Introduction
The Wilms tumor suppressor gene WT1 was first
identified due to its essential role in the normal development of
the human genitourinary system (1). WT1 functions as a transcription
factor regulating target gene expression (1). In addition, WT1 was revealed to
regulate the expression of genes involved in the Wnt signaling
pathway via a genome-wide screening analysis (2).
Wilms tumor 1 associated protein (WTAP) was
demonstrated to interact with WT1 using a yeast two-hybrid
screening (3). WTAP and WT1 were
observed to localize to the nucleoplasm and nuclear speckles, where
they were partially co-localized with splicing factors (3). WTAP is the mammalian homolog of the
Drosophila gene female-lethal(2) D
[fl(2)D], and fl(2)D is involved in activating
female-specific patterns of alternative splicing of sex-lethal and
transformer pre-mRNA (4,5). Previous studies demonstrated that
WTAP-null and heterozygous mice succumbed between embryonic day 6.5
and 10.5, and exhibited marked defects in cell proliferation, which
resulted in defects in endoderm and mesoderm formation (6,7).
Furthermore, it has been demonstrated in mice that WTAP is required
for G2/M cell cycle transition by stabilizing the cyclin
A2 mRNA and, thus, is vital in early development (6). In addition, WTAP is involved in the
function of human spliceosomes (8). Recently, it was demonstrated that
WTAP and virilizer are subunits of the N6-methyladenosine
methylation complex, which regulates mRNA stability (9,10).
A limited number of studies have been performed on
the role of WTAP in tumor genesis. WTAP is overexpressed in
cholangiocarcinoma and WTAP expression was observed to correlate
with metastasis (11). In
addition, WTAP was demonstrated to be overexpressed in
glioblastoma, and regulated glioblastoma cell migration and
invasion (12). Bansal et
al (13) identified WTAP as an
oncogenic protein in acute myeloid leukemia. Carbonic anhydrase 4,
a novel tumor suppressor in colorectal cancer, inhibited the Wnt
signaling pathway by targeting the WTAP-WT1-transducin β-like 1
axis (14). The aim of the present
study was to investigate the role of WTAP in tumor formation by
identifying novel WTAP genes in vertebrate genomes. The expression
of these genes in healthy and tumor tissue samples was determined,
and functionally relevant single nucleotide polymorphisms (SNPs)
and somatic mutations in WTAP were identified. Conserved
transcription factor binding sites within the promoter region of
the human WTAP gene were identified. Furthermore, meta-analysis of
the prognostic value of WTAP gene expression in various cancers was
performed.
Materials and methods
Identification of novel WTAP genes in
vertebrate genomes and transcription factor-binding sites
The DNA and amino acid sequences of novel vertebrate
WTAPs were obtained by searching Ensembl genome databases
(ensembl.org/index.html) using
orthologous and paralogous associations. The prospective WTAP
sequences were confirmed using the Basic Local Alignment Search
Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) (15–18).
Conserved transcription factor-binding sites in the promoter
regions of the human WTAP gene were identified from the
SABiosciences' proprietary database, DECODE (Qiagen, Inc.,
Valencia, CA, USA), which combines text mining with the University
of California, Santa Cruz genome browser data (19–21).
Comparative proteomic analyses of WTAP
proteins
The amino acid sequences of identified vertebrate
WTAPs were aligned using ClustalW (ebi.ac.uk/Tools/msa/). A maximum likelihood (ML) tree
of vertebrate WTAPs was constructed using Molecular Evolutionary
Genetics Analysis version 5.05 (megasoftware.net/) with the optimal model (Kimura
2-parameter model). The relative support of internal nodes was
determined by bootstrap analyses with 1,000 replications for ML
reconstructions (22). The
program, codeml within the Phylogenetic Analysis by ML version 4.7
software package (abacus.gene.ucl.ac.uk/software/paml.html) was used to
investigate whether WTAP proteins were positively selected
(23).
Identification of functionally relevant
SNPs in the human WTAP gene and somatic mutations in human
cancer
The functionally relevant SNPs of the human WTAP
gene were extracted from the Ensembl genome databases and the Short
Genetic Variations database (ncbi.nlm.nih.gov/snp), as previously described
(15–21). The SNPs causing missense mutations
were then identified. The somatic mutations of the human WTAP gene
in cancer tissues were extracted from the Catalogue of Somatic
Mutations in Cancer (COSMIC) database (cancer.sanger.ac.uk/cosmic), which mines somatic
mutations in complete cancer genomes (24).
In silico expression analyses of the
human WTAP gene
Expression profiles of the human WTAP gene in normal
tissues were obtained from the GeneAnnot (genecards.weizmann.ac.il/geneannot/index.shtml)
(25) and ArrayExpress (ebi.ac.uk/arrayexpress/) databases (26).
Meta-analysis of the prognostic value of
human WTAP gene expression in cancer tissues
The expression of the WTAP gene and the biological
association between gene expression and prognosis were determined
by inputting human WTAP gene (NP_001257460) into the PrognoScan
database (prognoscan.org/) (27).
