Introduction
Human glioma, which is the most common and fatal
primary brain tumor, originates from glial cells and accounts for
70% of malignant primary brain tumors (1). In the United States, the estimated
incidence rate of glioma is 20,000 new cases per year (2). Glioma is classified into three major
histological groups, as follows: Well-differentiated low grade
diffuse astrocytoma; anaplastic astrocytoma; and glioblastoma
multiforme, according to the 2007 World Heath Organization (WHO)
classification (3). Despite
progress being made in improving the treatment of glioma, including
the use of aggressive surgery combined with chemotherapy,
radiotherapy and biological therapy, the final prognosis of glioma
remains extremely poor because the tumors are highly proliferative
and invasive (4,5). The average 5-year survival rate of
glioma is 4–5% and mean survival time following diagnosis is 12–15
months (6,7). Therefore, it is crucial to explore
the molecular mechanisms underlying glioma, in order to improve the
prognosis for patients with glioma.
MicroRNAs (miRNAs, miR) have been demonstrated to
alter the expression of genes involved in gliomagenesis (8). miRNAs are a group of endogenous,
small non-coding RNAs 22–25 nucleotides in length, which are
derived from pri- and pre-miRNAs (9). miRNAs regulate the expression of
their target mRNAs through partial complementarity to seed
sequences in the 3′-untranslated region (UTR) of the target gene,
resulting in translational suppression or mRNA cleavage (10). Notably, studies have demonstrated
that miRNAs regulate several important normal biological processes
and pathological processes, including cell proliferation,
apoptosis, differentiation, metabolism and metastasis (11,12).
Calin et al (13)
demonstrated that ~50% of miRNAs are located at fragile sites and
cancer susceptibility locations, thus indicating the potential
functions of miRNAs in tumor formation. Furthermore, it has been
suggested that the expression of miRNAs are often down- or
upregulated in numerous types of cancer, and can function as tumor
suppressors or oncogenes in various tumors (14), including glioma. Therefore, further
exploration of the functions and target mRNAs of miRNAs may provide
insight into the mechanisms of glioma development and progression.
It has also been suggested that miRNAs may be targets for cancer
therapy.
The expression levels of miR-610 were previously
reported to be downregulated in gastric cancer and hepatocellular
carcinoma (15,16). However, to the best of our
knowledge, there have been no studies of miR-610 in glioma. In the
present study, the expression and function of miR-610 in glioma was
examined. The results of the present study demonstrated that
miR-610 was downregulated in human glioma tissues compared with
their normal adjacent tissues (NATs) and normal brain tissues. In
addition, the low expression levels of miR-610 were associated with
WHO grade and the Karnofsky performance status (KPS) of patients
with glioma. Furthermore, miR-610 suppressed cell proliferation,
migration and invasion by directly targeting MDM2 proto-oncogene E3
ubiquitin protein ligase (MDM2). The findings of the present study
have therapeutic implications and may be exploited for the
treatment of glioma.
Materials and methods
Clinical specimens
The tumor specimens used in the present study were
obtained from patients that had undergone surgery at the Second
Xiangya Hospital, Central South University (Changsha, China).
Normal brain tissues (n=5) used in the present study were obtained
from patients with traumatic brain injury requiring a partial
resection of brain tissues to decrease the intracranial pressure.
Informed consent was obtained from all patients. Of the samples
from patients with glioma, 29 cases were low-grade (WHO I and WHO
II) and 32 cases were high-grade (WHO III and WHO IV). Tissues were
snap-frozen in liquid nitrogen and were stored at −80°C. This study
was approved by the institutional review board of the Second
Xiangya Hospital.
Cell lines and cell transfection
The human U251 and U87 glioma cell lines were
purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China). The cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% v/v fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a cell
incubator containing 5% CO2.
Mature miR-610 mimics, miRNA mimics negative control
(NC), miR-610 inhibitor, miRNA mimics negative control inhibitor
(NC inhibitor) and the luciferase reporter plasmid were designed,
synthesized and validated by GeneChem Co., Ltd. (Shanghai, China).
PGL3-MDM2-3′UTR Wt and PGL3-MDM2-3′UTR Mut luciferase reporter
plasmid were designed, synthesized and confirmed by Shanghai
GenePharma Co., Ltd., (Shanghai, China). Cell transfection and
co-transfection was conducted using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The pcDNA3.1-MDM2 plasmid was synthesized by GeneChem
Co., Ltd., and MDM2 cDNA was cloned into a pcDNA plasmid purchased
from GeneChem Co., Ltd. A total of 24 h after co-transfection, the
cells were used for rescue experiments.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from homogenized tissues and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RT was
conducted using Moloney Murine Leukemia Virus Reverse Transcription
system (Promega Corporation, Madison, WI, USA). The reverse
transcription was performed in a 25 µl reaction volume. The
temperature protocol was as follows: 95°C for 2 min; 20 cycles of
94°C for 1 min, 55°C for 1 min and 72°C for 2 min; and 72°C for 5
min. RT-qPCR was performed using a SYBR Premix Ex Taq kit
(Takara Biotechnology Co., Ltd., Dalian, China) and Applied
Biosystems 7500 Real-Time PCR system (Thermo Fisher Scientific,
Inc.) according to the manufacturers' protocols. SYBR Green PCR
master mix (Takara Biotechnology Co., Ltd.) was used to measure the
mRNA expression levels of MDM2. The 20 µl reaction system
contained 10 µl SYBR Green I mix, 2 µl forward
primer, 2 µl reverse primer and 4 µl double distilled
water. The cycling conditions were as follows: 95°C for 10 min; 40
cycles at 95°C for 15 sec; and 60°C for 1 min. U6 and
glyceraldehyde 3-phosphate dehydrogenase were used as reference
genes for miR-610 and MDM2 mRNA expression, respectively. Each
sample was analyzed in triplicate. Primers were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Relative expression
fold changes were calculated using the 2−ΔΔCq method
(17).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
MTT assay was performed to investigate whether
miR-610 had a role in glioma cell proliferation. Post-transfection
with miR-610 or NC mimics and inhibitors, cells were collected and
seeded in 96-well plates at 3,000 cells/well. MTT assay was
performed every 24 h for 6 days. Briefly, 20 µl MTT solution
(Sigma-Aldrich, St. Louis, MO, USA) was added to each well and
incubated for 4 h at 37°C. The culture medium containing MTT
solution was then removed, and 200 µl dimethyl sulfoxide was
added to each well. The absorbance was measured at 490 nm using an
enzyme-linked immunosorbent assay reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). All experiments were analyzed in
triplicate.
