Introduction
Endometrial adenocarcinoma is the fourth most
frequent gynecological malignancy in Germany (1), ~11,300 women are newly diagnosed with
this cancer annually (2). The risk
of developing endometrial carcinoma increases with age, and may be
augmented by estrogen-based hormonal therapies (3), diabetes mellitus, nulliparity and
former carcinomas (4). The most
common symptom is sudden bleeding, in peri- and postmenopausal
women (5). In contrast to other
tumors, preventive screening is often ineffective. A specific
diagnosis may only be made by histological tissue examination
subsequent to hysteroscopy (6).
Therapeutic interventions primarily consist of surgery, followed by
radiation therapy (7).
Chemotherapy is rarely administered (8). Regardless of medical treatment, in
~25% of all cases, patients develop remote metastasis (8), resulting in necessary follow-up care
(9).
The aim of the present study is to predict the
formation of metastasis, in order to prevent the processes
associated with metastatic outgrowth. Metastasis develop from
single cells that have dissociated from the primary solid tumor,
circulate through lymph vessels and the blood stream before
settling in different sites of the body, thus allowing for
metastatic formation. These single tumor cells are termed
circulating tumor cells (CTCs) (10). In cases where CTCs infiltrate the
bone marrow, they are able to persist for years without doing any
harm and become disseminated tumor cells (DTCs) (11–16).
The occurrence of CTCs and DTCs is often an indicator of an
unfavorable prognosis for patients (10,17),
and therefore are included in international tumor staging systems
(18,19).
The detection of DTCs is exhausting and a
time-consuming procedure for patients, whereas the detection of
CTCs from blood samples is advantageous, as this biomaterial is
more easily accessible. One disadvantage in the detection of CTCs
from blood samples is that the number of CTCs obtained is small in
comparison to the surrounding white blood cells (20); therefore, highly sensitive methods
for detection are required. The primary detection system currently
available is Cell Search® System, distributed by Veridex
LLC (Raritan, NJ, USA), which is approved by the US Food and Drug
Administration for metastatic breast cancer. It is based on an
immunomagnetic enrichment of tumor cells with fluorescent staining
of tumor-specific cell surface epitopes. However, this method is
expensive and laborious.
The current study aimed to identify a method for the
detection of CTCs using the quantitative polymerase chain reaction
(qPCR) method. This is based on the fact that as the primary tumor
is of epithelial origin and expresses an epithelial gene panel.
CTCs may also express these epithelial genes. Therefore, they may
be distinguished from blood cells, which are mesenchymal cells, by
gene expression. The advantage of this method is that it is more
sensitive, less expensive than other methods in the field and may
be performed quickly in nearly every laboratory. However, it is
challenging to find suitable marker genes for the detection of
CTCs, which are able to distinguish between tumor and blood cells
(21–23). Therefore, the expression of a set
of genes was compared between cells isolated from healthy
endometrium and endometrial adenocarcinoma cell lines. To the best
of our knowledge, there are only a few studies describing the
presence of CTCs in patients with endometrial adenocarcinoma. In
particular, a high-risk group of patients with high-grade
endometrial adenocarcinomas (24),
demonstrated a correlation of the occurrence of CTCs with stemness
was demonstrated in a previous study and CTCs were recognized as
possible therapeutic targets (25).
The marker genes used in the present study were
cyclin E1 (CCNE), cytokeratin 19 (CK19), claudin 4 (CLDN-4),
G-protein coupled estrogen receptor (GPER), human epidermal growth
factor receptor 2 (Her-2), luteinizing hormone/choriogonadotropin
receptor (LHCGR), T-cell differentiation protein 2 (MAL2),
mammaglobin (MGL), migration inducing protein 7 (MIG7) and vascular
endothelial growth factor receptor 2 (VEGFR2) (Table I). CCNE is often overexpressed in
endometrial adenocarcinomas (26)
and has been used in qPCR detection of CTCs from blood samples of
different malignant entities. CCNE and MAL2 have been identified as
useful markers in endometrial carcinoma tissue (27). MIG7 was described as a marker gene,
which often predicts poor prognosis in patients with endometrial
adenocarcinoma (28). CK19 is a
marker of epithelial cells and is also used in the alkaline
phosphatase-anti-alkaline phosphatase test, which is routinely used
in cancer diagnosis (29,30). CLDN-4 has been recognized as a
biomarker in the treatment of patients with endometrial
adenocarcinoma (31). MGL is
frequently used as a marker gene for breast malignancies; however,
it may have an important function in malignant endometrial tissues
(32), and was therefore also
selected for the set of marker genes to be tested by the present
study. The expression of GPER is dysregulated in malignant
endometrium and is involved in steroid hormone signaling (33). Her-2 is particularly prevalent in
early endometrial tumorigenesis, in coherence with Cox-1 and -2
(34). VEGFR2 is an important
factor in metastasis formation and neoangiogenesis, ensuring blood
supply in the newly growing tumor mass (35). LHCGR is also associated with tumor
staging (36). It is also
important for cell proliferation (37,38)
and may be correlated with grading of endometrial carcinomas
(39).
