categorization method, nearly half the lncRNAs (593/1,545)
(Fig. 2B) belonged to the
intergenic lncRNA category, of which a large proportion may have
important functions due to their clear conservation across
mammalian species and their association with chromatin-modifying
complexes and gene expression (24).
lncRNAs are significantly correlated with
mRNAs
While the differential expression of lncRNAs and
protein-coding mRNAs indicated that this subset of lncRNAs may be
associated with the local transcriptional activity of
protein-coding mRNAs, a lncRNA-mRNA co-expression network was
constructed to ascertain the correlation between DE lncRNAs and DE
mRNAs in GSC differentiation. A total of 19,642 lncRNA-mRNA pairs,
composed of 1,087 DE lncRNAs and 1,928 DE mRNAs, were identified.
In the network, single lncRNAs were associated with multiple
(between 1 and 10) mRNAs and vice versa (data not shown). The
lncRNA-mRNA network was large and complex; therefore, to present
the association between lncRNAs and mRNAs more clearly, DE lncRNAs
and DE mRNAs with fold-change values >3.0 and P<0.05 were
selected to generate a 'core' sub-network map (data not shown).
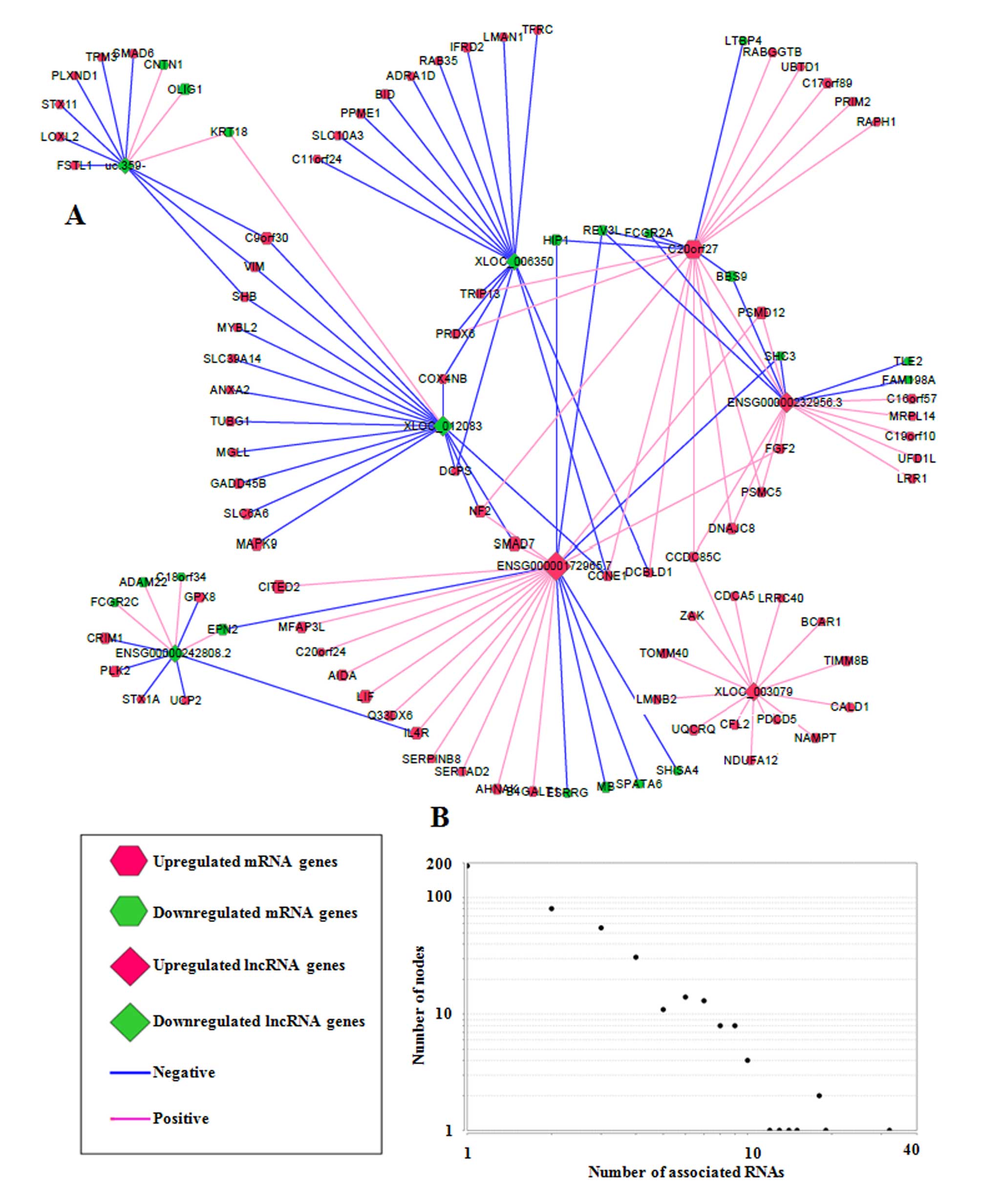
From the core sub-network, a schematic of eight RNA hub nodes (RNA
nodes were designated as hub nodes if they were associated with
multiple RNAs, implying their important functional roles) and their
associated RNAs was drawn (Fig.