Results
Comparative proteomics of WTAP proteins
identified in vertebrate genomes
WTAP DNA and protein sequences were collected from
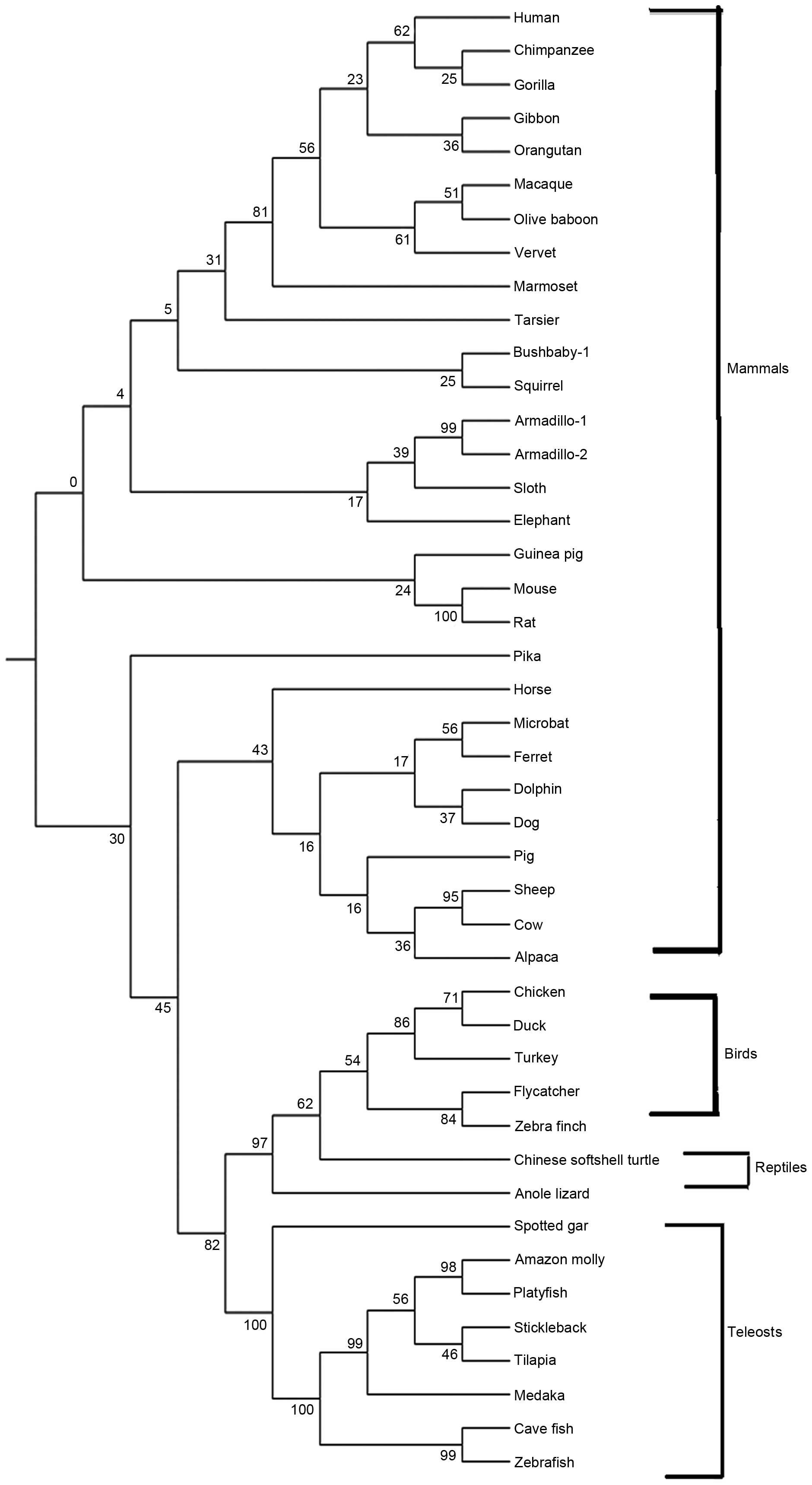
the Ensembl genome database and confirmed by BLASTing. Completed
WTAP genes were identified in the following genomes: Human,
chimpanzee, gibbon, macaque, gorilla, orangutan, olive baboon,
vervet monkey, marmoset, tarsier, bush baby, armadillo, sloth,
squirrel, elephant, guinea pig, mouse, rat, pika, horse, microbat,
ferret, dolphin, dog, pig, sheep, cow, alpaca, chicken, duck,
turkey, flycatcher, zebra finch, Chinese softshell turtle, anole
lizard, spotted gar, Amazon molly, platyfish, stickleback, tilapia,
medaka, cave fish and zebrafish. In the armadillo genome, two WTAP
genes were identified. The maximum likelihood method was used to
construct the phylogenetic tree of vertebrate WTAPs (Fig. 1). The vertebrate WTAP genes
clustered into the primate, rodent and teleost lineages.
Furthermore, site-specific analysis for positive selection with six
models of codon substitution, M0 (one-ratio), M1a (nearly neutral),
M2a (positive selection), M3 (discrete), M7 (β), and M8 (β and ω)
were performed in vertebrate, mammalian, bird, reptile and teleost
lineages. No sites were identified under positive selection with
any models in the various WTAP groups. Therefore, it was concluded
that WTAP proteins were under purifying selection (data not
shown).
Expression profile of the human WTAP
gene
Investigation of available microarray data revealed
that the human WTAP gene was predominantly expressed in the
following tissues: Bone marrow, whole blood, lymph node, brain,
cerebellum, retina, spinal cord, heart, smooth muscle, skeletal
muscle, small intestine, colon, adipocyte, kidney, liver, lung,
pancreas, thyroid, salivary gland, adrenal gland, skin, ovary,
uterus, placenta, prostate and testis. In addition, the human WTAP
gene was expressed in the following types of cancer: Bladder,
blood, brain, breast, colorectal, esophagus, eye, head and neck,
lung, ovarian, prostate, skin and soft tissue.