 | Table ICharacterization of used marker genes
and their respective Taq-Man qPCR primers (Thermo Fisher
Scientific, Inc.). |
Table I
Characterization of used marker genes
and their respective Taq-Man qPCR primers (Thermo Fisher
Scientific, Inc.).
| Gene | Chromosomal
location | Primer cat.
no. |
Characteriaztion | Function |
|---|
| CCNE | 19q12 | Hs_00180319_m1 | Regulator of
CDK2 | Overexpression
results in chromosome instability |
| CK19 | 17q21.2 | Hs_00761767_m1 | Intermediate
filament protein | Structural
integrity of epithelial cells |
| CLDN-4 | 7q11.23 | Hs_00976831_s1 | Integral membrane
protein | Component of tight
junctions |
| GPER | 7p22.3 | Hs_00173506_m1 | Binds estrogen,
important for cellular signaling | Some splice
variants are known |
| Her-2 | 17q12 | Hs_00170433_m1 | Proto-oncogene | Overexpression
results in development and progression of aggressive cancer
types |
| LHCGR | 2q16.3 | Hs_00174885_m1 | G-protein coupled
receptor | Male secondary
sexual character development |
| MAL2 | 8q24 | Hs_00294541_m1 | Multispan
transmembrane protein | Involved in
polarized transport |
| MGL | 11q12.3 | Hs_00419570_m1 | Belongs to family
of secretoglobulins | Involved in cell
signalling, immune response and chemotaxis |
| MIG7 | 1p22.1 | Hs_00706258_m1 | Involved in cell
signalling | Limited to
embryonic/fetal cells and epithelial cancer cells |
| VEGFR2 | 4q12 | Hs_00911700_m1 | Mediator of
VEGF-induced endothelial proliferation, survival, migration and
morphogenesis | Angiogenesis and
vascular development |
Materials and methods
Cell lines and subcultivation
The endometrial adenocarcinoma cell lines AN3CA
(HTB-111), HEC-1-A (HTB-112), HEC-1-B (HTB-113), KLE (CRL-1622) and
RL-95-2 (CRL-1671) were purchased from American Type Culture
Collection (Manassas, VA, USA), Ishikawa (cat. no. 99040201) from
European Collection of Authenticated Cell Cultures (Salisbury, UK).
All cell lines were established from endometrial carcinoma patients
(see Table II) and were cultured
in Dulbecco's modified Eagle's medium (DMEM; Biochrom, Ltd.,
Berlin, Germany), supplemented with 10% fetal calf serum (FCS;
Biochrom, Ltd.) and 1% penicillin/streptomycin (P/S) (Biochrom,
Ltd.), except KLE, as they required DMEM-F12 (Biochrom, Ltd.), 10%
FCS and 1% P/S as culture medium. Cells were subcultured as
indicated by the supplier's protocol.
 | Table IIEndometrial adenocarcinoma cell
lines. |
Table II
Endometrial adenocarcinoma cell
lines.
| First author,
year | Cell line | Tumour type | Donor age
(years) | Donor
ethnicity | Depositor | Cell line
characteristics | Refs |
|---|
| Dawe, 1964 | AN3CA | Adenocarcinoma | 55 | Caucasian | CJ Dawe | Isolated from
metastatic lesion in lymph nodes | (43) |
| Kuramoto, 1972 | HEC-1-A | Adenocarcinoma | 71 | Caucasian | H Kuramoto | Cells from well
differentiated adenocarcinoma (grade II), expresses platelet
activated growth factor and c-fos, tumourigenic in nude mice | (44) |
| Kuramoto, 1972 | HEC-1-B | Adenocarcinoma | 71 | Caucasian | H Kuramoto | Substrain of
HEC-1-A, more flattened growth, tumourigenic in nude mice, diploid
to tetraploid | (44) |
| Lessey, 1996 | Ishikawa | Adenocarcinoma | 39 | Asian | A Taylor | Induces cancer in
nude mice, expresses ER and PR | (45) |
| Hendricks,
1997 | KLE | Adenocarcinoma | 64 | Caucasian | GR Richardson | Tumourigenic in
nude mice, cells have microvilli and junctional complexes, no
formation of glands observed | (46) |
| Way, 1983 | RL-95-2 | Carcinoma | 65 | Caucasian | DL Way | Expresses ER,
possesses α-keratin, cells have junctional complexes and surface
microvilli | (47) |
For RNA isolation, cells were washed with
phosphate-buffered saline (Biochrom, Ltd.), and TRIzol LS
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was
added. Cells disrupted by the addition of TRIzol were wiped off the
cell culture bottle by a cell scraper (Corning Inc., Corning, NY,
USA).
