Introduction
Heart defects account for the majority of human
birth defects and are the leading cause of birth defect-related
cases of mortality (1). Congenital
heart disease (CHD) is a defect in the structure of the heart and
great vessels that is present at birth. Approximately 9 in 1,000
people are born with a congenital heart defect (2). The heritability of risk for CHD is
estimated to be 55–65%, however, both genetic and environmental
factors are responsible for the onset of the disease (3–5).
During fetal development, a series of events including cell growth,
migration and programmed cell death results in the development of a
well-formed heart at birth. Disruption of any one of these
processes may result in a heart defect (6). It is therefore important to identify
the genes that function to regulate this process of cardiac
development.
The T-box 20 (TBX20) gene is a member of the
T-box family of transcription factors that share a highly conserved
DNA binding region (known as the T-box) and serve an essential role
in early heart development (7–10),
adult heart function (11) and CHD
in humans (12–16). During heart morphogenesis, TBX20
coordinates cardiac cell proliferation and differentiation, and
formation of cardiac chambers (8–10).
Tbx20 knockout mice have been observed to exhibit arrested
development at E9.0 and die at E10.5 (7), and increased Tbx20 expression leads
to congenital atrial septal defects, patent foramen ovale and
cardiac valve defects (14). One
study involving heterozygous mutations of Tbx20 in adult mice,
indicates that Tbx20 haploinsufficiency is associated with left
ventricular dilation, decreased heart wall thickness and
contractile dysfunction (9).
Ablation of Tbx20 in the adult mouse myocardium causes dilation of
the cardiac chambers and lethality within 15 days (17). Mechanistically, TBX20 physically
interacts with a number of major factors involved in the regulation
of cardiac development, such as GATA binding protein 4 (GATA4) and
NK2 homeobox 5 (NKX2-5) transcription factors (9,14).
Tbx20 also functions as a transcriptional repressor of T-box 2
(9) and ISL LIM homeobox 1
transcription factors (7) and is
an activator of myocyte enhancer factor 2C (10). Therefore, TBX20 serves a
crucial role in cardiac morphogenesis and functions by interacting
with other genes and regulating downstream targets.
In the present study, the expression levels of
TBX20 were investigated in cardiac tissue samples derived
from patients with sporadic types of CHD. Reduced TBX20
expression levels were observed in CHD tissue samples compared with
normal tissues. To determine whether reduced TBX20
expression leads to inhibition of cell proliferation and cell cycle
arrest, TBX20 small-interfering RNAs (siRNAs) were
transfected into H9c2(2-1) Rattus norvegicus myocardial
cells. Additionally, TBX20 short-hairpin RNAs (shRNAs) were
transfected into HEK293 human embryonic kidney cells to investigate
the effects of TBX20 knockdown in human cells.
Materials and methods
Patient samples and cell lines
Informed consent from patients or guardians was
first obtained prior to the collection of 24 cardiac tissue
samples, which were provided by the Shengjing Hospital of China
Medical University (Shenyang, China). This study received ethical
approval from the local Medical Ethics Committee of China Medical
University (Shenyang, China). Tissue specimens were obtained from
the free wall of the left ventricle or atrial appendage in 12
patients with CHD (patient group; gestational age, GA: 14–38
weeks), and 12 age and gender-matched autopsies (control group; GA:
22–32 weeks) that exhibited no structural or hemodynamic
abnormalities of the heart.
HEK293 human embryonic kidney cells and H9c2(2-1)
Rattus norvegicus myocardial cells were purchased from the
cell bank of Chinese Academy of Sciences (Shanghai, China). The
cell lines were cultured in Dulbecco's modified Eagle's medium
supplemented with 10% fetal bovine serum, and maintained in a
humidified 5% (v/v) CO2 incubator at 37°C.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cardiac tissue samples
and cell lines using the TRIzol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Carlsbad, CA, USA) according to the
manufacturer's instructions. cDNA was synthesized from 3 µg
of RNA using a Reverse Transcription system purchased from Promega
(Beijing) Biotech Co., Ltd. (Beijing, China) and PCR was performed
using β-actin as an internal control to analyze TBX20 mRNA
expression in cardiac tissue samples and the primers listed in
Table I. The relative expression
levels of mRNA were determined using the optical density ratio
(TBX20/β-actin) using AlphaImager 2200 software
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Analysis of
TBX20 expression in cell lines by qPCR was achieved using
the primers listed in Table I and
was performed using an Applied Biosystems 7500 Real-Time PCR system
(Thermo Fisher Scientific, Inc., Foster City, CA, USA). Reaction
mixtures consisted of 12.5 µl SYBR® Green PCR
Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.),
0.5 µl primer (10 mM/l) and 1 µl cDNA. Thermal
cycling conditions consisted of an initial denaturation step of
95°C for 10 min, followed by 40 cycles of denaturation at 95°C for
10 sec and annealing and extension at 60°C for 1 min. Fluorescence
measurements were collected at the end of each extension step. The
quantification cycles (Cq) were then determined and the
relative concentrations of mRNA were calculated and normalized
against the levels of β-actin or glyceraldehyde 3-phosphate
dehydrogenase (Gapdh) expression in each sample (18). Reactions were performed with
non-template controls. Melting curve analyses were conducted
following completion of the thermal cycling program using a
temperature ramp that increased the temperature from 45–95°C at a
rate of 0.5°C every 2 sec. During this time, fluorescence signals
were monitored continuously to determine the specificity of PCR
primers, which was subsequently confirmed by conventional gel
electrophoresis. For each sample, reactions were conducted in
triplicate to ensure the reproducibility of the results.