Comparative genomics on the human WTAP
gene
Signal transducer and activator of transcription 1
(STAT1), forkhead box protein O1 (FOXO1), interferon regulatory
factor 1 (IRF1), glucocorticoid receptor and peroxisome
proliferator-activated receptor γ (PPARγ) transcription factor
binding sites were identified in the upstream (promoter) region of
the WTAP gene.
Functionally relevant SNP identification
in the human WTAP gene
A total of 1,347 available SNPs were identified in
the human WTAP gene. Among these, 19 SNPs were functionally
relevant, causing missense mutations (Table I).
 | Table IFunctionally relevant SNP
identification in the human WTAP gene. |
Table I
Functionally relevant SNP
identification in the human WTAP gene.
| SNP | Chr 6 position
sequence | Sequence | Type | Amino acid |
|---|
| rs1543500 | 159,755,455(+) |
GGCAG(G/T)GAAAA | Missense | S (Ser) ⇒ R
(Arg) |
| rs112093927 | 159,753,489(+) |
GAAGT(A/G)TCGAA | Missense | C (Cys) ⇒ Y
(Tyr) |
| rs140439442 | 159,755,519(+) |
AGAAA(A/G)CAGTG | Missense | A (Ala) ⇒ T
(Thr) |
| rs146208471 | 159,755,499(+) |
CAGTC(A/G)TGACC | Missense | H (His) ⇒ R
(Arg) |
| rs144625269 | 159,755,303(+) |
ACAGG(A/G)AGGGC | Missense | E (Glu) ⇒ K
(Lys) |
| rs148080007 | 159,739,015(+) |
CTTCA(A/G)AGTTA | Missense | K (Lys) ⇒ R
(Arg) |
| rs149103382 | 159,755,447(+) |
CTCCC(A/G)CGGGC | Missense | T (Thr) ⇒ A
(Ala) |
| rs150215853 | 159,743,764(+) |
AGCAA(C/G)CAAGG | Missense | T (Thr) ⇒ S
(Ser) |
| rs187127278 | 159,755,555(+) |
GTTCC(C/T)GCCAC | Missense | R (Arg) ⇒ C
(Cys) |
| rs373161821 | 159,755,405(+) |
GTTAC(A/G)TAAAT | Missense | V (Val) ⇒ I
(Ile) |
| rs375163417 | 159,755,312(+) |
GCAAC(A/G)CAACC | Missense | T (Thr) ⇒ A
(Ala) |
| rs375840138 | 159,755,421(+) |
CAGTG(C/T)GGGGT | Missense | A (Ala) ⇒ V
(Val) |
| rs528166112 | 159,755,353(+) |
GGTAA(A/T)AAGTC | Missense | N (Asn) ⇒ K
(Lys) |
| rs532857576 | 159,748,230(+) |
CGAGC(A/G)TTGCC | Missense | V (Val) ⇒ I
(Ile) |
| rs543667028 | 159,755,190(+) |
AGCTT(C/T)TGAAC | Missense | S (Ser) ⇒ F
(Phe) |
| rs546830649 | 159,755,349(+) |
GAATG(A/G)TAATA | Missense | G (Gly) ⇒ D
(Asp) |
| rs558627827 | 159,742,104(+) |
ATGAA(A/G)CATAT | Missense | A (Ala) ⇒ T
(Thr) |
| rs560214098 | 159,748,326(+) |
GCATC(A/G)TCTGC | Missense | Q (Gln) ⇒ K
(Lys) |
| rs563264867 | 159,755,400(+) |
TAGTG(A/G)TTACG | Missense | G (Gly) ⇒ D
(Asp) |
Identification of somatic mutations of
the WTAP gene in human cancer
By searching the COSMIC database, 65 somatic
mutations of the human WTAP gene were identified in various types
of cancer tissue (Table II).
 | Table IISomatic mutations of WTAP in cancer
tissues. |
Table II
Somatic mutations of WTAP in cancer
tissues.