Cell migration and invasion assays
Migration and invasion of the human glioma cell
lines were assessed using 8-µm Transwell chambers (Costar,
Corning Incorporated, Corning, NY, USA). For the migration assay,
5×104 transfected cells were harvested and resuspended
in 200 µl serum-free RPMI 1640 medium (Gibco). The cells
were placed into the upper chamber of the Transwell. RPMI 1640
containing 20% FBS (0.5 ml) was added to the lower chamber as a
chemoattractant. For the invasion assay, 5×104
transfected cells were placed into the upper chamber, which was
coated with Matrigel (BD Biosciences, San Jose, CA, USA). RPMI 1640
containing 20% FBS (0.5 ml) was added to the lower chamber as a
chemoattractant. Cells were incubated in the Transwell chambers for
12 and 24 h for the migration and invasion assays, respectively.
The cells remaining on the upper surface of the membranes were
scraped off with cotton swabs and the cells adhering to the lower
surface were fixed with 100% methanol (Shanghai Macklin Biochemical
Co., Ltd., Shanghai, China) for 10 min, stained with 0.5% crystal
violet (Beyotime Institute of Biotechnology, Haimen, China) and
were then counted under an inverted microscope (×200; Olympus
Corporation, Tokyo, Japan) to calculate their relative numbers.
Each experiment was repeated at least three times.
Western blot analysis
A total of 72 h post-transfection, human glioma
cells were washed with ice cold phosphate-buffered saline and lysed
in cold radioimmunoprecipitation lysis buffer (Beyotime Institute
of Biotechnology). The concentrations of the proteins were
determined using a bicinchoninic acid protein assay kit (Beyotime
Institute of Biotechnology). Proteins (30 µg) were then
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and were transferred to a polyvinylidene difluoride
membranes (Beyotime Institute of Biotechnology). After blocking
with 5% skimmed milk in TBS/0.1% Tween (TBST; Beyotime Institute of
Biotechnology) at room temperature for 2 h, the membranes were
incubated with primary antibodies, including mouse anti-human MDM2
monoclonal antibody (1:1,000 dilution; cat no. ab3110) and mouse
anti-human β-actin monoclonal antibody (1:1,000 dilution; cat no.
ab6276; both AbCam, Cambridge, MA, USA), at 4°C for overnight. The
PVDF membranes were washed and then incubated for 1 h with the
corresponding horseradish peroxidase-conjugated secondary antibody
in TBST. The secondary antibody used was goat anti-mouse
horseradish peroxidase conjugated secondary antibody (1:5,000
dilution; ab6789; AbCam). Finally, protein bands were visualized
with an enhanced chemiluminescence kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA) and analyzed using Quantity One software
(version 4.62; Bio-Rad Laboratories, Inc.).
Luciferase assay
The human glioma cells, seeded in a 12-well plate at
~90% confluence, were transfected with the reporter plasmid, and
miR-610 mimics or NC using Lipofectamine 2000. Renilla and
firefly luciferase activity was measured using the Dual-Luciferase
Reporter Assay system (Promega Corporation) and a luminometer
(Tecan Group Ltd., Männedorf, Switzerland) 48 h post-transfection.