3A). Hub nodes were selected based on the degree distribution
graph (Fig. 3B; degree was used to
depict the number of RNAs associated with one RNA node, and one hub
node was defined when its degree was ≥10). Among them,
C20orf27 was a protein-coding mRNA, and the remaining 7 hub nodes
(uc.359-, XLOC_006350, ENSG00000232956.3, ENSG00000172965.7,
ENSG00000242808.2, ENSG00000172965.7 and XLOC_003079) were lncRNAs,
suggesting that lncRNAs were more likely to be executing functions
via the regulation of other RNAs due to their multiple connections
with other RNAs. These lncRNA hub nodes were associated with
protein-coding mRNAs, including leukemia inhibitory factor, OLIG1
and fibroblast growth factor 2, which have key roles in the
regulation of biological processes associated with stem cells.
Together, these results suggest that DE lncRNAs may potentially be
involved in GSC differentiation via associations with
protein-coding mRNAs.
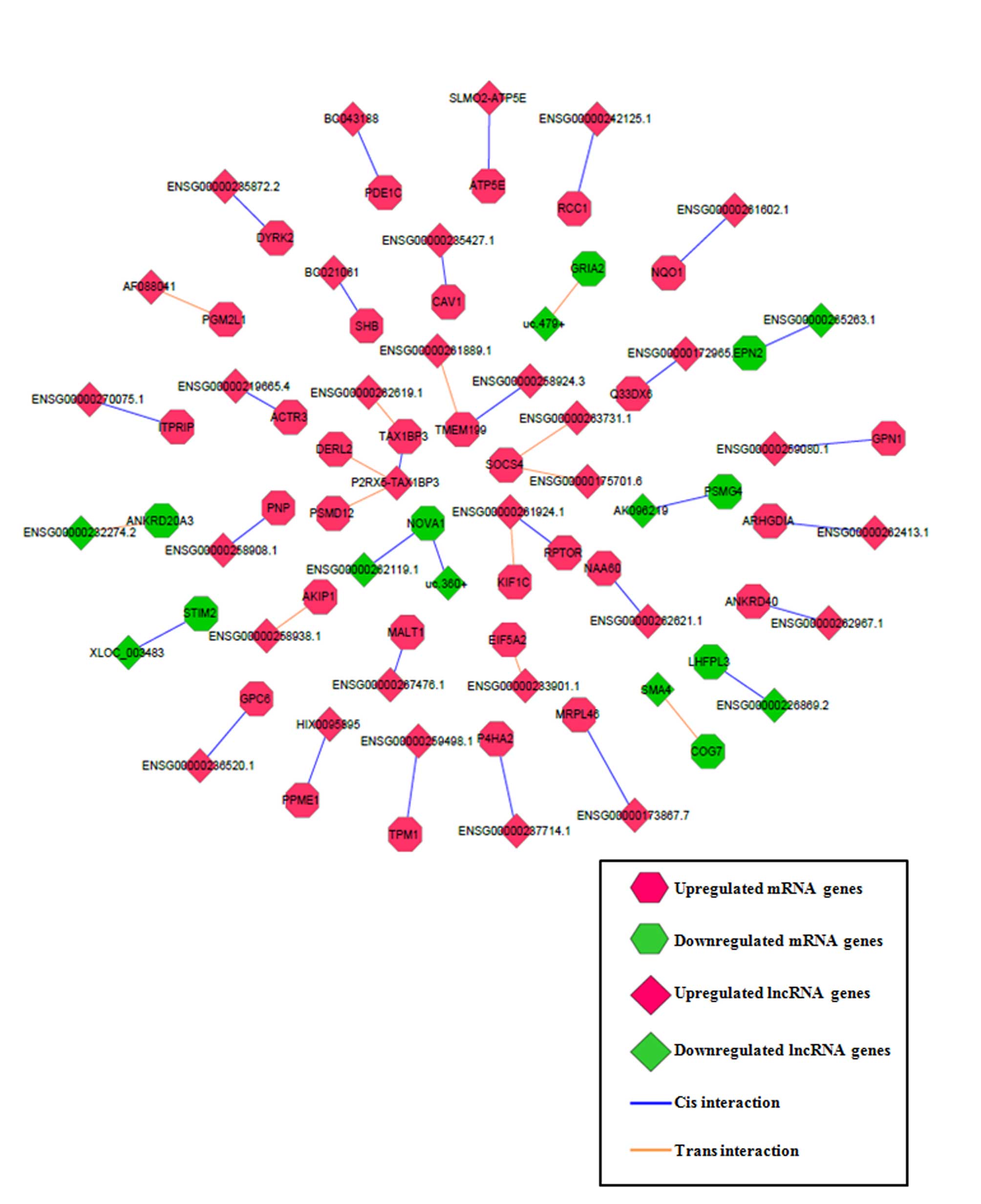
lncRNA functional prediction via cis- and
trans- target prediction
To improve the prediction accuracy of the functions
of the lncRNAs, cis- and trans- target prediction
programs were utilized based on the lncRNA-mRNA co-expression
network (containing 19,642 lncRNA-mRNA pairs, as described
previously) obtained. A total of 30 lncRNA-mRNA matched pairs were
fitted with cis- regulatory effects and 13 lncRNA-mRNA
matched pairs were fitted with trans- regulatory effects
(Fig. 4). Among the matched pairs,
lncRNA purinergic receptor P2X 5 (P2RX5)-Tax1 binding protein 3
(TAX1BP3) had 3 cis- or trans-genes (TAX1BP3, derlin
2 and proteasome 26S subunit, non-ATPase 12), lncRNA