| Position (AA) | Mutation (CDS) | Mutation (amino
acid) | Mutation ID
(COSM) | Count | Mutation type |
|---|
| 12 | c.34C>T | p.R12a | COSM207378 | 3 |
Substitution-nonsense |
| 30 | c.90G>A | p.W30a | COSM223136 | 1 |
Substitution-nonsense |
| 32 | c.96A>T | p.Q32H | COSM3622455 | 1 |
Substitution-missense |
| 33 | c.98A>G | p.Y33C | COSM4930374 | 1 |
Substitution-missense |
| 35 | c.104C>T | p.A35V | COSM4445039 | 1 |
Substitution-missense |
| 44 | c.132C>A | p.Y44a | COSM450822 | 1 |
Substitution-nonsense |
| 51 | c.152A>C | p.D51A | COSM450823 | 1 |
Substitution-missense |
| 54 | c.161G>A | p.G54D | COSM3860103 | 1 |
Substitution-missense |
| 66 | c.198G>A | p.Q66Q | COSM1075534 | 1 | Substitution-coding
silent |
| 71 | c.211C>T | p.R71C | COSM3860104 | 1 |
Substitution-missense |
| 79 | c.235C>T | p.R79a | COSM1441924 | 1 |
Substitution-nonsense |
| 79 | c.236G>A | p.R79Q | COSM450824 | 1 |
Substitution-missense |
| 86 | c.256G>A | p.E86K | COSM741338 | 1 |
Substitution-missense |
| 91 | c.271A>G | p.T91A | COSM1496256 | 1 |
Substitution-missense |
| 95 | c.283C>T | p.Q95a | COSM1621161 | 2 |
Substitution-nonsense |
| 103 | c.307C>T | p.P103S | COSM1075535 | 1 |
Substitution-missense |
| 104 | c.312C>T | p.S104S | COSM1441925 | 1 | Substitution-coding
silent |
| 108 | c.324G>C | p.L108L | COSM1311778 | 1 | Substitution-coding
silent |
| 116 | c.347C>T | p.A116V | COSM1311779 | 1 |
Substitution-missense |
| 117 | c.350T>A | p.I117N | COSM595210 | 1 |
Substitution-missense |
| 119 | c.355T>G | p.L119V | COSM1075536 | 1 |
Substitution-missense |
| 135 | c.405G>T | p.L135L | COSM450825 | 1 | Substitution-coding
silent |
| 141 | c.421G>A | p.E141K | COSM4806762 | 1 |
Substitution-missense |
| 147 | c.440T>C | p.F147S | COSM1075537 | 1 |
Substitution-missense |
| 148 | c.443C>T | p.T148M | COSM1232873 | 1 |
Substitution-missense |
| 150 | c.448G>A | p.D150N | COSM3023284 | 1 |
Substitution-missense |
| 150 | c.448G>C | p.D150H | COSM4828326 | 1 |
Substitution-missense |
| 154 | c.461G>A | p.G154E | COSM341987 | 2 |
Substitution-missense |
| 156 | c.466A>G | p.K156E | COSM5003572 | 1 |
Substitution-missense |
| 156 | c.467A>G | p.K156R | COSM420994 | 1 |
Substitution-missense |
| 157 | c.470T>C | p.L157S | COSM1545449 | 1 |
Substitution-missense |
| 159 | c.476C>A | p.A159E | COSM248361 | 1 |
Substitution-missense |
| 159 | c.476C>T | p.A159V | COSM3023287 | 1 |
Substitution-missense |
| 162 | c.485G>A | p.R162Q | COSM1441926 | 1 |
Substitution-missense |
| 162 | c.485G>C | p.R162P | COSM1487425 | 1 |
Substitution-missense |
| 163 | c.489G>A | p.M163I | COSM483632 | 1 |
Substitution-missense |
| 163 |
c.489_494delGCTTAT |
p.M163_L164delML | COSM242271 | 1 | Deletion-in
frame |
| 164 | c.491T>C | p.L164P | COSM1311780 | 1 |
Substitution-missense |
| 209 | c.627C>A | p.I209I | COSM3366370 | 1 | Substitution-coding
silent |
| 211 | c.632T>G | p.L211R | COSM4880307 | 1 |
Substitution-missense |
| 212 | c.635A>C | p.D212A | COSM595209 | 1 |
Substitution-missense |
| 226 | c.676C>T | p.Q226a | COSM4741494 | 1 |
Substitution-nonsense |
| 231 | c.693G>A | p.E231E | COSM207380 | 1 | Substitution-coding
silent |
| 248 | c.744C>T | p.A248A | COSM1270542 | 1 | Substitution-coding
silent |
| 250 | c.749G>C | p.S250T | COSM395321 | 1 |
Substitution-missense |
| 255 | c.763A>G | p.T255A | COSM4645558 | 1 |
Substitution-missense |
| 255 | c.765A>T | p.T255T | COSM741337 | 1 | Substitution-coding
silent |
| 271 | c.811A>T | p.S271C | COSM3023294 | 2 |
Substitution-missense |
| 305 | c.913T>C | p.S305P | COSM3622456 | 1 |
Substitution-missense |
| 308 | c.922G>T | p.G308W | COSM1441927 | 1 |
Substitution-missense |
| 309 | c.927T>G | p.N309K | COSM4160342 | 1 |
Substitution-missense |
| 314 | c.941C>G | p.S314C | COSM1645128 | 1 |
Substitution-missense |
| 334 | c.1001C>T | p.A334V | COSM3928204 | 2 |
Substitution-missense |
| 335 | c.1003G>T | p.G335W | COSM595208 | 1 |
Substitution-missense |
| 342 |
c.1020_1021delCT | p.P342fsa4 | COSM3732402 | 1 |
Deletion-frameshift |
| 343 | c.1029G>A | p.T343T | COSM4741495 | 1 | Substitution-coding
silent |
| 353 | c.1058C>G | p.S353a | COSM1545448 | 1 |
Substitution-nonsense |
| 367 | c.1099G>T | p.A367S | COSM1075538 | 1 |
Substitution-missense |
| 368 | c.1104G>C | p.V368V | COSM4832802 | 1 | Substitution-coding
silent |
| 374 | c.1120C>T | p.R374a | COSM4741496 | 1 |
Substitution-nonsense |
| 374 | c.1121G>A | p.R374Q | COSM1075539 | 1 |
Substitution-missense |
| 377 | c.1130G>C | p.G377A | COSM4980694 | 1 |
Substitution-missense |
| 384 | c.1150G>A | p.G384S | COSM4637994 | 1 |
Substitution-missense |
| 391 | c.1173A>G | p.V391V | COSM3023307 | 1 | Substitution-coding
silent |
| 393 | c.1178G>T | p.G393V | COSM595207 | 1 |
Substitution-missense |
Meta-analysis of the prognostic value of
human WTAP gene expression in cancer tissues
A total of 17 out of 328 microarrays identified an
association between WTAP gene expressions and cancer prognosis
(bladder cancers, 1/7; blood cancers, 0/37; brain cancers, 1/23;
breast cancers, 5/110; colorectal cancers, 3/48; esophagus cancers,
0/1; eye cancers, 2/5; head and neck cancers, 0/6; lung cancers,
4/56; ovarian cancers, 0/25; prostate cancers, 0/1; skin cancers,
0/6; and soft tissue cancers, 1/3), P<0.05 (Table III) (28–38).