The firefly luciferase activity was normalized to the
Renilla luciferase activity for each transfected well. All
the experiments were performed in triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation
and compared with Student's t-test or one-way analysis of variance
using Stata software version 10.0 (StataCorp LP, College Station,
TX, USA). The associations between miR-610 expression levels and
clinicopathological factors were analyzed using the Pearson's chi
square test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-610 expression in glioma tissues and
its association with clinicopathological factors
A total of 61 glioma samples were used in the
present study. cDNA from the samples was subjected to RT-qPCR
analysis to determine the expression levels of miR-610. As
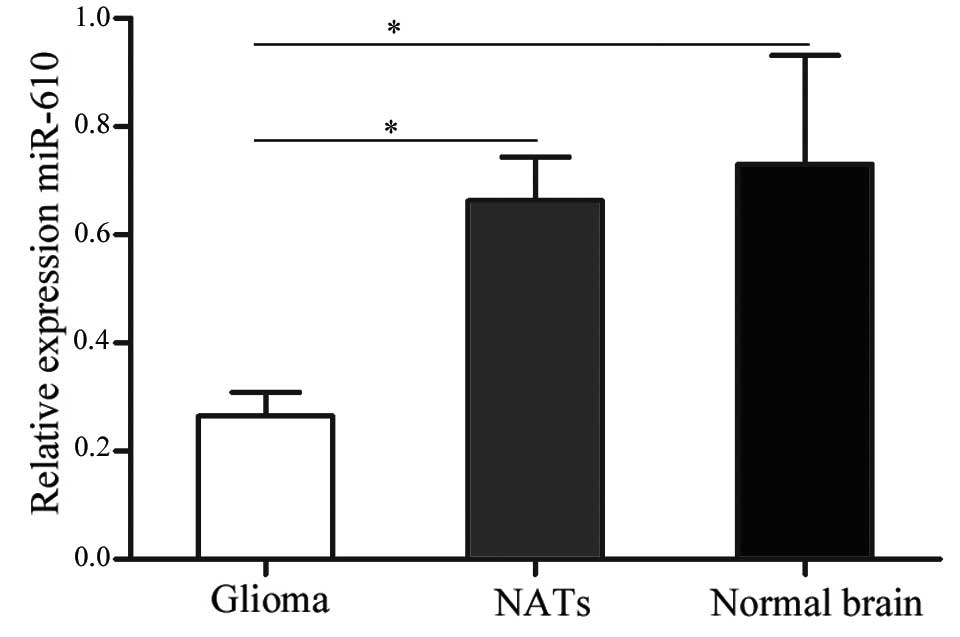
demonstrated in Fig. 1, miR-610
was significantly downregulated in glioma tissues compared with
NATs and normal brain tissues (P<0.05). These results indicate
that miR-610 may have an important function in human glioma.
The present study also determined whether the
expression levels of miR-610 were associated with gender, age,
extent of resection, WHO grade and KPS of patients with glioma. The
statistical analysis demonstrated that miR-610 expression was
significantly associated with WHO grade and KPS of patients with
glioma (P<0.05; Table I).