Isolation of endometrial stromal cells
from tissue samples
Tissue samples of healthy endometrium were obtained
from 10 patients undergoing endometrial examination in scope of
fertility and are included in this examination following an
unsuspicious pathological result as a negative control group.
Patients were informed and had signed consent, following the
Declaration of Helsinki (ethical vote LMU 148-12). Samples were
maintained in DMEM-F12, 10% FCS and 1% P/S at 4°C overnight or
processed immediately. Extraction of stromal cells was performed as
described in Fernandez-Shaw et al (40) and Zhang et al (41). Briefly, tissue was cut in 2–3 mm
pieces with a scalpel and incubated with 1 mg/ml collagenase
(Invitrogen; Thermo Fisher Scientific, Inc.) in complete DMEM
medium for 2 h at 37°C. The suspension was then filtered through
250 μm tissue strainers (Thermo Fisher Scientific, Inc.).
The liquid phase was placed onto 40 μm cell strainers
(Falcon; Thermo Fisher Scientific, Inc.). Stromal cells were
maintained in the liquid phase and spun down at 300 × g for 10 min
at 4°C. The supernatant was discarded, and RNA was isolated from
the cell pellet by addition of 1 ml TRIzol LS.
RNA isolation
Cell suspensions were already in TRIzol LS, and 0.2X
volume of chloroform (Merck Millipore, Darmstadt, Germany) was
added for RNA isolation. The cell suspension was then vigorously
vortexed and centrifuged at 12,000 × g and 4°C for 15 min. The
clear liquid phase was carefully aspirated and transferred into a
fresh reaction tube. Isopropanol (0.5 ml; Merck Millipore) was
added to each sample, vortexed again and incubated overnight at
−20°C.
The next day the suspension was centrifuged at
12,000 × g and 4°C for 10 min, the supernatant discarded and the
RNA pellet washed by the addition of 1 ml 75% ethanol (Merck
Millipore) and centrifuged at 12,000 × g and 4°C for 10 min. The
ethanol was subsequently removed, the pellet air-dried for 15 min
and dissolved in diethylpyrocarbonate-treated water. The
concentration and ratio of the isolated RNA was determined
photometrically at wavelengths 260 and 280 nm. Only RNA with a
ratio of 1.7–1.9 is used for further experiments.
Reverse transcription
A total of 4 μg of the isolated RNA in a
maximum volume of 6 μl were used for reverse transcription.
Reverse transcription was performed using SuperScript III First
Strand Synthesis Super Mix kit by Invitrogen (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Briefly, 1 μl Oligo-dTs and 1 μl First Strand buffer
were added to the RNA and incubated at 65°C for 5 min. Next, 10
μl 2× Reaction Mix and 2 μl reverse transcriptase
were added and the solution was incubated at 42°C for 50 min.
Reverse transcriptase was subsequently heat-inactivated by an
incubation at 85°C for 5 min. The cDNA produced was stored at −20°C
until it was used in qPCR.
qPCR
For qPCR, 2 μl of the respective cDNA sample
was pipetted into each well of a 96-well plate (Thermo Fisher
Scientific, Inc.). A mastermix for each gene was prepared, for the
number of samples, which were analyzed. Therefore, for each
reaction, 10 μl reaction mix (Thermo Fisher Scientific,
Inc.), 7 μl water and 1 μl of the respective
gene-specific probe (Table I) were
mixed and 18 μl of this mixture was added to the cDNA,
giving a total reaction volume of 20 μl. The plate was
sealed and centrifuged at 315 × g for 1 min. and placed in a qPCR
machine. The cycles were run in the following scheme: 20 sec at
95°C as an initial denaturation, followed by 40 cycles consisting
of 3 sec at 95°C and 30 sec at 60°C. Fluorescence was determined at
the end of each amplification cycle and the relative quantification
values (RQ values) were calculated using SDS-software (version
1.3.1) by the 2−ΔΔCq method (42). 18S was used as an internal
reference, and gene expression values of endometrial adenocarcinoma
cell lines were set in reference to healthy stromal cells isolated
from tissue samples as aforementioned. The reaction assays for each
gene and cell line were performed as quadruplicates.
