Introduction
In western countries, ovarian cancer can be
attributed to the primary cause of cancer-associated mortality.
Based on the statistical data of the American Cancer Society in
2013, 70–90% of women with ovarian cancer are susceptible to
relapse or metastasis, and ~30% of patients with advanced ovarian
cancer develop peritoneal metastasis (1). The importance of long non-coding RNAs
(lncRNAs) has been increasingly recognized, as they are widely
involved in carcinogenesis, metastasis and drug resistance. Thus,
it is particularly important to examine the association between
ovarian carcinogenesis and the abnormal expression of lncRNAs,
thereby elucidating the complexity of tumor biology.
At present, only a relatively small number of
lncRNAs have been identified, and they have been found to be widely
involved in gene expression at transcriptional and
post-transcriptional levels (2,3).
Previous studies have suggested that lncRNAs serve as competing
endogenous RNAs (ceRNAs) to sponge micro (mi)RNAs, thereby
regulating gene expression (4).
Linc-MD1 has been reported to function as an important ceRNA, which
acts as an effective decoy to protect MyoD mRNA from miRNA-mediated
degradation (5). Furthermore,
lnc-RoR may serve as an important ceRNA to regulate the expression
of pluripotency-associated genes. The liver specific lncRNA, HULC,
has also been found to act as an endogenous 'sponge', thereby
repressing the activities of certain miRNAs, including miR-372
(6). These previous studies
suggest that lncRNAs function as ceRNAs and are involved in ovarian
carcinogenesis.
As a transcript from the HOXC locus, Hox transcript
antisense intergenic RNA (HOTAIR) significantly inhibits the
transcription of HOXD in foreskin fibroblasts (7). In primary breast tumors and breast
cancer metastases, HOTAIR has been found to be significantly
upregulated, which is then involved in the invasiveness and
metastasis of the cancer (8). In
addition, in colorectal cancer, hepatocellular carcinoma and other
types of cancer, HOTAIR is positively associated with malignant
processes and poor outcome (9–12).
However, the specific role of HOTAIR in ovarian carcinoma remains
to be fully elucidated.
In the present study, the level of HOTAIR in ovarian
carcinoma was examined, and the role of HOTAIR in ovarian
carcinogenesis was investigated. The results suggested that HOTAIR
may act as a ceRNA to regulate the expression of Rab22a through the
competitive binding of miR-373. The present study also indicated a
positive correlation between HOTAIR and Rab22a, as well as the
interaction with miR-373, which may assist in elucidating the
molecular mechanism of ovarian carcinogenesis.
Materials and methods
Tissue collection
Ovarian cancer tissues and adjacent non-tumorous
ovarian samples were obtained from 30 Chinese patients (age,
45.8±10.5 years) at Tongji University Hospital affiliated to
Shanghai Tongji University (Shanghai, China) between 2008 and 2010.
Based on histopathological evaluation, all cases were carefully
reviewed as ovarian cancer. None of the patients received adjuvant
therapy and the tissues were collected during biopsy. The present
study was approved by the Research Ethics Committee of Tongji
University Hospital affiliated to Shanghai Tongji University.
Informed consent was obtained from each patient.
Cell lines and culture conditions
The human epithelial ovarian cancer (EOC) cells
(SKOV3, A2780 and CP70), the human ovarian immortalized
nontumorigenic ovarian surface epithelial (IOSE) cells, EOC cells
(HeyC2) and human embryonic kidney (HEK)293T cells were purchased
from American Type Culture Collection (Manassas, VA, USA). The
cells were cultured in RPMI-1640 medium containing 10% fetal bovine
serum (FBS). HEK293T cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) with 10% FBS and 1% penicillin-streptomycin and used for the
dual luciferase reporter assay. All cell lines were maintained at
37°C in a humidified atmosphere with 5% CO2.
RNA isolation and
reverse-transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis
Total RNA from the tissues and cells was extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
RNA quality and concentration were determined using a Nanodrop 2000
system (Thermo Fisher Scientific, Inc.). To analyze the levels of
HOTAIR, a reverse transcription kit (Applied Biosystems, Foster
City, CA, USA) was used, with GAPDH used as the reference. To
measure the level of HOTAIR, a SYBR Premix Ex Taq™ kit (Takara Bio,
Dalian, China) was used, and the expression of GAPDH was used as an
endogenous control. The reaction system contained 2X TaqMan
Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems;
Thermo Fisher Scientific, Inc.), 20X TaqMan MicroRNA Assay mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and template
cDNA. The primers were as follows: Forward, 5′-GCG CTG CAA GTG CTT
ACT GTGCA-3′ and reverse, 5′-CCG AGG TAT TCG CAC TGG ATAC-3′ for
HOTAIR; and forward, 5′-GTC GGT GTG AAC GGA TTTG-3′ and reverse,
5′-AAG ATG GTG ATG GGC TTCC-3′ for GAPDH. The thermal cycling
conditions were as follows: 95°C for 10 min; 40 cycles at 95°C for
15 sec and 60°C for 1 min. RT-qPCR was performed using the Applied
Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems). The
data were analyzed using the 2−ΔΔcq method (13).