In bladder, brain, eye and soft tissue cancers, reduced expression
of the WTAP gene was associated with poor survival. However, an
increased expression of the WTAP gene was associated with poor
survival in lung cancer. Of the six breast cancer microarrays,
reduced expression of the WTAP gene was associated with poor
survival in two cases from the same database (GSE2990) and in the
database GSE1456-GPL96, while increased expression of the WTAP gene
was associated with poor survival in the GSE1456-GPL96 and
GSE1456-GPL97 databases. Of the colorectal cancer microarrays,
reduced expression of the WTAP gene was associated with poor
survival in two cases (GSE17537 and GSE17538), while increased
expression of the WTAP gene was associated with poor survival in
the GSE14333 database.
 | Table IIIAssociations between microarray WTAP
expression and cancer prognosis. |
Table III
Associations between microarray WTAP
expression and cancer prognosis.
| Author, year | Database | Cancer type | Subtype | Patient number | Endpoint | Cutpoint | P-value | Prognosis | Ref. |
|---|
| Kim, 2010 | GSE5287 | Bladder | Transitional cell
carcinoma | 165 | Disease specific
survival | 0.11 | 0.025526 | 1 | 28 |
| Freije, 2004 | GSE4412-GPL97 | Brain | Glioma | 74 | Overall
survival | 0.14 | 0.000331 | 1 | 29 |
| Pawitan, 2005 | GSE1456-GPL96 | Breast | | 159 | Overall
survival | 0.43 | 0.003810 | 1 | 30 |
| Pawitan, 2005 | GSE1456-GPL96 | Breast | | 159 | Relapse free
survival | 0.78 | 0.023849 | 2 | 30 |
| Pawitan, 2005 | GSE1456-GPL97 | Breast | | 159 | Relapse free
survival | 0.86 | 0.047352 | 2 | 30 |
| Sotiriou, 2006 | GSE2990 | Breast | | 54 | Distant metastasis
free survival | 0.15 | 0.039634 | 1 | 31 |
| Sotiriou, 2006 | GSE2990 | Breast | | 62 | Relapse free
survival | 0.13 | 0.038288 | 1 | 31 |
| Jorissen, 2009 | GSE14333 | Colorectal | | 226 | Disease free
survival | 0.85 | 0.011858 | 2 | 32 |
| Smith, 2010 | GSE17537 | Colorectal | | 49 | Disease specific
survival | 0.24 | 0.010751 | 1 | 33 |
| Smith, 2010 | GSE17538 | Colorectal | | 55 | Overall
survival | 0.25 | 0.010567 | 1 | 33 |
| Laurent, 2011 | GSE22138 | Eye | Uveal melanoma | 63 | Distant metastasis
free survival | 0.49 | 0.049578 | 1 | 34 |
| Laurent, 2011 | GSE22138 | Eye | Uveal melanoma | 63 | Distant metastasis
free survival | 0.21 | 0.007475 | 1 | 34 |
| Tomida, 2009 | GSE13213 | Lung | Adenocarcinoma | 117 | Overall
survival | 0.89 | 0.000567 | 2 | 35 |
| Tomida, 2009 | GSE13213 | Lung | Adenocarcinoma | 117 | Overall
survival | 0.90 | 0.012852 | 2 | 35 |
| Okayama, 2012 | GSE31210 | Lung | Adenocarcinoma | 204 | Overall
survival | 0.88 | 0.043703 | 2 | 36 |
| Bild, 2006 | GSE3141 | Lung | NSCLC | 111 | Overall
survival | 0.89 | 0.002284 | 2 | 37 |
| Gobble, 2011 | GSE30929 | Soft tissue | Liposarcoma | 140 | Distant recurrence
free survival | 0.21 | 0.008282 | 1 | 38 |
Discussion
WT1 was first identified due to its essential role
in the normal development of the human genitourinary system
(1) and WTAP was identified as a
protein that interacted with WT1 (3). A total of 44 complete WTAP genes were
identified in the human, chimpanzee, gibbon, macaque, gorilla,
orangutan, olive baboon, vervet monkey, marmoset, tarsier, bush
baby, armadillo, sloth, squirrel, elephant, guinea pig, mouse, rat,
pika, horse, microbat, ferret, dolphin, dog, pig, sheep, cow,
alpaca, chicken, duck, turkey, flycatcher, zebra finch, Chinese
softshell turtle, anole lizard, spotted gar, Amazon molly,
platyfish, stickleback, tilapia, medaka, cave fish and zebrafish
genomes. It was observed that WTAP genes were widely expressed in
vertebrates, existing in fish, amphibians, birds and mammals. The
phylogenetic tree revealed that the vertebrate WTAP proteins were
clustered into the primate, rodent and teleost lineages. All
vertebrate WTAPs are conserved according to the analysis of
alignment and phylogenetic tree construction. Furthermore, the
vertebrate WTAPs were under purifying selection. These results
suggest that WTAP performs an essential physiological role in all
vertebrates.