ENSG00000261924.1 had 2 cis- or trans-genes [kinesin
family member 1C and regulatory associated protein of MTOR complex
1 (RPTOR)], and the remaining lncRNAs had 1 cis- or
trans-gene each. The functional roles of the target
protein-coding mRNAs, including RPTOR, in stem cell biology or
glioma malignancy (25,26) raised the possibility that lncRNAs
regulate GSC differentiation through regulation of their target
mRNAs. Thus, the results identified the most plausible functional
lncRNAs and their target mRNAs through a series of bioinformatics
filter strategies.
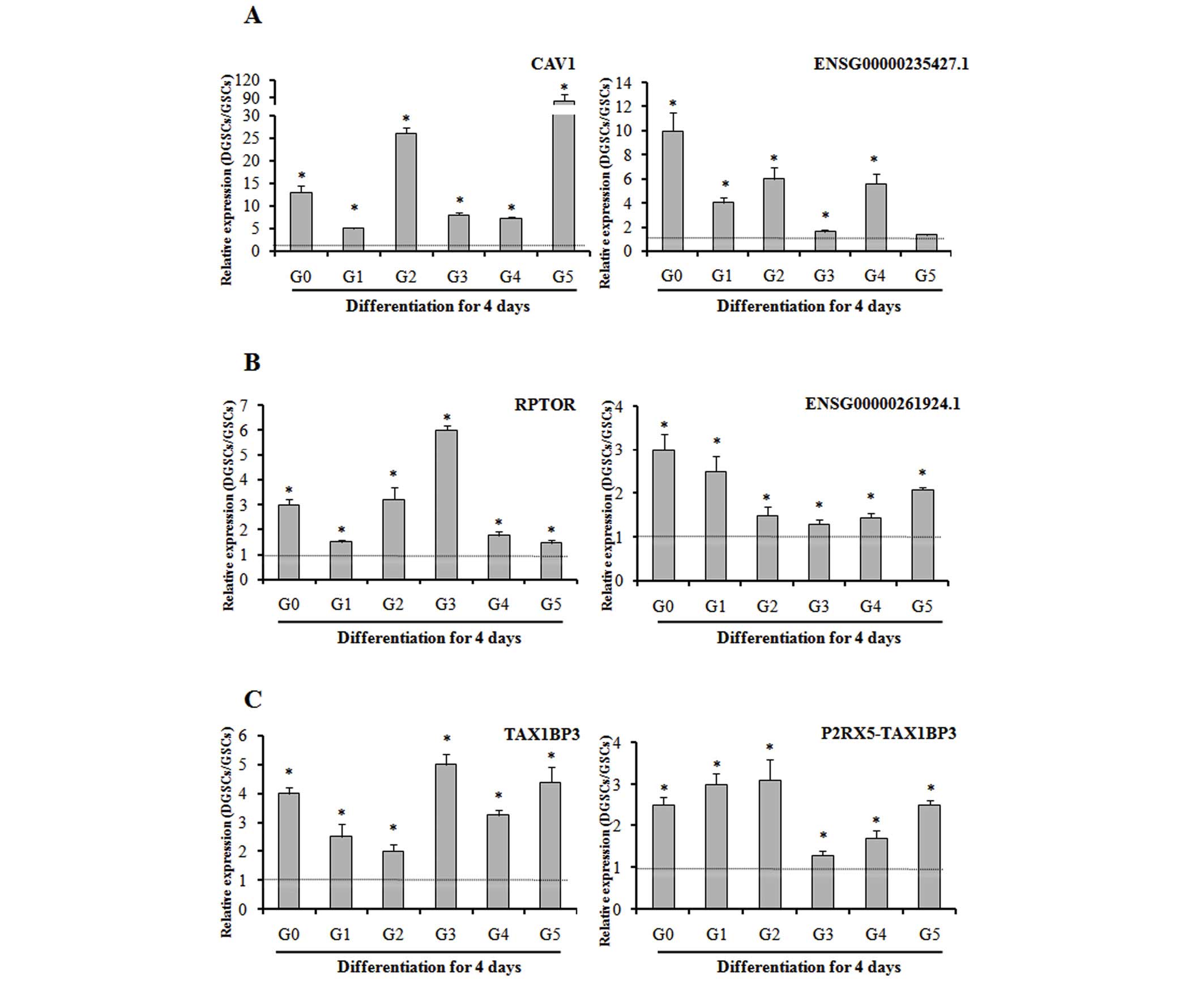
Validation of the lncRNAs and their
predicted protein-coding mRNA genes
To evaluate the consistency of the microarray and
confirm expression of lncRNAs and their target mRNAs, RT-qPCR was
performed in samples G0-G5 and their differentiated counterparts. A
total of three pairs [RPTOR-ENSG00000261924.1, cave ol i n 1 (CAV1)
-ENSG00000235427.1 a nd TAX1BP3-P2RX5-TAX1BP3] were selected for
investigation, according to fold-change values and reported gene
functions. Whilst the fold-change values of the lncRNAs and mRNAs
varied between samples, the expression trends were similar to the
microarray results. These data, presented in Fig. 5, verify the consistency of the
microarray and confirm the expression of lncRNAs and their target
mRNAs despite the tumor-specific genetic background. The P-values
for the expression level of DGSCs compared to that of GSCs for each
transcript in each sample were as follows: RPTOR G0, P=0.002; G1,
P=0.04; G2, P<0.001; G3, P<0.001; G4, P=0.004; G5, P=0.01;
ENSG00000261924.1 G0, P=0.008; G1, P=0.012; G2, P=0.04; G3, P=0.04;