Plasmid construction
The cDNA was cloned into a pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.), and miRNA precursors
were cloned into the modified pLL3.7 vector (Invitrogen; Thermo
Fisher Scientific, Inc.). In addition, the 3′-untranslated region
(UTR) of Rab22a and HOTAIR with the predicted potential miRNA
binding sites were cloned into a pLUC luciferase vector (Ambion,
Carlsbad, CA, USA).
Oligonucleotide transfection
The cells (1×106 cells/well) were plated
in a six-well plate 1 day prior to transfection. Using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
the cells were transfected with the plasmid vectors (50 nmol/l).
The HOTAIR small interfering (si)RNA (si-HOTAIR) and negative
control siRNA (si-NC) were purchased from Shanghai Jima Technology
Co., Ltd. The siRNAs were mixed with HiPerFect transfection reagent
(Qiagen GmbH, Hilden, Germany) and incubated at room temperature
for 10 min. The complex was transfected into cells for 48 h.
Dimethyl thiazolyl diphenyl tetrazolium
(MTT) assay
A total of 5,000 cells per well in 100 μl of
medium were seeded in 96-well plates. The cells were then
transfected with pcHOTAIR/pcDNA or miR-373 mimics/NC at a final
concentration of 50 nM. Following transfection, 20 μl of MTT
reagent (Solarbio, Beijing, China) was added into the wells and
incubated with the cells for 4 h at 37°C. The medium was removed
and washed with phosphate-buffered saline (PBS) three times. The
blue formazan was dissolved in 200 μl of dimethyl sulfoxide
(DMSO; Solarbio), and was measured at 550 nm. Wells with cells only
served as a blank control.
Hoechst 33258 staining
Following transfection with pcHOTAIR/pcDNA or
miR-373 mimics/NC for 48 h, the cells were washed with PBS three
times. Subsequently, the cells were incubated with Hoechst 33258
(10 μg/ml; Solarbio) for 5 min. The cells were then washed
with cold PBS three times and observed under a fluorescent
microscope.
Quantification of apoptotic cells
An Annexin V-fluorescein-5-isothiocyanate (FITC)
apoptosis detection kit (BioVision, Inc., Paroo, CA, USA) was
applied to detect apoptotic cells. Following transfection with
miR-373 mimics or NC for 48 h, the cells were placed in a 5 ml
tube. The cells were then washed with cold PBS and resuspended in
1X Annexin V binding buffer containing 10 mM HEPES/NaOH (pH 7.4)
(1×106 cells/ml). The cells were mixed with FITC-Annexin
V (5 μl) and propidium iodide for 15 min at room
temperature. Following washing with PBS three times, the samples
were analyzed using flow cytometry.
Bioinformatics methods
The potential miRNA binding sites of HOTAIR were
predicted using the following websites: 132.77.150.113/pubs/mir07/mir07_prediction.html,
regrna.mbc.nctu.edu.tw/html/prediction.html and
www.microRNA.org.
Cell migration assay
The cells were grown to confluence as a monolayer in
6-well plates. To initiate migration, the cell layer was scratched
using a pipette tip. The cells were subsequently transfected with
miR-373 mimics or negative control. Cell migration was captured
under a microscope (CX21; Olympus Corporation, Tokyo, Japan).
Statistical analysis
The data are expressed as the mean ± standard error
of the mean and statistical significance was analyzed using
Student's t-test (two-tailed). All statistical analyses were
performed using SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) or the
GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla CA, USA)
software packages. P<0.05 was considered to indicate a
statistically significant difference.