WTAP was predominantly expressed in bone marrow,
whole blood, lymph node, brain, cerebellum, retina, spinal cord,
heart, smooth muscle, skeletal muscle, small intestine, colon,
adipocyte, kidney, liver, lung, pancreas, thyroid, salivary gland,
adrenal gland, skin, ovary, uterus, placenta, prostate and testis.
The expression pattern of WTAP appeared to be ubiquitous, which is
indicative of a housekeeping role. By contrast, WT1 is expressed at
low levels in only the spleen, heart, gonad and kidney (3). A total of 19 SNPs that cause missense
mutations were identified in the human WTAP gene; however, it
remains unclear whether these SNPs affect the physiological or
pathological functions of WTAP.
WTAP and WT1 partially co-localize with splicing
factors, and are distributed together in the nucleoplasm and in
nuclear speckles (3). It has been
demonstrated in numerous tumors that WT1 is a tumor suppressor,
exerting effects including inhibiting cell proliferation and
enhancing apoptosis (39–42). However, WTAP is an oncogene, which
is overexpressed in cholangiocarcinoma (11), glioblastoma (12) and acute myeloid leukemia (13). In the present study, it was
demonstrated that WTAP was expressed in bladder, blood, brain,
breast, colorectal, esophagus, eye, head and neck, lung, ovarian,
prostate, skin and soft tissue cancers. Of a total of 328
microarrays, 17 revealed an association between microarray WTAP
expression and cancer prognosis (bladder cancers, 1; brain cancers,
1; breast cancers, 6; colorectal cancers, 3; eye cancers, 2; lung
cancers, 4; and soft tissue cancers, 1). The majority of
microarrays did not reveal an association between microarray WTAP
expression and cancer prognosis. This may be due to a lack of WTAP
expression information in the database. Notably, WTAP was not
involved in all tumor types. In addition, it is notable that the
association between WTAP expression and prognosis varied between
the different cancer types, and even in identical cancers from
separate databases. This suggests that the function of WTAP in
these tumors may not be solely as an oncogene, but may be
multidimensional (11–13). The differing WTAP expression in
various tumors may be due to the distinct oncogenes or tumor
suppressors stabilized by WTAP in particular tumors (9,10).
Furthermore, 65 somatic mutations of WTAP were
identified in cancer tissues. The effects of these mutations on
tumor formation remain to be elucidated and require future
investigation. The results of the present study suggest that WTAP
has a comprehensive and complex role in tumor formation. STAT1,
FOXO1, IRF1, glucocorticoid receptor and PPARγ transcription factor
binding sites were identified in the upstream (promoter) region of
the WTAP gene. STAT1 is a cytoplasmic protein, which functions as a
signal messenger and transcription factor in cellular responses to
cytokines and growth factors (43). It exhibits anti-tumor functions via
control of the immune system and promotion of tumor immune
surveillance (44–46). FOXO1 is an important
transcriptional regulator of cell proliferation and is considered
to be essential for tumor growth and progression (47). Deregulation of FOXO1 promotes cell
proliferation and tumorigenesis, and has thus become a primary
target of tumorigenesis prevention (48,49).
IRF1 is involved in the regulation of interferon α and β
transcription, and it has been demonstrated that IRF1 gene deletion
or rearrangement correlates with the development of human cancers
(50,51). The glucocorticoid receptor is a
member of the nuclear receptor family, which acts as a
ligand-dependent transcription factor to regulate gene expression.
In addition, the estrogen and androgen receptors are members of the
nuclear receptor family. In breast cancer, the estrogen receptor
drives cell growth, proliferation and metastasis, and the androgen
receptor has a similar role in prostate cancer (52,53).
These tumor-associated transcriptional factors may affect the
expression of WTAP and contribute to tumor formation (12–14).
In conclusion, 44 complete WTAP genes were
identified in vertebrate genomes. The vertebrate WTAP proteins
clustered into the primate, rodent and teleost lineages. The
association between WTAP gene expression and prognosis varied in
distinct cancers, and even in identical cancers from separate
microarray databases. Furthermore, a total of 65 somatic mutations
were identified in the human WTAP gene from cancer tissue samples.
The results of the present study suggest that the function of WTAP
in tumor formation may be multidimensional.
Acknowledgments
The present study was supported by Anhui Science and
Technology Research Projects (grant no. 130zc04065), the National
Natural Science Foundation of China (grant nos. 81372828 and
810001329), and the Priority Academic Program Development of
Jiangsu Higher Education Institutions.
References
|
1
|
Kreidberg JA, Sariola H, Loring JM, Maeda
M, Pelletier J, Housman D and Jaenisch R: WT-1 is required for
early kidney development. Cell. 74:679–691. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim MK, McGarry TJ, O Broin P, Flatow JM,
Golden AA and Licht JD: An integrated genome screen identifies the
Wnt signaling pathway as a major target of WT1. Proc Natl Acad Sci
USA. 106:11154–11159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Little NA, Hastie ND and Davies RC:
Identification of WTAP, a novel Wilms' tumour 1-associating
protein. Hum Mol Genet. 9:2231–2239. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Granadino B, Penalva LO and Sánchez L: The
gene fl(2)d is needed for the sex-specific splicing of transformer
pre-mRNA but not for double-sex pre-mRNA in Drosophila
melanogaster. Mol Gen Genet. 253:26–31. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Penalva LO, Ruiz MF, Ortega A, Granadino
B, Vicente L, Segarra C, Valcárcel J and Sánchez L: The drosophila
fl(2) d gene, required for female-specific splicing of Sxl and tra
pre-mRNAs, encodes a novel nuclear protein with a HQ-rich domain.