G4, P=0.02; G5, P<0.001; CAV1 G0, P<0.001; G1, P<0.001;
G2, P<0.001; G3, P<0.001; G4, P<0.001; G5, P<0.001;
ENSG00000235427.1 G0, P<0.001; G1, P<0.001; G2, P<0.001;
G3, P=0.003; G4, P<0.001; G5, P=0.08; TAX1BP3 G0, P<0.001;
G1, P=0.008; G2, P=0.01; G3, P<0.001; G4, P<0.001; G5,
P<0.001; P2RX5-TAX1BP3 G0, P=0.001; G1, P<0.001; G2, P=0.004;
G3, P=0.02; G4, P=0.01; G5, P<0.001.
Identification of DE lncRNAs regulated by
a set of core TFs
A set of core TFs (POU3F, SOX2, SALL2 and OLIG2)
have previously been demonstrated to bind to, and activate,
GSC-specific regulatory elements, and thus determine and reprogram
the differentiation status of the GSCs (21). In the present study, microarray and
RT-qPCR revealed that the expression of all four TFs decreased upon
differentiation of GSCs to DGSCs, which was consistent with a
previous study (21). A total of
262 lncRNAs were revealed to contain at least one TF binding site
in their regulatory regions or gene body, and 21 lncRNAs contained
binding sites for all four TFs. The results are summarized as a
Venn diagram in Fig. 6A. In
addition, a representative schematic of the binding of the four TFs
in the ENSG00000242808 regions is presented in Fig. 6B. Furthermore, as evidence has
suggested that conservation often indicates functionality (27), the sequence conservation of the 21
lncRNAs was then evaluated with the phastCons score, with 10
lncRNAs confirmed to be conserved among vertebrates (data not
shown). Taken together, the present results suggest that the DE
lncRNAs are regulated by a set of core TFs, and that they may be
integrated into known pluripotency TF networks regulating GSC
differentiation.