Results
In order to identify a suitable set of marker genes
for CTC-detection from blood samples of patients with endometrial
adenocarcinoma, qPCR was performed on mRNA/cDNA obtained from 6
human endometrial adenocarcinoma cell lines (AN3 CA, HEC-1-A,
HEC-1-B, Ishikawa, KLE, RL95-2) and from healthy endometrial
tissue, with 10 different genes, which were previously described in
the literature as qPCR marker genes or were associated with
endometrial carcinoma and metastasis formation.
RQ values of >1 were deemed to be a indicator of
upregulation of gene expression in comparison to the expression
levels of the same gene in the reference tissue (healthy
endometrium). RQ values <1 indicated that the gene was expressed
at a lower level compared with the control sample.
The present study found low RQ values for LHCGR,
VEGFR2 and MGL, which in all cell lines have RQ values <1, with
the exception of VEGFR2 in KLE-cells, where the RQ value is 4.742.
In RL95-2 cells, LHCGR had a very low expression level, and even
after 40 cycles of PCR no PCR-product was fluorescently detected,
resulting in a not detected (nd) RQ value. The remaining seven
genes tested via qPCR had different expression levels in the
various cell lines tested. High expression levels were observed for
CLDN-4 and CK19, especially in RL95-2 cells (317.51 and 501.911,
respectively). This was also evident in the remaining five cell
lines; however, not to the same extent. For Her-2, CCNE and MAL2
intermediate expression levels were found. GPER and MIG7 were
upregulated in comparison with healthy tissues in the majority of
the cell lines investigated, with exception of MIG7 in RL95-2 cells
(0.897) and GPER in Ishikawa (0.211) and KLE (nd) (Fig. 1; Table III).
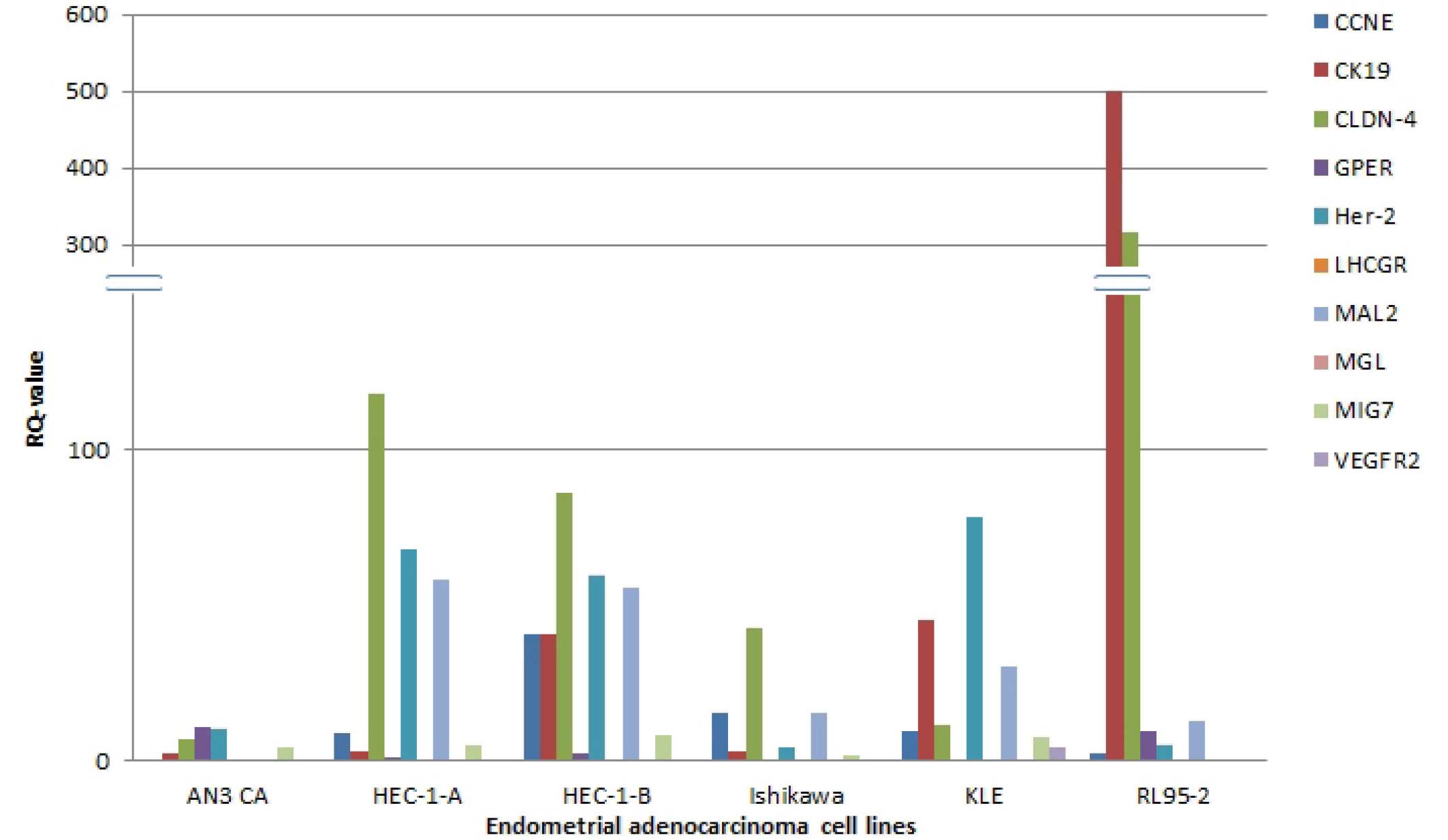 | Figure 1Comparison of the expression of the
different genes in various endometrial adenocarcinoma cell lines.