Genetics. 155:129–139. 2000.PubMed/NCBI
|
|
6
|
Horiuchi K, Umetani M, Minami T, Okayama
H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo
T and Kodama T: Wilms' tumor 1-associating protein regulates G2/M
transition through stabilization of cyclin A2 mRNA. Proc Natl Acad
Sci USA. 103:17278–17283. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naruse C, Fukusumi Y, Kakiuchi D and Asano
M: A novel gene trapping for identifying genes expressed under the
control of specific transcription factors. Biochem Biophys Res
Commun. 361:109–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Horiuchi K, Kawamura T, Iwanari H, Ohashi
R, Naito M, Kodama T and Hamakubo T: Identification of Wilms' tumor
1-associating protein complex and its role in alternative splicing
and the cell cycle. J Biol Chem. 288:33292–33302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jo HJ, Shim HE, Han ME, Kim HJ, Kim KS,
Baek S, Choi KU, Hur GY and Oh SO: WTAP regulates migration and
invasion of cholangiocarcinoma cells. J Gastroenterol.
48:1271–1282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin DI, Lee SW, Han ME, Kim HJ, Seo SA,
Hur GY, Jung S, Kim BS and Oh SO: Expression and roles of Wilms'
tumor 1-associating protein in glioblastoma. Cancer Sci.
103:2102–2109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bansal H, Yihua Q, Iyer SP, Ganapathy S,
Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et
al: WTAP is a novel oncogenic protein in acute myeloid leukemia.
Leukemia. 28:1171–1174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang
K, Go MY, Ng SC, Chan FK, Sung JJ and Yu J: Carbonic anhydrase IV
inhibits colon cancer development by inhibiting the Wnt signalling
pathway through targeting the WTAP-WT1-TBL1 axis. Gut. pii:
gutjnl-2014-308614. 2015.Epub ahead of print.
|
|
15
|
Ding Z, Yang HW, Xia TS, Wang B and Ding
Q: Integrative genomic analyses of the RNA-binding protein, RNPC1,
and its potential role in cancer prediction. Int J Mol Med.
36:473–484. 2015.PubMed/NCBI
|
|
16
|
Wang B, Xu W, Tan M, Xiao Y, Yang H and
Xia TS: Integrative genomic analyses of a novel cytokine,
interleukin-34 and its potential role in cancer prediction. Int J
Mol Med. 35:92–102. 2015.
|
|
17
|
Yang L, Luo Y and Wei J: Integrative
genomic analyses on Ikaros and its expression related to solid
cancer prognosis. Oncol Rep. 24:571–577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Luo Y, Wei J and He S: Integrative
genomic analyses on IL28RA, the common receptor of
interferon-lambda1, -lambda2 and -lambda3. Int J Mol Med.
25:807–812. 2010.PubMed/NCBI
|
|
19
|
Yang L, Wei J and He S: Integrative
genomic analyses on interferon-lambdas and their roles in cancer
prediction. Int J Mol Med. 25:299–304. 2010.PubMed/NCBI
|
|
20
|
Wang M, Wei X, Shi L, Chen B, Zhao G and
Yang H: Integrative genomic analyses of the histamine H1 receptor
and its role in cancer prediction. Int J Mol Med. 33:1019–1026.
2014.PubMed/NCBI
|
|
21
|
Wang B, Chen K, Xu W, Chen D, Tang W and
Xia TS: Integrative genomic analyses of secreted protein acidic and
rich in cysteine and its role in cancer prediction. Mol Med Rep.
10:1461–1468. 2014.PubMed/NCBI
|
|
22
|
Kumar S, Nei M, Dudley J and Tamura K:
MEGA: A biologist-centric software for evolutionary analysis of DNA
and protein sequences. Brief Bioinform. 9:299–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z: PAML: A program package for
phylogenetic analysis by maximum likelihood. Comput Appl Biosci.
13:555–556. 1997.PubMed/NCBI
|
|
24
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39(Database issue):
D945–D950. 2011. View Article : Google Scholar :
|
|
25
|
Chalifa-Caspi V, Yanai I, Ophir R, Rosen
N, Shmoish M, Benjamin-Rodrig H, Shklar M, Stein TI, Shmueli O,
Safran M and Lancet D: GeneAnnot: Comprehensive two-way linking
between oligonucleotide array probesets and GeneCards genes.
Bioinformatics. 20:1457–1458. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parkinson H, Sarkans U, Shojatalab M,
Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG,
Holloway E, Kapushesky M, et al: ArrayExpress-a public repository
for microarray gene expression data at the EBI. Nucleic Acids Res.
33(Database issue): D553–D555. 2005. View Article : Google Scholar
|
|
27
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Freije WA, Castro-Vargas FE, Fang Z,
Horvath S, Cloughesy T, Liau LM, Mischel PS and Nelson SF: Gene
expression profiling of gliomas strongly predicts survival. Cancer
Res. 64:6503–6510. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pawitan Y, Bjöhle J, Amler L, Borg AL,
Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al: Gene
expression profiling spares early breast cancer patients from
adjuvant therapy: Derived and validated in two population-based
cohorts. Breast Cancer Res. 7:R953–R964. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sotiriou C, Wirapati P, Loi S, Harris A,
Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al:
Gene expression profiling in breast cancer: Understanding the
molecular basis of histologic grade to improve prognosis. J Natl
Cancer Inst. 98:262–272. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jorissen RN, Gibbs P, Christie M, Prakash
S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhøffer M,
et al: Metastasis-associated gene expression changes: Predict poor
outcomes in patients with dukes stage B and C colorectal cancer.