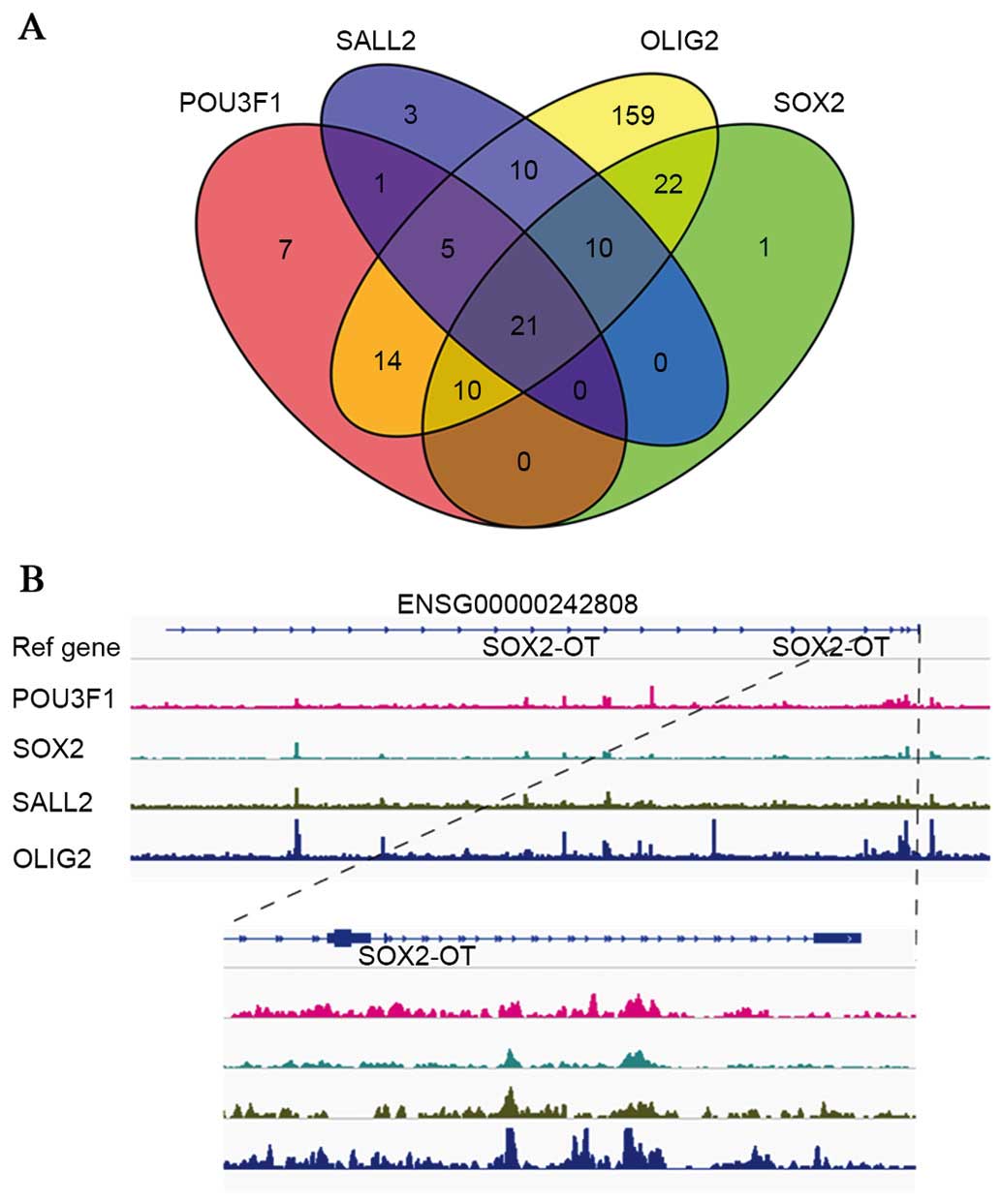 | Figure 6ChIP-Seq analysis reveals that
lncRNAs are regulated by core TFs (POU3F, SOX2, SALL2 and OLIG2).