However, there was no correlation between miR-610 expression and
the other measured clinicopathological factors (gender, age and
extension of resection).
 | Table ICorrelation between miR-610 and
clinicopathological features in patients with glioma. |
Table I
Correlation between miR-610 and
clinicopathological features in patients with glioma.
| | miR-610 level
| |
|---|
| Clinical feature | No. cases | Low (n=39) | High (n=22) | P-value |
|---|
| Gender | | | | 0.791 |
| Male | 34 | 21 | 13 | |
| Female | 27 | 18 | 9 | |
| Age (years) | | | | 0.282 |
| <55 | 26 | 19 | 7 | |
| ≥55 | 35 | 20 | 15 | |
| Extension of
resection | | | | 0.588 |
| Subtotal | 22 | 13 | 9 | |
| Total | 39 | 26 | 13 | |
| KPS | | | | 0.014 |
| ≥80 | 25 | 11 | 14 | |
| <80 | 36 | 28 | 8 | |
| WHO grade | | | | 0.004 |
| I–II | 29 | 13 | 16 | |
| III | 32 | 26 | 6 | |
miR-610 suppresses cell proliferation in
U251 and U87 glioma cells
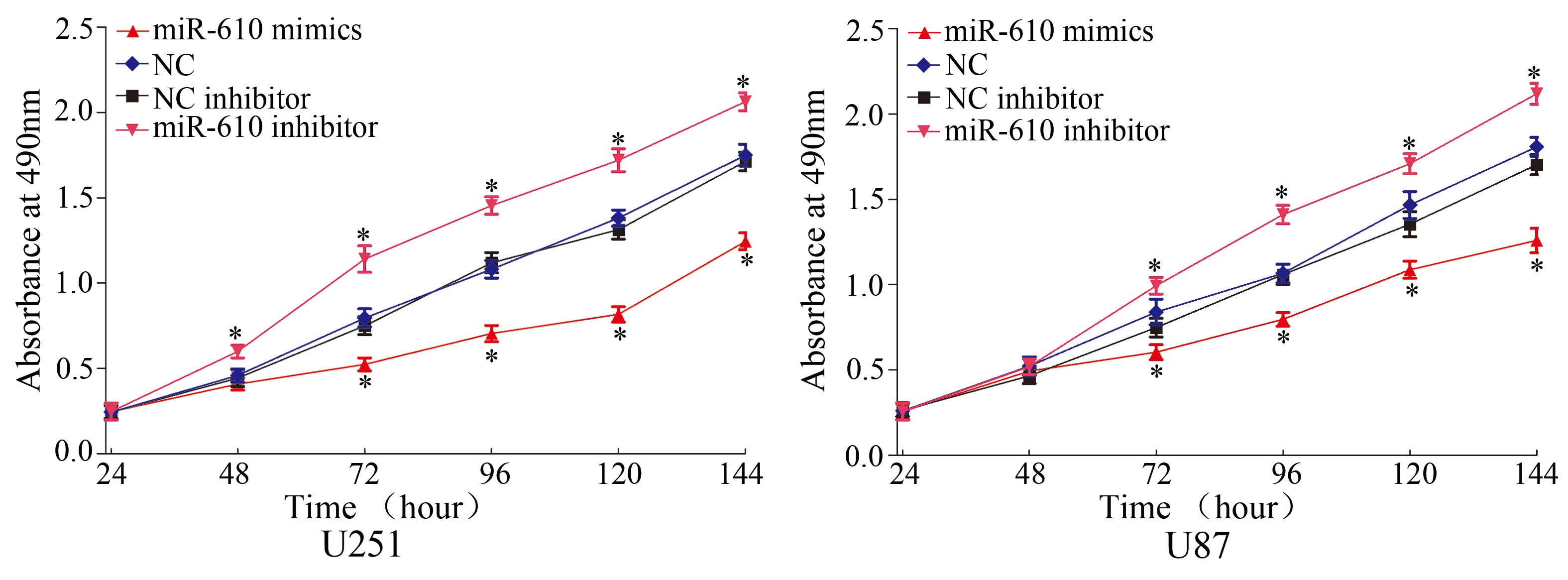
To verify the effects of miR-610 on cell
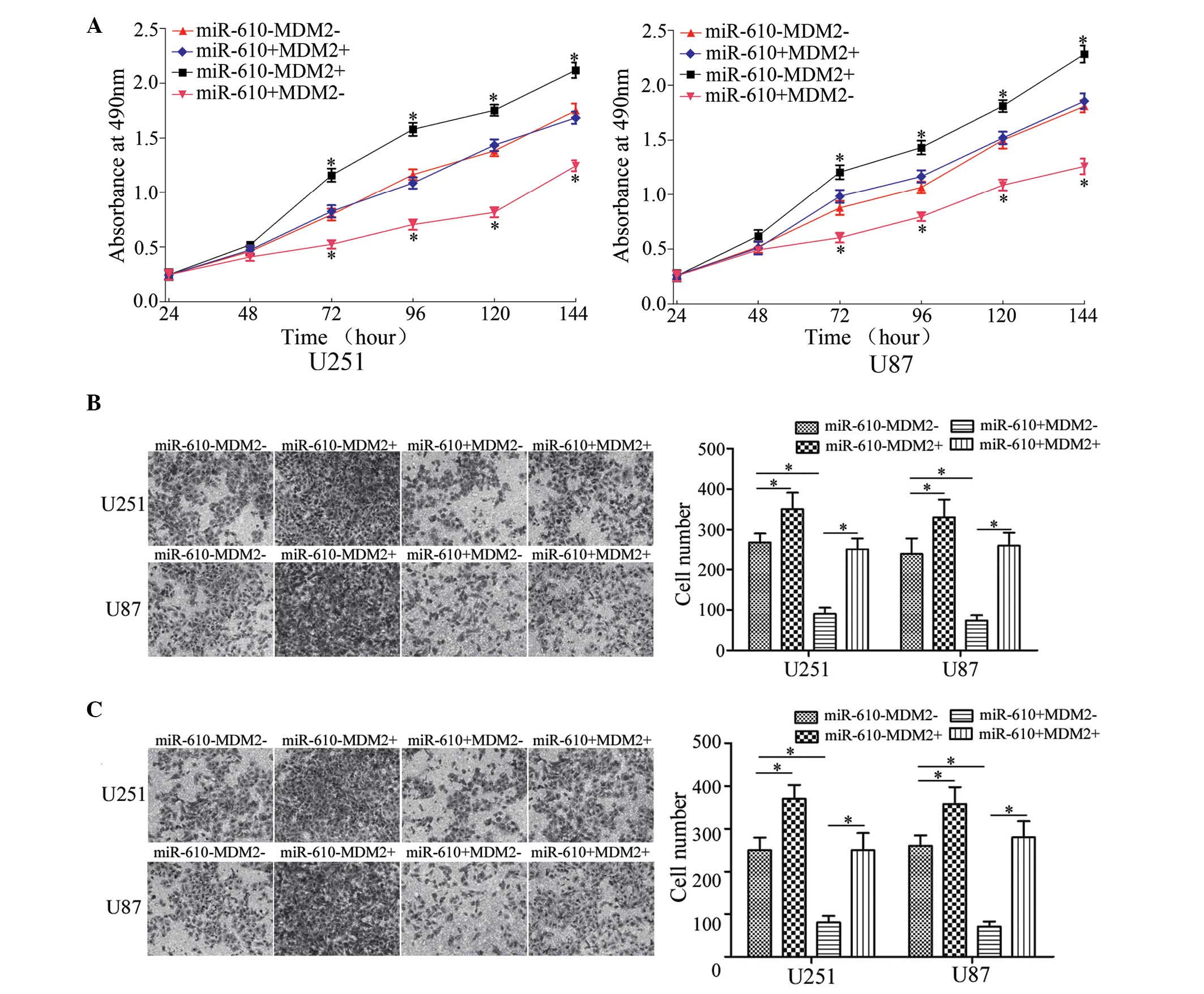
proliferation, an MTT assay was performed. As demonstrated in
Fig. 2, miR-610 mimics
significantly inhibited cell proliferation in U251 and U87 cells
compared with NC (P<0.05). Conversely, miR-610 inhibitor
enhanced cell proliferation in U251 and U87 cells compared with NC
inhibitor (P<0.05). These results suggest that miR-610 functions
as a tumor growth suppressor in glioma.