CCNE, cyclin E1; CK19, cytokeratin 19; CLDN-4, Claudin 4; GPER,
G-protein coupled estrogen receptor; Her-2, human epidermal growth
factor receptor 2; LHCGR, luteinizing hormone/choriogonadotropin
receptor; MAL2, T-cell differentiation protein 2; MGL, mammaglobin;
MIG7, migration inducing protein 7; VEGFR2, vascular endothelial
growth factor receptor 2. |
 | Table IIIRelative quantification values for
all genes and cell lines examined. |
Table III
Relative quantification values for
all genes and cell lines examined.
| AN3 CA | HEC-1-A | HEC-1-B | Ishikawa | KLE | RL95-2 |
|---|
| CCNE | 0.045 | 8.938 | 40.558 | 15.215 | 9.807 | 2.582 |
| CK19 | 2.716 | 3.069 | 40.581 | 3.305 | 45.305 | 501.911 |
| CLDN-4 | 7.063 | 0.345 | 86.115 | 42.771 | 11.693 | 317.510 |
| GPER | 10.682 | 1.002 | 2.345 | 0.211 | nd | 9.547 |
| Her-2 | 10.095 | 68.220 | 59.618 | 4.491 | 78.536 | 5.297 |
| LHCGR | 0.005 | 0.016 | 0.201 | 0.018 | 0.328 | nd |
| MAL2 | 0.003 | 58.343 | 55.510 | 15.292 | 30.297 | 12.726 |
| MGL | 0.015 | 0.010 | 0.003 | 0.008 | 0.005 | 0.011 |
| MIG7 | 4.513 | 4.974 | 8.131 | 1.781 | 7.839 | 0.897 |
| VEGFR2 | 0.002 | 0.003 | 0.003 | 0.010 | 4.742 | <0.001 |
Discussion
In the present study, markedly different gene
expression values were observed between the different genes, and
also between the cell lines. The differences observed for one gene
among the different cell lines to a certain degree reflects the
situation, that would occur when using patient samples. For
example, the gene expression levels in the HEC-1-A and HEC-1-B cell
lines are similar, possibly due to their common origin. By
contrast, the Ishikawa cell line has a different gene expression
pattern, which is rather different compared with the remaining cell
lines. This could be due to it being the only cell line that was
obtained from an Asian patient, whereas the rest were from
Caucasian patients; therefore, it is possible that the carcinomas
developed in different genetic backgrounds. Furthermore, the donor
of the Ishikawa cell line was younger, possibly premenopausal,
whereas the donors of the other cell lines were older and
presumably postmenopausal, and this may additionally have
contributed to the variations in genetic background for
tumorigenesis. Therefore, in order to successfully detect CTCs from
blood samples of patients with endometrial adenocarcinoma, the
primary challenge is to establish a suitable set of marker genes,
which would enable the detection of CTCs with a high sensitivity.
These results are in accordance with previous findings using breast
cancer cell line cells and blood samples from patients with breast
cancer for CTC detection (21–23).
CLDN-4 and CK19 were identified to be highly
suitable, confirming the recent results of Pan et al
(31), which used CLDN-4 as a
biomarker for endometrial adenocarcinoma. By contrast, CK19 is an
established marker gene and has been used in tumor cell diagnostics
routinely (29,30). CK19 is a typical epithelial marker,
therefore it is not unusual that it is expressed in CTCs.
Furthermore, Her-2 and MAL2 may also be suitable marker genes, with
high expression levels in the majority of cell lines investigated
in the present study. MAL2 was also previously identified as a
marker gene in endometrial adenocarcinoma (27). A previous study has determined that
the expression levels of CCNE are upregulated in endometrial
adenocarcinoma (26).
Additionally, as Her2 also exhibits high expression levels in the
majority of the cell lines, and may be easily detected by qPCR, it
represent a potential marker gene in patient samples, as it has
been previously identified as a marker of early tumorigenesis
(34).