Results
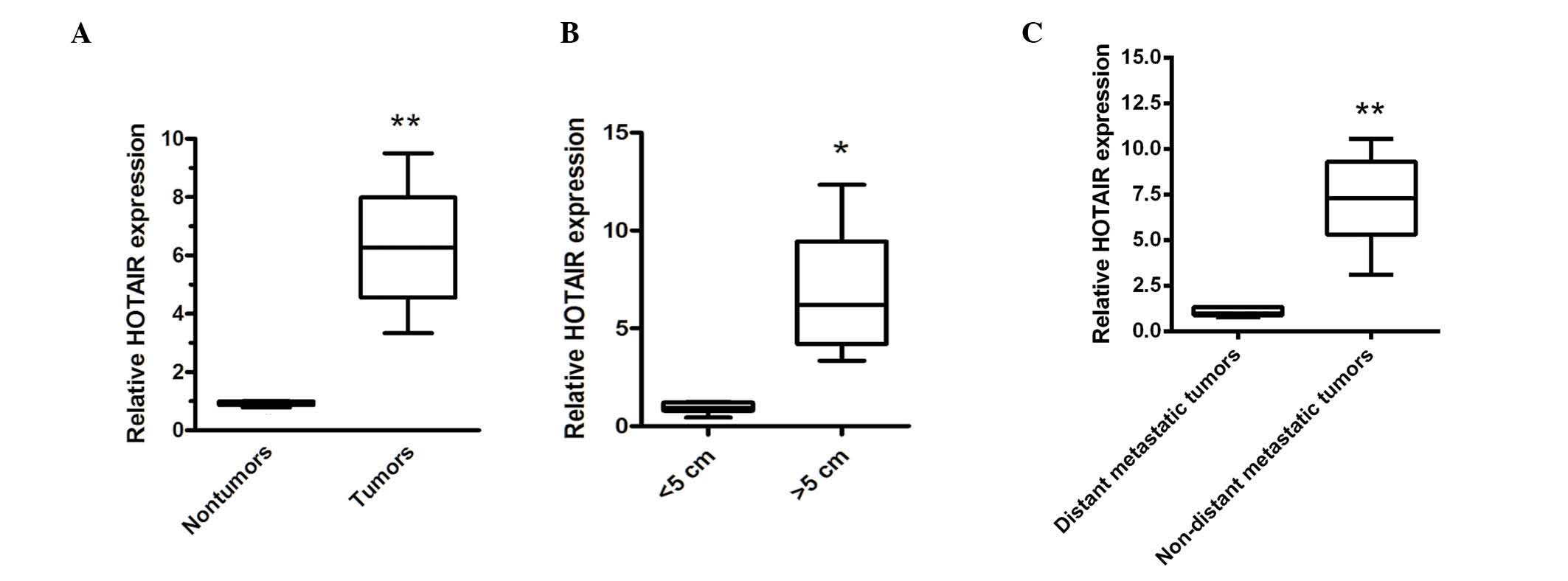
Expression of HOTAIR is upregulated in
human gastric cancer tissues
RT-qPCR was used to examine the expression of HOTAIR
in 30 paired ovarian cancer samples and adjacent, histologically
normal tissues. Compared with the adjacent, histologically normal
tissues, the expression of HOTAIR was significantly enhanced
(Fig. 1A). Furthermore, the
upregulation of HOTAIR was positively correlated with increased
tumor size and distant metastasis (Fig. 1B and C). These data indicated that
the upregulation of HOTAIR was involved in ovarian carcinogenesis
or distant metastasis.
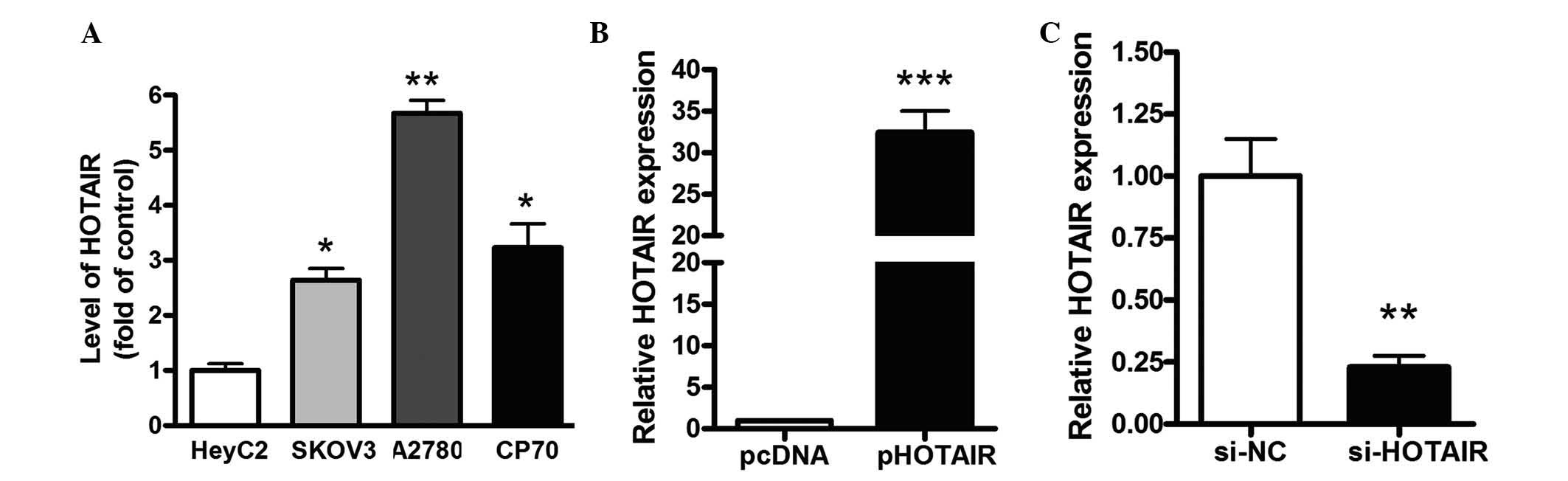
Manipulation of HOTAIR levels in gastric
cancer cells
The expression level of HOTAIR was also examined in
different ovarian cancer cells, including SKOV3, A2780 and CP70
cells, as well as IOSE and EOC cells (HeyC2). As shown in Fig. 2A, the level of HOTAIR in the
ovarian cancer cells was significantly increased, compared with the
normal human EOCs. To examine the role of HOTAIR on cell viability,
a pCDNA/HOTAIR vector was transfected into HeyC2 cells. As shown in
Fig. 2B, the levels of HOTAIR were
increased by >108-fold in the HeyC2 cells transfected with the
pHOTAIR vector, compared with the pcDNA vector. siRNAs targeting
HOTAIR were also transfected into HeyC2 cells. At 48 h
post-transfection, the level of HOTAIR was inhibited by 82% in the
si-HOTAIR2 group, which was identified as the most effective siRNA
targeting HOTAIR (Fig. 2C).