miR-610 suppresses cell migration and
invasion in U251 and U87 glioma cells
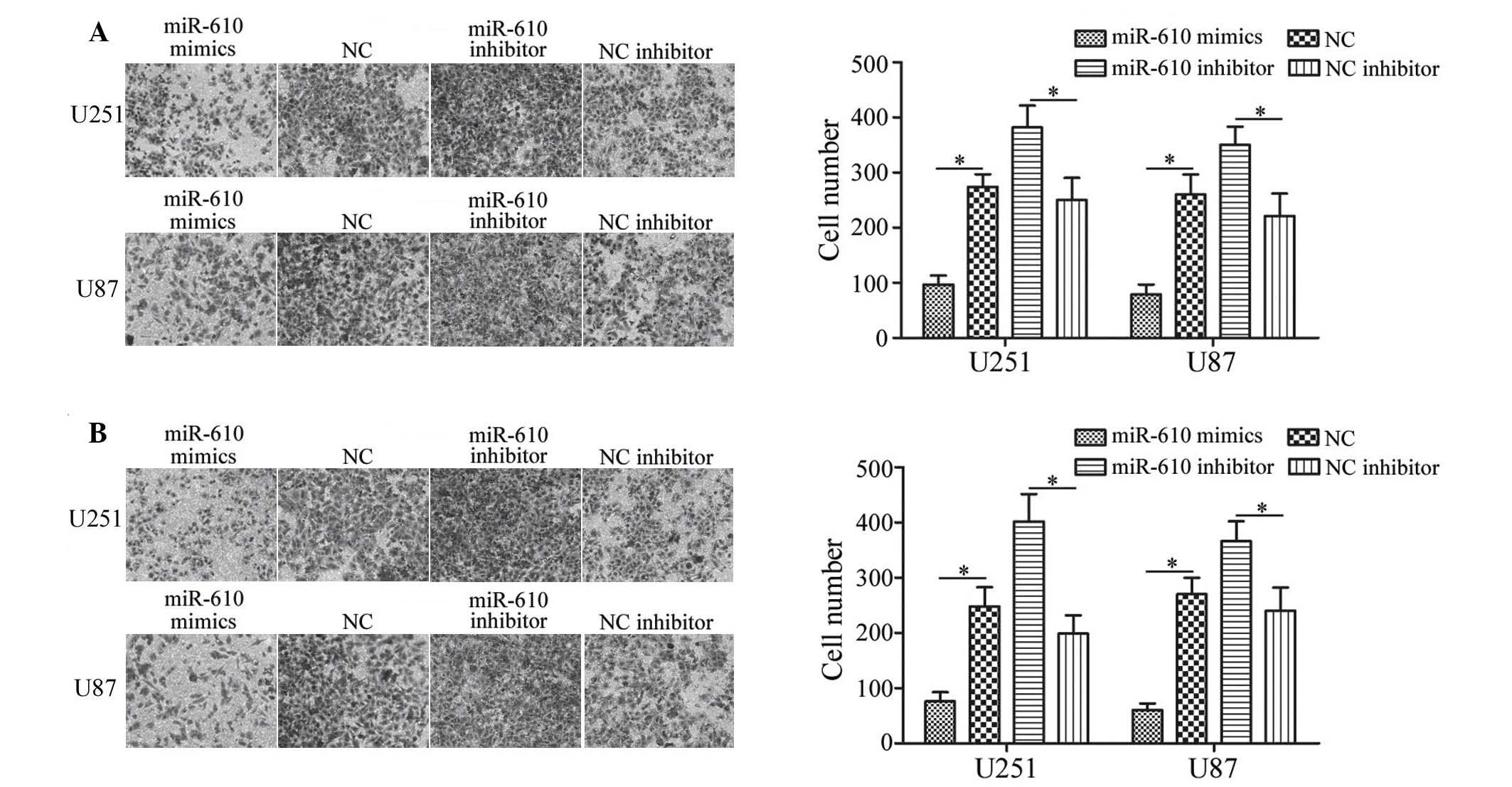
To determine the effects of miR-610 on tumor cell
migration and invasion, Transwell chamber assays were conducted. As
demonstrated in Fig. 3A, miR-610
mimics inhibited U251 and U87 cell migration compared with NC
(P<0.05), whereas miR-610 inhibitor increased the migration of
U251 and U87 cells compared with NC inhibitor. As demonstrated by
invasion assays, the invasiveness of glioma cells transfected with
miR-610 mimics was significantly decreased compared with cells
transfected with NC (P<0.05; Fig.
3B). However, downregulation of miR-610 induced significant
U215 and U87 cell invasion compared with NC inhibitor transfected
cells (P<0.05; Fig. 3B). These
results indicate that miR-610 reduces the migratory and invasive
abilities of glioma cells.