(A) Venn diagram depicting the number of lncRNAs bound by the four
TFs. (B) Representative graphs of POU3F, SOX2, SALL2 and OLIG2
binding on the ENSG00000242808 region. lncRNA, long non-coding RNA;
TF, transcription factor; POU3F, POU domain, class 3, transcription
factor; SOX2, sex determining region Y-box 2; SALL2, spalt-like
transcription factor; OLIG2, oligodendrocyte lineage transcription
factor 2; SOX2-OT, SOX2 overlapping transcript. |
Discussion
There is a growing understanding of the importance
of lncRNAs in the regulation of pluripotency and differentiation in
stem cells, including cancer stem cells (14,28).
In addition, certain classical lncRNAs, including HOX transcript
antisense RNA (HOTAIR), metastasis associated lung adenocarcinoma
transcript 1 (MALAT-1) and long intergenic non-protein coding RNA,
regulator of reprogramming (Linc-RoR), whose roles in numerous
biological processes have been widely investigated, have been
reported to possess regulatory roles in stemness maintenance and
differentiation in various cancer stem cells (29–31).
However, no significant alterations in HOTAIR, MALAT-1 and Linc-RoR
expression were observed in the present study, which may be partly
explained by the differences between cell lines of different types
of cancer. Based on transcriptome microarray analysis and deep
sequencing technologies, a number of novel lncRNAs were observed to
regulate stem cell stemness and differentiation in the present
study, notably LncTCF7 and pnky. Previously, LncTCF7 was reported
to maintain the stemness of human liver cancer stem cells via the
activation of Wnt signaling (32).
Pnky, a conserved lncRNA, has been demonstrated to interact with
and regulate the neuronal differentiation of embryonic and
postnatal neural stem cells (33).
Despite a number of lncRNAs being examined in a
variety of cancer stem cells, few studies have reported lncRNA
profile and their roles in GSC differentiation. In the current
study, the use of an lncRNA-mRNA human gene expression microarray
platform identified for the first time a total of 1,545 lncRNA
profile changes during the differentiation of patient-derived GSCs.
Using a series of bioinformatics strategies for lncRNA prediction,
it was observed that DE lncRNAs may interact with protein-coding
mRNAs by associating with them and in addition through cis-
or trans-targeting. Certain protein-coding mRNAs that are
regulated by lncRNAs possess significant biological functions in
cancer or stem cells. For example, RPTOR, a regulatory protein that
forms the stoichiometric mammalian target of rapamycin complex 1
(mTORC1), is indispensable for the kinase activity of mTORC1,
through which it may exert its functions (25). A previous study demonstrated that
RPTOR-deficient mesenchymal stem cells have impaired mTORC1
signaling and a reduced capacity to form lipid-laden adipocytes
(34). Furthermore, a
differentiation regulation capacity for mTORC1 in GSCs has been
suggested (26). In addition to
RPTOR, certain mRNAs, including CAV1, tropomyosin 1, TAX1BP3 and
NADPH quinone dehydrogenase 1, have previously been reported to be
involved in stem cell differentiation or glioma malignancy
(35–39). These results highlight the roles of
lncRNAs in regulating GSC differentiation through associating with,
and targeting, a series of functional protein-coding mRNAs.
A previous study has suggested that pluripotent
lncRNAs may be regulated by TFs known to regulate pluripotency
(40). It is well-established that
the GSC differentiation process may be artificially manipulated via
the induction of combinations of a set of core pluripotent TFs
including POU3F2, SOX2, SALL2 and OLIG2 (21). The partially identified functional
targets to which the four core TFs bind are responsible for
reprograming the differentiation process in GSCs (21). In the present study, DE lncRNAs
were integrated into the core pluripotency TF network to provide a
more comprehensive network involved in the regulation of GSC
differentiation. The results of the present study not only reveal
that a large proportion of DE lncRNAs are regulated by TFs, but in
addition provide an alternative method to interpret the functions
of the core TFs that bind to and activate DE lncRNAs and thus
direct the GSC differentiation process.
In conclusion, the present study identified a lncRNA
and mRNA expression profile that clearly distinguished GSCs from
DGSCs. Using a series of bioinformatics strategies for lncRNA
prediction, it was observed that DE lncRNAs were able to regulate
the GSC differentiation process through an association with
protein-coding mRNAs and additionally through cis- or
trans-targeting. Furthermore, the DE lncRNAs were regulated
by a set of core pluripotency TFs. The results of the present study
provide a potential direction for future research via the
comprehensive integration of lncRNAs and mRNAs, and the
identification of a set of lncRNAs as candidates for further
study.