Knockdown of HOTAIR reduces cell
viability and promotes cell apoptosis
In order to determine the effect of HOTAIR on cell
viability, HeyC2 cells were transfected with pHOTAIR or pcDNA for
24, 48 or 72 h. As shown in Fig.
3A, the upregulation of HOTAIR enhanced cell viability in the
HeyC2 cells by 20 and 30% at 48 and 72 h, respectively, whereas the
downregulation of HOTAIR decreased cell viability by 25 and 30% at
48 and 72 h, respectively (Fig.
3B). The effect of HOTAIR on cell apoptosis was also examined.
The results showed that the inhibition of HOTAIR enhanced cell
apoptosis by almost 4-fold, compared with the NC in HeyC2 cells
(14.32±0.78, vs. 3.56±0.32%; Fig.
3C). Cell morphology was also examined using Hoechst 33342
staining. As shown in Fig. 3D, the
number of apoptotic cells increased in the HeyC2 cells transfected
with siRNA targeting HOTAIR. These data suggested that HOTAIR
modulated HeyC2 cell viability and that the downregulation of
HOTAIR induced HeyC2 cell apoptosis.
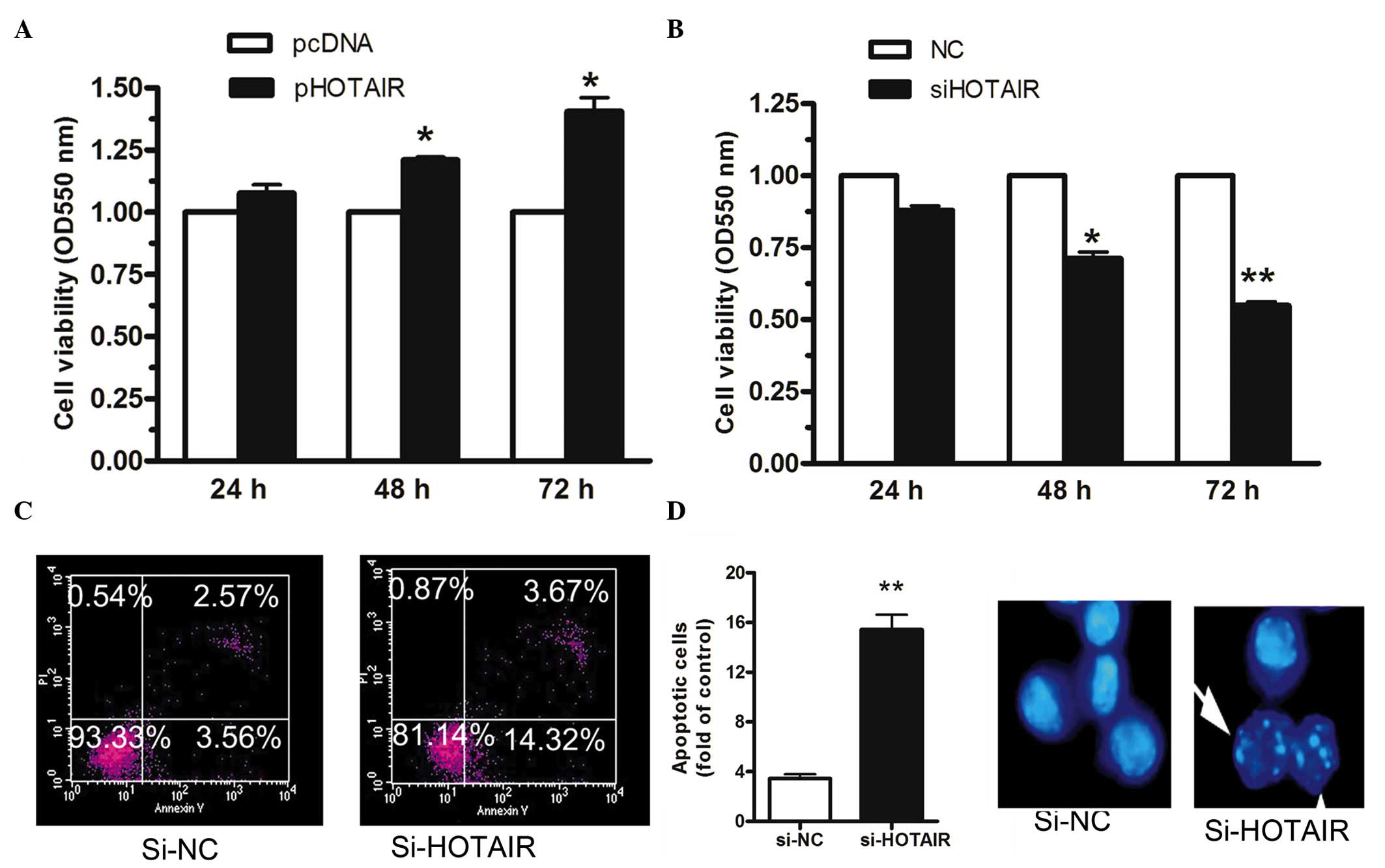 | Figure 3Knockdown of HOTAIR reduces cell
growth and promotes cell apoptosis. The HeyC2 cells were
transfected with pHOTAIR or pcDNA for 24, 48 or 72 h. (A)
Upregulation of HOTAIR enhanced cell viability in HeyC2 cells by 20
and 30% at 48 and 72 h, respectively. (B) Downregulation of HOTAIR
decreased cell viability by 25 and 30% at 48 and 72 h,
respectively. (C) Inhibition of HOTAIR enhanced cell apoptosis by
almost 4-fold, compared with the NC in HeyC2 cells, determined