MDM2 is a direct target gene of miR-610
in U251 and U87 glioma cells
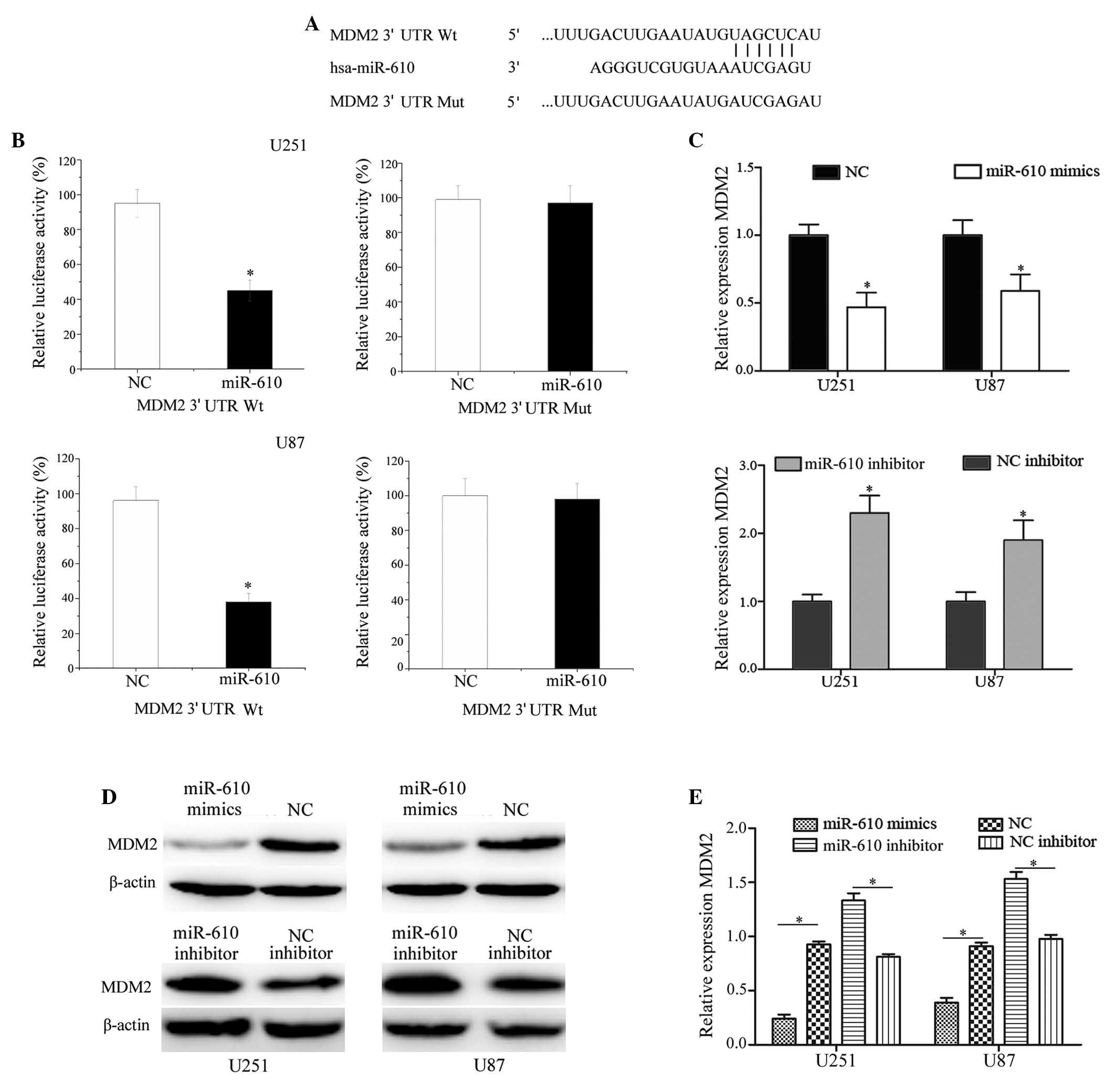
To identify the targets of miR-610 in glioma, a
public database (TargetScan; www.targetscan.org) was used. MDM2 was predicted to be
a target of miR-610 (Fig. 4A). To
verify whether miR-610 directly targets MDM2 mRNA, luciferase
reporter assays were performed. As demonstrated in Fig. 4B, miR-610 significantly inhibited
the luciferase activity of MDM2 wild-type 3′-UTR (P<0.05), but
not MDM2 mutant 3′-UTR compared with NC in U251 and U87 cells.
RT-qPCR and western blot analysis were performed to
establish whether MDM2 was downregulated at the mRNA and protein
level post-transfection of U251 and U87 glioma cells with miR-610
mimics and a miR-610 inhibitor. As presented in Fig. 4C, RT-qPCR analysis demonstrated
that MDM2 was significantly downregulated at the mRNA level in U251
and U87 cells post-transfection with miR-610 mimics compared with
cells transfected with NC (P<0.05). In addition, transfection
with the miR-610 inhibitor significantly increased the MDM2 mRNA
expression levels in U251 and U87 cells compared with the NC
inhibitor (P<0.05). As demonstrated in Fig. 4D and E, compared with NC, the
protein expression levels of MDM2 were significantly downregulated
in U251 and U87 cells post-transfection with miR-610 mimics.
Conversely, the MDM2 protein expression levels were upregulated in
miR-610 inhibitor-transfected U251 and U87 cells compared with
cells transfected with the NC inhibitor (P<0.05). These results
suggest that MDM2 is a direct target gene of miR-610 in
vitro.
miR-610 inhibits glioma cell
proliferation, migration and invasion by targeting MDM2
To explore whether miR-610 inhibited glioma cell
proliferation, migration and invasion by targeting MDM2, U251 and
U87 cells were co-transfected with miR-610 mimics or NC, and MDM2
plasmid or control. As shown in Fig.
5A, transfection with the MDM2 plasmid significantly enhanced
U251 and U87 cell proliferation, compared with the control.