Clin Cancer Res. 15:7642–7651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
34
|
Laurent C, Valet F, Planque N, Silveri L,
Maacha S, Anezo O, Hupe P, Plancher C, Reyes C, Albaud B, et al:
High PTP4A3 phosphatase expression correlates with metastatic risk
in uveal melanoma patients. Cancer Res. 71:666–674. 2011.
View Article : Google Scholar
|
|
35
|
Tomida S, Takeuchi T, Shimada Y, Arima C,
Matsuo K, Mitsudomi T, Yatabe Y and Takahashi T: Relapse-related
molecular signature in lung adenocarcinomas identifies patients
with dismal prognosis. J Clin Oncol. 27:2793–2799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar
|
|
37
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar
|
|
38
|
Gobble RM, Qin LX, Brill ER, Angeles CV,
Ugras S, O'Connor RB, Moraco NH, Decarolis PL, Antonescu C and
Singer S: Expression profiling of liposarcoma yields a multigene
predictor of patient outcome and identifies genes that contribute
to liposarcomagenesis. Cancer Res. 71:2697–2705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Wang J, Li X, Jia Y, Huai L, He K,
Yu P, Wang M, Xing H, Rao Q, et al: Role of the Wilms' tumor 1 gene
in the aberrant biological behavior of leukemic cells and the
related mechanisms. Oncol Rep. 32:2680–2686. 2014.PubMed/NCBI
|
|
40
|
Lee SY, Choe YJ, Park JY, Lee SS, Kim YH,
Shin SJ, Chung YJ and Kim HS: Wilms' tumor gene 1 enhances
nutlin-3-induced apoptosis. Oncol Rep. 31:131–136. 2014.
|
|
41
|
Liu Y and Liu S: Berberine inhibits Wilms'
tumor cell progression through upregulation of Wilms' tumor gene on
the X chromosome. Mol Med Rep. 8:1537–1541. 2013.PubMed/NCBI
|
|
42
|
Li M, Cai MY, Lu JB, Hou JH, Wu QL and Luo
RZ: Clinicopathological investigation of four cases of desmoplastic
small round cell tumor. Oncol Lett. 4:423–428. 2012.
|
|
43
|
Ding Y, Yang M, She S, Min H, Xv X, Ran X,
Wu Y, Wang W, Wang L, Yi L, et al: iTRAQ-based quantitative
proteomic analysis of cervical cancer. Int J Oncol. 46:1748–1758.
2015.PubMed/NCBI
|
|
44
|
Kaewpiboon C, Srisuttee R, Malilas W, Moon
J, Oh S, Jeong HG, Johnston RN, Assavalapsakul W and Chung YH:
Upregulation of Stat1-HDAC4 confers resistance to etoposide through
enhanced multidrug resistance 1 expression in human A549 lung
cancer cells. Mol Med Rep. 11:2315–2321. 2015.
|
|
45
|
Chen S, Li C, Wu B, Zhang C, Liu C, Lin X,
Wu X, Sun L, Liu C, Chen B, et al: Identification of differentially
expressed genes and their subpathways in recurrent versus primary
bone giant cell tumors. Int J Oncol. 45:1133–1142. 2014.PubMed/NCBI
|
|
46
|
Li W, Wei Q and Liang J: Phosphorylated
signal transducer and activator of transcription 1 is a potential
predictor of interferon response in patients with advanced renal
cell carcinoma. Mol Med Rep. 9:1929–1934. 2014.PubMed/NCBI
|
|
47
|
Zhao Z, Li C, Xi H, Gao Y and Xu D:
Curcumin induces apoptosis in pancreatic cancer cells through the
induction of forkhead box O1 and inhibition of the PI3K/Akt
pathway. Mol Med Rep. 12:5415–5422. 2015.PubMed/NCBI
|
|
48
|
Gao F and Wang W: MicroRNA-96 promotes the
proliferation of colorectal cancer cells and targets tumor protein
p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1)
and FOXO3a. Mol Med Rep. 11:1200–1266. 2015.
|
|
49
|
Zhu H: Targeting forkhead box
transcription factors FOXM1 and FOXO in leukemia (Review). Oncol
Rep. 32:1327–1334. 2014.PubMed/NCBI
|
|
50
|
Xu Y, Wang W, Gou A, Li H, Tian Y, Yao M
and Yang R: Effects of suppressor of cytokine signaling 1 silencing
on human melanoma cell proliferation and interferon-γ sensitivity.
Mol Med Rep. 11:583–588. 2015.
|
|
51
|
Chen FF, Jiang G, Xu K and Zheng JN:
Function and mechanism by which interferonregulatory factor-1
inhibits oncogenesis. Oncol Lett. 5:417–423. 2013.PubMed/NCBI
|
|
52
|
Pufall MA: Glucocorticoids and Cancer. Adv
Exp Med Biol. 872:315–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Volden PA and Conzen SD: The influence of
glucocorticoid signaling on tumor progression. Brain Behav Immun.
30(Suppl): S26–S31. 2013. View Article : Google Scholar
|