using an Annexin V and PI kit. (D) Numbers of apoptotic cells
increased in the HeyC2 cells transfected with siRNA targeting
HOTAIR, as examined by Hoechst 33342 staining (magnification, ×40).
The white arrow indicate apoptotic cells. Data are presented as the
mean ± standard error of the mean (n=3). *P<0.05; and
**P<0.01, vs. control. HOTAIR, HOX transcript
antisense intergenic RNA; si, small interfering; NC, negative
control; PI, propidium iodide; OD, optical density. |
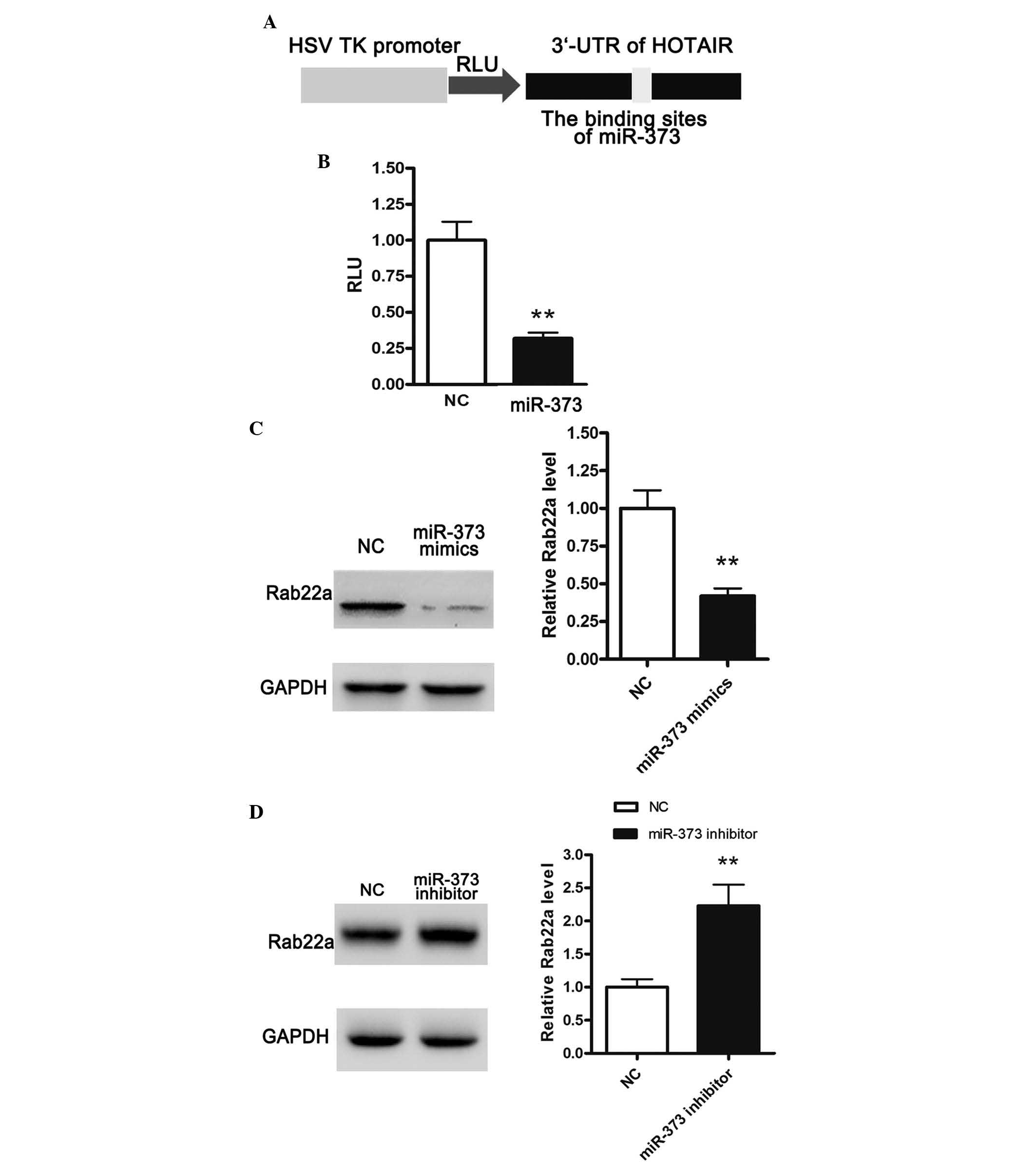
HOTAIR is a target of miR-373 and
positively regulates the expression of Rab22a
Previous studies have indicated that miR-331-3p and
miR-124 can directly bind to the 3′UTR of HOTAIR (14). According to bioinformatics
analysis, a novel miRNA binding site, miR-373 was identified in the
present study. The 3′UTR of HOTAIR was cloned downstream of the
luciferase gene and termed RLuc-HOTAIR (Fig. 4A), which was transfected into cells
together with the miRNA-373-coding plasmid. As shown in Fig. 4B, cotransfection of RLuc-HOTAIR and
pllmiR-373 significantly reduced luciferase activity, suggesting
that HOTAIR was a target gene of miR-373. miR-373 has been reported
to inhibit the expression of Rab22a, a member of the Rab family of
small GTPases (15). Thus, the
present study further examined the effects of miR-373 on the
protein expression of Rab22a. The HeyC2 cells were transfected with
miR-373 mimic or NC. Overexpression of miR-373 significantly
suppressed the expression of Rab22a, while inhibition of miR-373