Furthermore, as presented in Fig. 5B
and C, MDM2 plasmid significantly increased the migration and
invasion of U251 and U87 compared with the control plasmid. These
results suggest that the effects of MDM2 plasmid on the growth,
migration and invasion of U251 and U87 cells were similar to those
exerted by the miR-610 inhibitor, indicating that MDM2 is a
functional target of miR-610 in glioma.
Rescue experiments demonstrated that, compared with
miR-610 mimic-transfected cells, glioma cell proliferation,
migration and invasion was restored to normal levels by
co-transfection with miR-610 mimics and MDM2 plasmid (P<0.05).
These results indicate that miR-610 regulates glioma cell
proliferation, migration and invasion by targeting MDM2.
Discussion
miRNAs are important regulators of cell development,
differentiation and carcinogenesis (18). The understanding of the functional
importance of specific miRNAs is steadily increasing; however, it
remains insufficient for numerous identified miRNAs (19). Increasing evidence indicates that
miRNAs are aberrantly expressed in various types of human cancer,
including human glioma; however, their function and molecular
mechanisms in tumorigenesis remain unclear (20–22).
The current study demonstrated that the levels of miR-610 were
downregulated in human glioma and were significantly associated
with WHO grade and the KPS of patients with glioma. In addition,
the present study demonstrated that miR-610 suppresses the
proliferation, migration and invasion of glioma cells.
Zeng et al (16) observed that miR-610 expression was
downregulated in human hepatocellular carcinoma tissues and cell
lines, and was correlated with hepatocellular carcinoma progression
and patient survival. Furthermore, overexpression of miR-610
reduced hepatocellular carcinoma cell proliferation and
Wnt/β-catenin activity via directly suppressing LDL receptor
related protein 6 and transducin β-like protein 1. Wang et
al (15) demonstrated that the
expression levels of miR-610 were reduced in gastric cancer.
Furthermore, miR-610 directly targeted vasodilator-stimulated
phosphoprotein (VASP) and suppressed its expression, resulting in
the inhibition of VASP-mediated cell migration and invasion in
gastric cancer. Therefore, upregulation of miR-610 or the use of
analogous pharmaceutical compounds that exogenously increase
miR-610 expression may be considered effective cancer therapies for
tumors caused by the activation or overexpression of these
oncogenes.
To the best of our knowledge, the results of the
present study provide the first evidence that miR-610 is
downregulated in human glioma, which indicates that miR-610 may
have a tumor suppressive function in gliomagenesis and progression.
In addition, the present study demonstrated that miR-610 reduces
cell proliferation, migration and invasion, also suggesting that
miR-610 has a tumor suppressive function. Therefore, the results
may have future clinical implications.
The present study demonstrated an important
molecular association between miR-610 and MDM2. TargetScan
predicted that MDM2 was a direct target gene of miR-610. It was
demonstrated that MDM2 mRNA contains a miR-610 six-nucleotide seed
match at position 189–195 of the MDM2 3′-UTR. In addition, a
luciferase activity assay revealed that miR-610 directly targeted
the MDM2 3′-UTR, as predicted by bioinformatics. Furthermore, the
present study demonstrated that miR-610 negatively regulates MDM2
expression at the mRNA and protein level in glioma cell lines.
Finally, transfection with the MDM2 plasmid enhanced glioma cell
proliferation, migration and invasion. Rescue experiments
demonstrated that glioma cell proliferation, migration and invasion
were completely restored by MDM2 restoration post-transfection with
miR-610 mimics. Taken together, the findings of the present study
demonstrated that miR-610 is a tumor-suppressive molecule in
gliomagenesis and progression.
The MDM2 gene is a proto-oncogene located on
chromosome 12q13-14, which encodes a 90-kDa nuclear protein
(23). MDM2 was originally
isolated as an amplified sequence in a spontaneously transformed
murine cell line (24). To date,
the overall MDM2 gene amplification frequency is 7%, with the
highest frequency of amplification in soft tissue tumors (20%)
(25,26). It has previously been demonstrated
to be important during tumorigenesis by transfection studies using
the genomic DNA sequence (27–29).
MDM2 appears to act via p53-dependent and -independent mechanisms
of tumorigenesis in human tumors (26). MDM2 was previously observed to be
significantly upregulated in human glioma samples compared with the
control group, thus indicating that overexpression of MDM2 may be
important during gliomagenesis and progression. The upregulation of
MDM2 was previously demonstrated to be an early event in the
malignant transformation of glioma (30). Therefore, MDM2 deserves close
scrutiny as a potential target for inhibition in human glioma. The
results of the present study suggested that miR-610 suppresses the
proliferation, migration and invasion of glioma cells by directly
targeting MDM2, indicating that miR-610 may be investigated as a
targeted therapy for glioma treatment.
In conclusion, to the best of our knowledge, the
present study is the first to demonstrate that miR-610 is
downregulated in glioma, and associated with WHO grade and the KPS
of patients with glioma. The current study also demonstrated that
miR-610 suppresses cell proliferation, migration and invasion by
directly targeting MDM2 in glioma cells. The identification of a
target gene of miR-610 may further the understanding of the
carcinogenic mechanisms of glioma. The findings of the present
study indicated that miR-610 may be important as a diagnostic
marker in glioma and may be exploited for the treatment of glioma.
Further work is required to assess the potential of miR-610 as a
treatment for glioma and other types of cancer.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar
|
|
2
|
Davis FG and McCarthy BJ: Current
epidemiological trends and surveillance issues in brain tumors.