Acknowledgments
The authors would like to thank Dr Anchen Guo at the
Laboratory of Clinical Medicine Research, Beijing Tiantan Hospital
for providing the GSC-propagating medium. The present study was
supported by a grant from the China National Clinical Research
Center for Neurological Diseases, the Training Plan for Beijing
High-Level Healthcare Personnel (grant no. 2011-3-28).
References
|
1
|
Quick A, Patel D, Hadziahmetovic M,
Chakravarti A and Mehta M: Current therapeutic paradigms in
glioblastoma. Rev Recent Clin Trials. 5:14–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Orza A, Soriţău O, Tomuleasa C, Olenic L,
Florea A, Pana O, Bratu I, Pall E, Florian S, Casciano D and Biris
AS: Reversing chemoresistance of malignant glioma stem cells using
gold nanoparticles. Int J Nanomedicine. 8:689–702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Friedmann-Morvinski D: Glioblastoma
heterogeneity and cancer cell plasticity. Crit Rev Oncog.
19:327–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jackson M, Hassiotou F and Nowak A:
Glioblastoma stem-like cells: At the root of tumor recurrence and a
therapeutic target. Carcinogenesis. 36:177–185. 2015. View Article : Google Scholar
|
|
5
|
Nakano I: Stem cell signature in
glioblastoma: Therapeutic development for a moving target. J
Neurosurg. 122:324–330. 2015. View Article : Google Scholar
|
|
6
|
Binello E and Germano IM: Targeting glioma
stem cells: A novel framework for brain tumors. Cancer Sci.
102:1958–1966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stockhausen MT, Kristoffersen K, Stobbe L
and Poulsen HS: Differentiation of glioblastoma multiforme
stem-like cells leads to downregulation of EGFR and EGFRvIII and
decreased tumorigenic and stem-like cell potential. Cancer Biol
Ther. 15:216–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang YJ and Bikle DD: LncRNA profiling
reveals new mechanism for VDR protection against skin cancer
formation. J Steroid Biochem Mol Biol. 144:87–90. 2014. View Article : Google Scholar
|
|
9
|
Thum T and Condorelli G: Long noncoding
RNAs and microRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Curtale G and Citarella F: Dynamic nature
of noncoding RNA regulation of adaptive immune response. Int J Mol
Sci. 14:17347–17377. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin N, Chang KY, Li Z, Gates K, Rana ZA,
Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al: An
evolutionarily conserved long noncoding RNA TUNA controls
pluripotency and neural lineage commitment. Mol Cell. 53:1005–1019.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and Leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eades G, Zhang YS, Li QL, Xia JX, Yao Y
and Zhou Q: Long non-coding RNAs in stem cells and cancer. World J
Clin Oncol. 5:134–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Jin Y, Zheng X, Wu Y and Fu H: The long noncoding RNA expression
profile of hepatocellular carcinoma identified by microarray
analysis. PLoS One. 9:e1017072014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aldaz B, Sagardoy A, Nogueira L, Guruceaga
E, Grande L, Huse JT, Aznar MA, Díez-Valle R, Tejada-Solís S,
Alonso MM, et al: Involvement of miRNAs in the differentiation of
human glioblastoma multiforme stem-like cells. PLoS One.
8:e770982013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rheinbay E, Suvà ML, Gillespie SM,
Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL,
Rivera MN, et al: An aberrant transcription factor network
essential for Wnt signaling and stem cell maintenance in
glioblastoma. Cell Rep. 3:1567–1579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katsushima K and Kondo Y: Non-coding RNAs
as epigenetic regulator of glioma stem-like cell differentiation.
Front Genet. 5:142014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clark JD, Gebhart GF, Gonder JC, Keeling
ME and Kohn DF: Special Report: The 1996 guide for the care and use
of laboratory animals. ILAR J. 38:41–48. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nogueira L, Ruiz-Ontañon P,
Vazquez-Barquero A, Lafarga M, Berciano MT, Aldaz B, Grande L,
Casafont I, Segura V, Robles EF, et al: Blockade of the NFκB
pathway drives differentiating glioblastoma-initiating cells into
senescence both in vitro and in vivo. Oncogene. 30:3537–3548. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suvà ML, Rheinbay E, Gillespie SM, Patel
AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et
al: Reconstructing and reprogramming the tumor-propagating
tumor-propagating potential of glioblastoma stem-like cells. Cell.