markedly enhanced the protein expression of Rab22a (Fig. 4C and 4D).
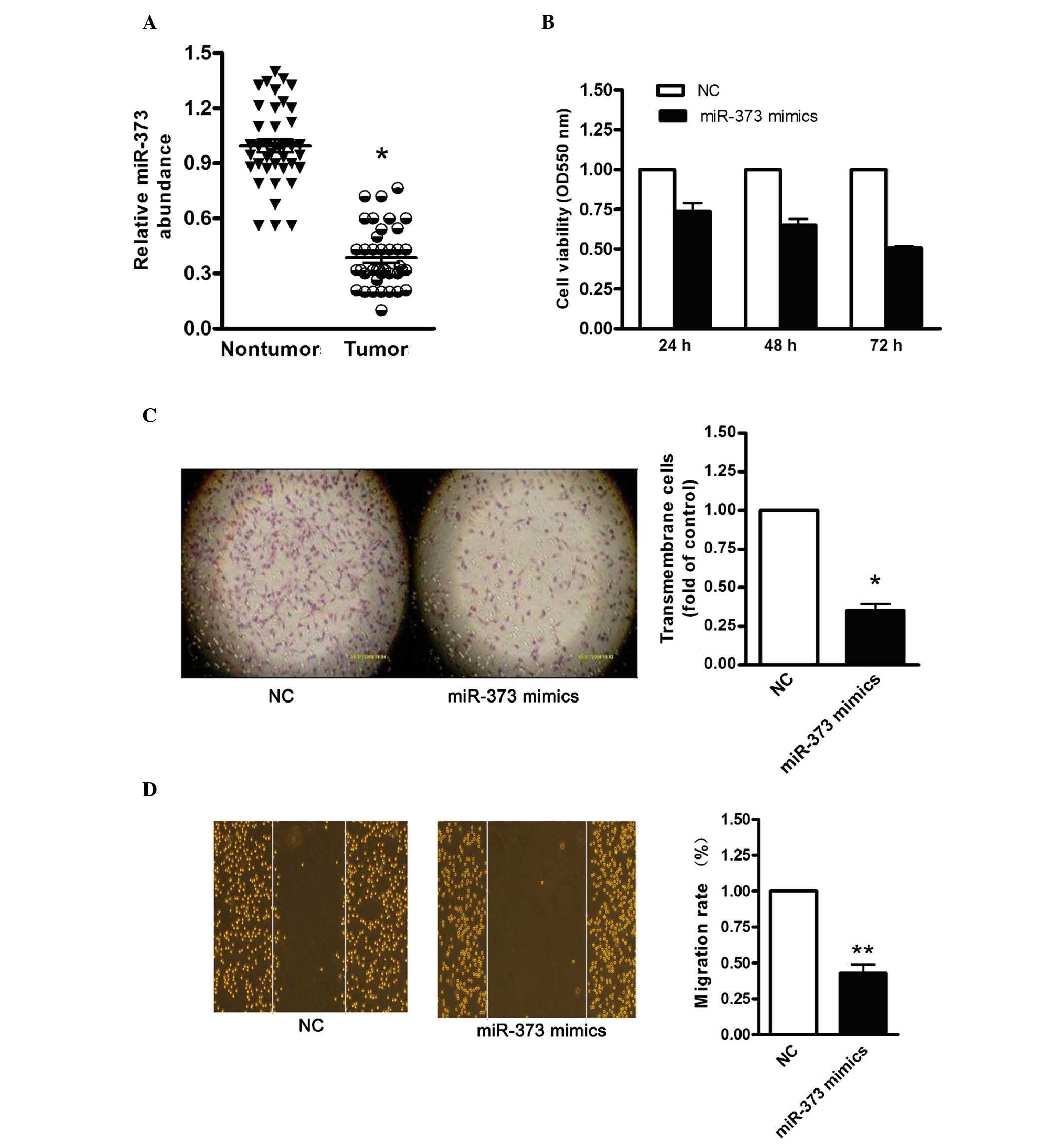
miR-373 suppresses ovarian cancer cell
proliferation
To investigate whether HOTAIR served as an
endogenous sink for target miRNAs, the level of HOTAIR was compared
with the level of miR-373. RT-qPCR analysis demonstrated that the
level of miR-373 was significantly reduced in the 30 pairs of
advanced ovarian cancer tissue (Fig.
5A). To determine the effect of miR-373 on ovarian cancer cell
proliferation, miR-373 was overexpressed in HeyC2 cells. MTT and
colony-formation assays were then performed. As shown in Fig. 5B, cell viability was decreased in
the miR-373-overexpressing cells. In addition, the colony-formation
capacity was significantly reduced following transfection with the
pllmiR-373 vector (Fig. 5C). It
was also found that transfection with the miR-373 mimics
significantly inhibited HeyC2 cell migration (Fig. 5D). These data indicated that the
upregulation of miR-373 significantly reduced ovarian cancer cell
proliferation and migration, which were in accordance with the
results obtained following HOTAIR knockdown in the HeyC2 cells.
Discussion
LncRNAs are >200 nucleotides in length with
limited protein-coding capacity. In disease or developmental
stages, lncRNAs are often differentially expressed, indicating the
importance of lncRNAs in disease pathology (16–18).
The importance of the dysregulation of lncRNAs has been
increasingly reported. For example, through regulating gene