LHCGR, VEGFR2 and MGL exhibited a consistently lower
expression across the tumor cell lines investigated. However, this
does not mean that these genes are not important for tumorigenesis
and the formation of remote metastasis. It is possible that the
mRNAs of those genes are degraded quickly following protein
translation and therefore that their expression may not be
determined using qPCR. In order to overcome this obstacle, a
potential approach may be to investigate marker genes with long RNA
half-lives, thus increasing the time available for detection.
In conclusion, qPCR is a potential method for the
diagnosis of CTCs in endometrial adenocarcinoma. Therefore, it may
aid in the refinement of treatment options, and indicate whether a
patient has a particular potential for metastasis, as CTCs are
present in the blood stream. The present study indicates a
potential set of reliable marker genes which may be used with this
methodology, however, further studies are required to confirm and
expand upon this. It will be important to clarify whether
particular levels of gene expression can be correlated to specific
numbers of tumor cells in a blood sample. Therefore standard curves
would be required to be generated, using varying quantities of
tumor cells diluted in blood samples from healthy individuals, an
approach already in use for breast cancer (23). A potential limitation of the method
is that for particular genes additional methods may be required, in
order to clarify their expression, as identified by the present
study for LHCGR, MGL and VEGFR2. qPCR is a fast, cost-efficient and
easy to perform method, which may be conducted in the majority of
laboratories, and may be a useful tool for CTC detection in various
types of cancer.
Acknowledgments
The current study was supported by the Funding
Program for Research and Teaching at of LMU Munich and Dr Alexandra
Kölbl was supported by the Engelhorn-Foundation for Medical
Research.
Abbreviations:
|
CCNE
|
cyclin E1
|
|
CK19
|
cytokeratin 19
|
|
CLDN-4
|
claudin-4
|
|
CTCs
|
circulating tumour cells
|
|
DEPC
|
diethylpyrocarbonat
|
|
DTCs
|
disseminated tumour cells
|
|
GPER
|
g-protein coupled estrogen
receptor
|
|
Her-2
|
human epithelial growth factor
receptor
|
|
LHCGR
|
luteinizing hormone/choriogonadotropin
receptor
|
|
MAL2
|
T-cell differentiation protein 2
|
|
MGL
|
mammaglobin
|
|
MIG7
|
migration inducing gene 7
|
|
VEGFR
|
vascular endothelial growth factor
receptor
|
References
|
1
|
Emons G and Mallmann P; acting for the
Uterus Commission of AGO: Recommendations for the Diagnosis and
Treatment of Endometrial Cancer, Update 2013. Geburtshilfe
Frauenheilkd. 74:244–247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Husmann G, Kaatsch P, Katalinic A, Bertz
J, Haberland J, Kraywinkel K and Wolf U: Cancer in Germany
2005/2006. Incidence and Trends. 7th edition. Robert Koch Institute
and Association of Population-based Cancer Registries in Germany;
Berlin: 2010
|
|
3
|
Emons G and Heyl W: Hormonal treatment of
endometrial cancer. J Cancer Res Clin Oncol. 126:619–623. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hecht JL and Mutter GL: Molecular and
pathologic aspects of endometrial carcinogenesis. J Clin Oncol.
24:4783–4791. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moodley M and Roberts C: Clinical pathway
for the evaluation of postmenopausal bleeding with an emphasis on
endometrial cancer detection. J Obstet Gynaecol. 24:736–741. 2004.
View Article : Google Scholar
|
|
6
|
Marsden DE and Hacker NF: Optimal
management of endometrial hyperplasia. Best Pract Res Clin Obstet
Gynaecol. 15:393–405. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tumorzentrum M: Malignome des Corpus
Uteri. PD Dr. PMKaPRK Christian Dann ecker : W. Zuckerschwendt
Verlag; Munich, Vienna, New York: 2007, In German.
|
|
8
|
Amant F, Moerman P, Neven P, Timmerman D,
Van Limbergen E and Vergote I: Treatment modalities in endometrial
cancer. Curr Opin Oncol. 19:479–485. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beckmann K, Iosifidis P, Shorne L,
Gilchrist S and Roder D: Effects of variations in hysterectomy
status on population coverage by cervical screening. Aust N Z J
Public Health. 27:507–512. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Franken B, de Groot MR, Mastboom WJ,
Vermes I, van der Palen J, Tibbe AG and Terstappen LW: Circulating
tumor cells, disease recurrence and survival in newly diagnosed
breast cancer. Breast Cancer Res. 14:R1332012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bragado P, Sosa MS, Keely P, Condeelis J
and Aguirre-Ghiso JA: Microenvironments dictating tumor cell
dormancy. Recent Results Cancer Res. 195:25–39. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Diel IJ, Solomayer EF, Costa SD, Gollan C,
Goerner R, Wallwiener D, Kaufmann M and Bastert G: Reduction in new
metastases in breast cancer with adjuvant clodronate treatment. N
Engl J Med. 339:357–363. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pantel K and Woelfle U: Micrometastasis in
breast cancer and other solid tumors. J Biol Regul Homeost Agents.