157:580–594. 2014. View Article : Google Scholar
|
|
22
|
Siepel A, Bejerano G, Pedersen JS,
Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW,
Richards S, et al: Evolutionarily conserved elements in vertebrate,
insect, worm, and yeast genomes. Genome Res. 15:1034–1050. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim DH and Sabatini DM: Raptor and mTOR:
Subunits of a nutrient-sensitive complex. Curr Top Microbiol
Immunol. 279:259–270. 2004.
|
|
26
|
Jhanwar-Uniyal M, Jeevan D, Neil J,
Shannon C, Albert L and Murali R: Deconstructing mTOR complexes in
regulation of Glioblastoma Multiforme and its stem cells. Adv Biol
Regul. 53:202–210. 2013. View Article : Google Scholar
|
|
27
|
Roberts TC, Morris KV and Wood MJ: The
role of long non-coding RNAs in neurodevelopment, brain function
and neurological disease. Philos Trans R Soc Lond B Biol Sci.
369:pii: 20130507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo M, Jeong M, Sun D, Park HJ, Rodriguez
BA, Xia Z, Yang L, Zhang X, Sheng K, Darlington GJ, et al: Long
non-coding RNAs control hematopoietic stem cell function. Cell Stem
Cell. 16:426–438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiao F, Hu H, Han T, Yuan C and Wang L,
Jin Z, Guo Z and Wang L: Long noncoding RNA MALAT-1 enhances stem
cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci.
16:6677–6693. 2015. View Article : Google Scholar :
|
|
30
|
Pádua Alves C, Fonseca AS and Muys BR:
Brief report: The lincRNA Hotair is required for
epithelial-to-mesenchymal transition and stemness maintenance of
cancer cell lines. Stem Cells. 31:2827–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou X, Gao Q, Wang J, Zhang X, Liu K and
Duan Z: Linc-RNA-RoR acts as a 'sponge' against mediation of the
differentiation of endometrial cancer stem cells by microRNA-145.
Gynecol Oncol. 133:333–339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ramos AD, Andersen RE, Liu SJ, Nowakowski
TJ, Hong SJ, Gertz CC, Salinas RD, Zarabi H, Kriegstein AR and Lim
DA: The long noncoding RNA Pnky regulates neuronal differentiation
of embryonic and postnatal neural stem cells. Cell Stem Cell.
16:439–447. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Martin SK, Fitter S, Dutta AK, Matthews
MP, Walkley CR, Hall MN, Ruegg MA, Gronthos S and Zannettino AC:
Brief report: The differential roles of mTORC1 and mTORC2 in
mesenchymal stem cell differentiation. Stem Cells. 33:1359–1365.
2015. View Article : Google Scholar
|
|
35
|
Cosset EC, Godet J, Entz-Werlé N, Guérin
E, Guenot D, Froelich S, Bonnet D, Pinel S, Plenat F, Chastagner P,
et al: Involvement of the TGFβ pathway in the regulation of α5 β1
integrins by caveolin-1 in human glioblastoma. Int J Cancer.
131:601–611. 2012. View Article : Google Scholar
|
|
36
|
Luanpitpong S, Wang L, Stueckle TA, Tse W,
Chen YC and Rojanasakul Y: Caveolin-1 regulates lung cancer
stem-like cell induction and p53 inactivation in carbon
nanotube-driven tumorigenesis. Oncotarget. 5:3541–3554. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du HQ, Wang Y, Jiang Y, Wang CH, Zhou T,
Liu HY and Xiao H: Silencing of the TPM1 gene induces
radioresistance of glioma U251 cells. Oncol Rep. 33:2807–2814.
2015.PubMed/NCBI
|
|
38
|
Wang H, Han M, Whetsell W Jr, Wang J, Rich
J, Hallahan D and Han Z: Tax-interacting protein 1 coordinates the
spatiotemporal activation of Rho GTPases and regulates the
infiltrative growth of human glioblastoma. Oncogene. 33:1558–1569.
2014. View Article : Google Scholar :
|
|
39
|
Okamura T, Kurisu K, Yamamoto W, Takano H
and Nishiyama M: NADPH/quinone oxidoreductase is a priority target
of glioblastoma chemotherapy. Int J Oncol. 16:295–303.
2000.PubMed/NCBI
|
|
40
|
Ng SY, Johnson R and Stanton LW: Human
long non-coding RNAs promote pluripotency and neuronal
differentiation by association with chromatin modifiers and
transcription factors. EMBO J. 31:522–533. 2012. View Article : Google Scholar :
|