Expert Rev Anticancer Ther. 1:395–401. 2001. View Article : Google Scholar
|
|
3
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gu JJ, Gao GZ and Zhang SM: miR-218
inhibits the migration and invasion of glioma U87 cells through the
Slit2-Robo1 pathway. Oncol Lett. 9:1561–1566. 2015.PubMed/NCBI
|
|
5
|
Caruso C, Carcaterra M and Donato V: Role
of radiotherapy for high grade gliomas management. J Neurosurg Sci.
57:163–169. 2013.PubMed/NCBI
|
|
6
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang BC and Ma J: Role of microRNAs in
malignant glioma. Chin Med J (Engl). 128:1238–1244. 2015.
View Article : Google Scholar
|
|
9
|
Wang LG, Ni Y, Su BH, Mu XR, Shen HC and
Du JJ: MicroRNA-34b functions as a tumor suppressor and acts as a
nodal point in the feedback loop with Met. Int J Oncol. 42:957–962.
2013.PubMed/NCBI
|
|
10
|
Shen J, Niu W, Zhou M and Zhang H, Ma J,
Wang L and Zhang H: MicroRNA-410 suppresses migration and invasion
by targeting MDM2 in gastric cancer. PLoS One. 9:e1045102014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuhn AR, Schlauch K, Lao R, Halayko AJ,
Gerthoffer WT and Singer CA: MicroRNA expression in human airway
smooth muscle cells: Role of miR-25 in regulation of airway smooth
muscle phenotype. Am J Respir Cell Mol Biol. 42:506–513. 2010.
View Article : Google Scholar :
|
|
12
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
96(Suppl): R40–R44. 2007.PubMed/NCBI
|
|
13
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Zhang J, Wu J, Luo D, Su K, Shi W,
Liu J, Tian Y and Wei L: MicroRNA-610 inhibits the migration and
invasion of gastric cancer cells by suppressing the expression of
vasodilator-stimulated phosphoprotein. Eur J Cancer. 48:1904–1913.
2012. View Article : Google Scholar
|
|
16
|
Zeng XC, Liu FQ, Yan R, Yi HM, Zhang T,
Wang GY, Li Y and Jiang N: Downregulation of miR-610 promotes
proliferation and tumorigenicity and activates Wnt/β-catenin
signaling in human hepatocellular carcinoma. Mol Cancer.
13:2612014. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Ichimi T, Enokida H, Okuno Y, Kunimoto R,
Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama
K, et al: Identification of novel microRNA targets based on
microRNA signatures in bladder cancer. Int J Cancer. 125:345–352.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dyrskjøt L, Ostenfeld MS, Bramsen JB,
Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL,
Andersen CL, Zieger K, et al: Genomic profiling of microRNAs in
bladder cancer: miR-129 is associated with poor outcome and
promotes cell death in vitro. Cancer Res. 69:4851–4860. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu XP, Mou KJ, Xu QF, Tang JH, Huang GH,
Xu JP, Li GH, Ai SJ, Hugnot JP, Zhou Z and Lv SQ: Microarray
analysis of the aberrant microRNA expression pattern in gliomas of
different grades. Oncol Rep. 34:318–324. 2015.PubMed/NCBI
|
|
21
|
Xu D, Ma P, Gao G, Gui Y, Niu X and Jin B:
MicroRNA-383 expression regulates proliferation, migration,
invasion, and apoptosis in human glioma cells. Tumour Biol.
36:7743–7753. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun JY, Xiao WZ, Wang F, Wang YQ, Zhu YH,
Wu YF, Miao ZL and Lin YC: MicroRNA-320 inhibits cell proliferation
in glioma by targeting E2F1. Mol Med Rep. 12:2355–2359.
2015.PubMed/NCBI
|
|
23
|
Nakayama T, Toguchida J, Wadayama B, Kanoe
H, Kotoura Y and Sasaki MS: MDM2 gene amplification in bone and
soft-tissue tumors: Association with tumor progression in
differentiated adipose-tissue tumors. Int J Cancer. 64:342–346.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haines DS, Landers JE, Engle LJ and George
DL: Physical and functional interaction between wild-type p53 and
mdm2 proteins. Mol Cell Biol. 14:1171–1178. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Momand J, Wu HH and Dasgupta G: MDM2 -
master regulator of the p53 tumor suppressor protein. Gene.
242:15–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Momand J, Jung D, Wilczynski S and Niland
J: The MDM2 gene amplification database. Nucleic Acids Res.
26:3453–3459. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Finlay CA: The mdm-2 oncogene can overcome
wild-type p53 suppression of transformed cell growth. Mol Cell
Biol. 13:301–306. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leach FS, Tokino T, Meltzer P, Burrell M,
Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW and Vogelstein
B: p53 Mutation and MDM2 amplification in human soft tissue
sarcomas. Cancer Res. 53:2231–2234. 1993.PubMed/NCBI
|
|
29
|
Oliner JD, Kinzler KW, Meltzer PS, George
DL and Vogelstein B: Amplification of a gene encoding a
p53-associated protein in human sarcomas. Nature. 358:80–83. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang AL, Liu ZX, Li G and Zhang LW:
Expression and significance of P53 protein and MDM-2 protein in
human gliomas. Chin Med J (Engl). 124:2530–2533. 2011.
|