18:120–125. 2004.PubMed/NCBI
|
|
14
|
Riethdorf S and Pantel K: Disseminated
tumor cells in bone marrow and circulating tumor cells in blood of
breast cancer patients: Current state of detection and
characterization. Pathobiology. 75:140–148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ring A, Smith IE and Dowsett M:
Circulating tumor cells in breast cancer. Lancet Oncol. 5:79–88.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smerage JB and Hayes DF: The measurement
and therapeutic implications of circulating tumor cells in breast
cancer. Br J Cancer. 94:8–12. 2006. View Article : Google Scholar
|
|
17
|
Graves H and Czerniecki BJ: Circulating
tumor cells in breast cancer patients: An evolving role in patient
prognosis and disease progression. Patholog Res Int.
2011:6210902011.PubMed/NCBI
|
|
18
|
Hermanek P, Hutter RV, Sobin LH and
Wittekind C: International union against cancer. Classification of
isolated tumor cells and micrometastasis. Cancer. 86:2668–2673.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singletary SE, Patel-Parekh L and Bland
KI: Treatment trends in early-stage invasive lobular carcinoma: A
report from the national cancer data base. Ann Surg. 242:281–289.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ghossein RA, Carusone L and Bhattacharya
S: Molecular detection of micrometastases and circulating tumor
cells in melanoma prostatic and breast carcinomas. In Vivo.
14:237–250. 2000.PubMed/NCBI
|
|
21
|
Andergassen U, Hofmann S, Kölbl AC,
Schindlbeck C, Neugebauer J, Hutter S, Engelstädter V, Ilmer M,
Friese K and Jeschke U: Detection of tumor cell-specific mRNA in
the peripheral blood of patients with breast cancer-evaluation of
several markers with real-time reverse transcription-PCR. Int J Mol
Sci. 14:1093–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andergassen U, Kölbl AC, Hutter S, Friese
K and Jeschke U: Detection of circulating tumor cells from blood of
breast cancer patients via RT-qPCR. Cancers (Basel). 5:1212–1220.
2013. View Article : Google Scholar
|
|
23
|
Zebisch M, Kölbl AC, Schindlbeck C,
Neugebauer J, Heublein S, Ilmer M, Rack B, Friese K, Jeschke U and
Andergassen U: Quantification of breast cancer cells in peripheral
blood samples by real-time rt-PCR. Anticancer Res. 32:5387–5391.
2012.PubMed/NCBI
|
|
24
|
Bogani G, Liu MC, Dowdy SC, Cliby WA, Kerr
SE, Kalli KR, Kipp BR, Halling KC, Campion MB and Mariani A:
Detection of circulating tumor cells in high-risk endometrial
cancer. Anticancer Res. 35:683–687. 2015.PubMed/NCBI
|
|
25
|
Alonso-Alconada L, Muinelo-Romay L,
Madissoo K, Diaz-Lopez A, Krakstad C, Trovik J, Wik E, Hapangama D,
Coenegrachts L, Cano A, et al: Molecular profiling of circulating
tumor cells links plasticity to the metastatic process in
endometrial cancer. Mol Cancer. 13:2232014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cassia R, Moreno-Bueno G,
Rodriguez-Perales S, Hardisson D, Cigudosa JC and Palacios J:
Cyclin E gene (CCNE) amplification and hCDC4 mutations in
endometrial carcinoma. J Pathol. 201:589–595. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Obermayr E, Sanchez-Cabo F, Tea MK, Singer
CF, Krainer M, Fischer MB, Sehouli J, Reinthaller A, Horvat R,
Heinze G, et al: Assessment of a six gene panel for the molecular
detection of circulating tumor cells in the blood of female cancer
patients. BMC Cancer. 10:6662010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Phillips TM and Lindsey JS: Carcinoma
cell-specific Mig-7: A new potential marker for circulating and
migrating cancer cells. Oncol Rep. 13:37–44. 2005.