expression, lncRNAs are widely involved in uncontrolled tumor
growth (19–21). Thus, elucidating the role of
lncRNAs may improve current understanding of tumor regulatory
networks.
HOTAIR is an lnRNA, which is defined at the HOXD
locus. RNA fluorescence in situ hybridization assays have
demonstrated that HOTAIR locates to the nucleus and the cytoplasm
(9). A previous study suggested
that, through suppressing polycomb repressive complex 2 (PRC2),
HOTAIR in the nucleus can widely alter gene expression in the whole
genome (9). For example, through
the trimethylation of histone H3 lysine-27 of the HOXC locus,
HOTAIR retargets PRC2 and inhibits several metastasis suppressor
genes, thereby promoting the metastasis of breast cancer cells
(8). In the cytoplasm, HOTAIR can
act as a scaffold to induce ubiquitin-mediated proteolysis
(22). In the present study, the
levels of HOTAIR in ovarian cancer tissue samples and adjacent
non-tumor tissue samples were analyzed. It was found that HOTAIR
was markedly increased in ovarian cancer. In accordance with the
above-mentioned studies, the upregulation of HOTAIR was positively
correlated with ovarian tumor size and metastasis. In addition, the
results of the present study revealed that the upregulation of
HOTAIR significantly enhanced ovarian epithelial cell
proliferation, whereas the inhibition of HOTAIR repressed cell
viability and promoted cell apoptosis. These data indicated that
HOTAIR was important in controlling ovarian carcinogenesis.
Previous studies have indicated that lncRNAs may be
involved in the ceRNA regulatory network (4). In this case, lncRNAs function as
ceRNAs to sponge miRNAs, thereby de-repressing miRNA targets and
affecting post-transcriptional regulation. In human gastric cancer
tissues, HOTAIR is also significantly upregulated; as a sink for
miR-331-3p, HOTAIR positively regulates the expression of human
epithelial growth factor receptor 2 and facilitates the malignant
phenotype of the gastric tumor (14). In the present study, whether HOTAIR
functions as a ceRNA by directly interacting with miRNAs was
investigated. It was found that miR-373 was able to bind with the
3′UTR of HOTAIR, and the RLuc-HOTAIR reporter assay suggested that
miR-373 had a translational repressive role on the expression of
HOTAIR. As an miRNA sponge, the level of HOTAIR is expected to be
higher or comparable to the expression level of miRNA. A previous
study suggested the reduced expression of miR-373 in ovarian cancer
(15), therefore, a negative
correlation was identified between the levels of HOTAIR and
miR-373. In addition, the present study found that the
overexpression of miR-373 significantly repressed ovarian cancer
proliferation, which mimicked the effects of HOTAIR inhibition in
ovarian cancer cells. These results indicated that HOTAIR may be
correlated with miRNA to be involved in ovarian carcinogenesis.
Rab22a is a small GTPase, which belongs to the Rab
family endocytic pathway (23).
Through the recruitment of Rabex-5, Rab22 interacts with Rabex-5
and Rab5, which further modulates cancer cell proliferation through
the integrin-mediated signaling pathway (24–26).
The activation of Rab5 has also been found to enhance transforming
growth factor-β signaling (27),
leading to the epithelial-mesenchymal transition process in the
progression of cancer. The overexpression of miR-373 reduced the
level of Rab22a, whereas the reduction in miR-373 increased the
expression of Rab22a. The present study also identified a positive
correlation between HOTAIR and Rab22a, indicating the interactive
role of miR-373 and HOTAIR in ovarian cancer.
In the present study, specific crosstalk between the
lncRNA HOTAIR and miR-373 was identified. Consistent with HOTAIR
sequestration of miR-373, it was found that the inhibition of
HOTAIR reduced the expression of Rab22a, whereas the overexpression
of HOTAIR enhanced the protein level of Rab22a. These data
suggested that the HOTAIR lncRNA positively regulated the protein
expression of Rab22a through competition for miR-373 binding. In
conclusion, the positive regulation between HOTAIR and Rab22a may
be partially attributed to the ceRNA regulatory network via
miR-373, indicating a potential therapeutic target for ovarian
cancer.
References
|
1
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar
|
|
11
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
12
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo J, Li M, Meng X, Sui J, Dou L, Tang W,
Huang X, Man Y, Wang S and Li J: MiR-291b-3p induces apoptosis in
liver cell line NCTC1469 by reducing the level of RNA-binding
protein HuR. Cell Physiol Biochem. 33:810–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amaral PP, Neyt C, Wilkins SJ,
Askarian-Amiri ME, Sunkin SM, Perkins AC and Mattick JS: Complex
architecture and regulated expression of the Sox2ot locus during
vertebrate development. RNA. 15:2013–2027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ravasi T, Suzuki H, Pang KC, Katayama S,
Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al:
Experimental validation of the regulated expression of large
numbers of non-coding RNAs from the mouse genome. Genome Res.
16:11–19. 2006. View Article : Google Scholar :
|
|
18
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15 (INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar
|
|
20
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoon JH, Abdelmohsen K, Kim J, Yang X,
Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL,
Kreft SG, et al: Scaffold function of long non-coding RNA HOTAIR in
protein ubiquitination. Nat Commun. 4:29392013. View Article : Google Scholar
|
|
23
|
Mesa R, Salomón C, Roggero M, Stahl PD and
Mayorga LS: Rab22a affects the morphology and function of the
endocytic pathway. J Cell Sci. 114:4041–4049. 2001.PubMed/NCBI
|
|
24
|
Zech T and Machesky L: Rab5 and rac team
up in cell motility. Cell. 134:18–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mendoza P, Ortiz R, Diaz J, Quest AF,
Leyton L, Stupack D and Torres VA: Rab5 activation promotes focal
adhesion disassembly, migration and invasiveness in tumor cells. J
Cell Sci. 126:3835–3847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Torres VA, Mielgo A, Barbero S, Hsiao R,
Wilkins JA and Stupack DG: Rab5 mediates caspase-8-promoted cell
motility and metastasis. Mol Biol Cell. 21:369–376. 2010.
View Article : Google Scholar :
|
|
27
|
Hu H, Milstein M, Bliss JM, Thai M,
Malhotra G, Huynh LC and Colicelli J: Integration of transforming
growth factor beta and RAS signaling silences a RAB5 guanine
nucleotide exchange factor and enhances growth factor-directed cell
migration. Mol Cell Biol. 28:1573–1583. 2008. View Article : Google Scholar
|