|
|
29
|
Kurec AS, Baltrucki L, Mason DY and Davey
FR: Use of the APAAP method in the classification and diagnosis of
hematologic disorders. Clin Lab Med. 8:223–236. 1988.PubMed/NCBI
|
|
30
|
Noack F, Schmitt M, Bauer J, Helmecke D,
Krüger W, Thorban S, Sandherr M, Kuhn W, Graeff H and Harbeck N: A
new approach to phenotyping disseminated tumor cells:
Methodological advances and clinical implications. Int J Biol
Markers. 15:100–104. 2000.PubMed/NCBI
|
|
31
|
Pan XY, Li X, Che YC, Li HY, Li X, Zhang Y
and Yang X: Overexpression of claudin-4 may be involved in
endometrial tumorigenesis. Oncol Lett. 5:1422–1426. 2013.PubMed/NCBI
|
|
32
|
Classen-Linke I, Moss S, Gröting K, Beier
HM, Alfer J and Krusche CA: Mammaglobin 1: Not only a
breast-specific and tumor-specific marker, but also a
hormone-responsive endometrial protein. Histopathology. 61:955–965.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Plante BJ, Lessey BA, Taylor RN, Wang W,
Bagchi MK, Yuan L, Scotchie J, Fritz MA and Young SL: G
protein-coupled estrogen receptor (GPER) expression in normal and
abnormal endo-metrium. Reprod Sci. 19:684–693. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sugimoto T, Koizumi T, Sudo T, Yamaguchi
S, Kojima A, Kumagai S and Nishimura R: Correlative expression of
cyclooxygenase-1 (Cox-1) and human epidermal growth factor receptor
type-2 (Her-2) in endometrial cancer. Kobe J Med Sci. 53:177–187.
2007.
|
|
35
|
Pugh CW and Ratcliffe PJ: Regulation of
angiogenesis by hypoxia: Role of the HIF system. Nat Med.
9:677–684. 2003. View Article : Google Scholar
|
|
36
|
Arcangeli A, Noci I, Fortunato A and
Scarselli GF: The LH/hCG axis in endometrial cancer: A new target
in the treatment of recurrent or metastatic disease. Obstet Gynecol
Int. 2010:pii: 486164. 2010.PubMed/NCBI
|
|
37
|
Davies S, Bax CM, Chatzaki E, Chard T and
Iles RK: Regulation of endometrial cancer cell growth by
luteinizing hormone (LH) and follicle stimulating hormone (FSH). Br
J Cancer. 83:1730–1734. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pike MC, Peters RK, Cozen W, Probst-Hensch
NM, Felix JC, Wan PC and Mack TM: Estrogen-progestin replacement
therapy and endometrial cancer. J Natl Cancer Inst. 89:1110–1116.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Noci I, Pillozzi S, Lastraioli E, Dabizzi
S, Giachi M, Borrani E, Wimalasena J, Taddei GL, Scarselli G and
Arcangeli A: hLH/hCG-receptor expression correlates with in vitro
invasiveness in human primary endometrial cancer. Gynecol Oncol.
111:496–501. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fernández-Shaw S, Shorter SC, Naish CE,
Barlow DH and Starkey PM: Isolation and purification of human
endometrial stromal and glandular cells using immunomagnetic
microspheres. Hum Reprod. 7:156–161. 1992.PubMed/NCBI
|
|
41
|
Zhang L, Rees MC and Bicknell R: The
isolation and long-term culture of normal human endometrial
epithelium and stroma. Expression of mRNAs for angiogenic
polypeptides basally and on oestrogen and progesterone challenges.
J Cell Sci. 108:323–331. 1995.PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
43
|
Dawe CJ, Banfield WG, Morgan WD, Slatick
MS and Curth HO: Growth in continuous culture, and in hamsters, of
cells from a neoplasm associated with acanthossi nigricans. J Natl
Cancer Inst. 33:441–456. 1964.PubMed/NCBI
|
|
44
|
Kuramoto H: Studies of the growth and
cytogenetic properties of human endometrial adenocarcinoma in
culture and its development into an established line. Acta Obstet
Gynaecol Jpn. 19:47–58. 1972.PubMed/NCBI
|
|
45
|
Lessey BA, Ilesanmi AO, Castelbaum AJ,
Yuan L, Somkuti SG, Chwalisz K and Satyaswaroop PG:
Characterization of the functional progesterone receptor in an
endometrial adenocarcinoma cell line (Ishikawa):
Progesterone-induced expression of the alpha1 integrin. J Steroid
Biochem Mol Biol. 59:31–39. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hendricks DT, Taylor R, Reed M and Birrer
MJ: FHIT gene expression in human ovarian, endometrial, and
cervical cancer cell lines. Cancer Res. 57:2112–2115.
1997.PubMed/NCBI
|
|
47
|
Way DL, Grosso DS, Davis JR, Surwit EA and
Christian CD: In Vitro. 19:147–158. 1983. View Article : Google Scholar : PubMed/NCBI
|