 | Table IDetails of primer sequences used for
reverse transcription-quantitative polymerase chain reaction. |
Table I
Details of primer sequences used for
reverse transcription-quantitative polymerase chain reaction.
| Species | Primers | Sequence
(5′−3′) | Product size
(bp) |
|---|
| Homo
sapiens | TBX20
(1) | F:
AGGAGGCGACGGAGAACA | 286 |
| | R:
CTGCCCGACTTGGTGATG | |
| TBX20
(2) | F:
CATCCAGATTCTCCTTTTACCG | 272 |
| | R:
TTCAGCTTCGTTATCAGTTGATTC | |
| P27 | F:
AGCGACCTGCAACCGACGATTC | 120 |
| | R:
GGCCAGGCTTCTTGGGCGTC | |
| Cyclin
B1 | F:
TCTGGATAATGGTGAATGGACA | 157 |
| | R:
CGATGTGGCATACTTGTTCTTG | |
| NKX2-5 | F:
CAAGTGTGCGTCTGCCTTT | 105 |
| | R:
GCGCACAGCTCTTTCTTTTC | |
| GATA4 | F:
CGGAAGCCCAAGAACCTGA | 176 |
| | R:
CTGCTGTGCCCGTAGTGAG | |
| β-actin
(1) | F:
CTCTTCCAGCCTTCCTTCCT | 511 |
| | R:
CACCTTCACCGTTCCAGTTT | |
| β-actin
(2) | F:
ATAGCACAGCCTGGATAGCAACGTAC | 158 |
| | R:
CACCTTCTACAATGAGCTGCGTGTG | |
| Rattus
norvegicus | Tbx20 | F:
AGCAGTCACAGCCTACCAGA | 187 |
| | R:
ATGCCAAGGAAGACGAGTT | |
| p27 | F:
GCGGCAAGAGAGGCGAGGC | 129 |
| | R:
CGGAAGGCTTGGGGTGCTCG | |
| Cyclin
b1 | F:
GGCGCTCAGGGTCACTAGGAACA | 173 |
| | R:
GGGGTATTCTTGACTGTTCGCTGAC | |
| Nkx2-5 | F:
GATGCCACAGGGCAATTC | 104 |
| | R:
TCTCCTAAAGGTGGGAGTCG | |
| Gata4 | F:
CACTATGGGCACAGCAGCTCC | 186 |
| | R:
TTGGAGCTGGCCTGCGATGTC | |
| Gapdh | F:
CCCACTCGTAGCCCCTCTG | 289 |
| | R:
TGCTGAGTATGTCGTGGAGT | |
Western blotting analysis
Total protein was extracted from 24 frozen cardiac
tissue samples and cultured cells using a lysis buffer containing
protease inhibitors (KeyGen Biotechnology, Co., Ltd., Nanjing,
China). Protein concentrations of sample lysates were determined
using a bicinchoninic acid kit (KeyGen Biotechnology, Co., Ltd.)
according to the manufacturer's instructions. Samples (20
µg) were denatured by adding 5X SDS-PAGE sample loading
buffer (Beyotime Institute of Biotechnology, Jiangsu, China) and
incubating for 10 min at 95°C. Sample proteins were then separated
by 12% SDS-PAGE and electroblotted onto a polyvinylidene fluoride
membrane. Membranes were blocked using non-fat dry milk (5%) in
phosphate-buffered saline (PBS, 0.05%) and 0.05% Tween-20 at room
temperature for 2 h. This was followed by incubation with rabbit
anti-TBX20 (catalog no. sc-134061) or rabbit anti-α-tubulin
(catalog no. sc-5546) at a dilution of 1:500 (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The
following day, membranes were washed three times with PBS
containing 0.05% Tween-20 for 15 min, and incubated with the
secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit
IgG antibody (1:2,000; catalog no. sc-2004; Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature. Protein bands
were visualized using enhanced chemiluminescence detection, and the
membranes were exposed to X-ray film. α-Tubulin was used as the
internal control. The relative expression levels of protein were
determined using the optical density ratio (TBX20/α-Tubulin) using
AlphaImager 2200 software (Bio-Rad Laboratories, Inc.).
Design of shRNA and siRNA duplexes
shRNA and siRNA duplexes targeting TBX20 were
designed according to the characterization of the TBX20 gene
by Hammer et al (19).
TBX20 has two splice variants, TBX20A and
TBX20B; both isoforms share six identical exons, while
TBX20A has two additional exon sequences. Therefore, shRNA
and siRNA duplexes used for the purposes of this study, were
designed to target TBX20B. Similarly, rat Tbx20 has
two splice variants, Tbx20a and Tbx20b. shRNAs that
target human TBX20B (Ensembl Transcript ID: ENST00000492961;
www.ensembl.org) and siRNAs that targeted rat
Tbx20b (Ensembl Transcript ID: ENSRNOT00000064783) were
designed by GenePharma Co., Ltd., (Shanghai, China). A total of
three green-fluorescent protein (GFP)-tagged shRNA sequences were
designed to target human TBX20 mRNA transcripts at the
nucleotide positions 845–864, 1094–1113, and 1152–1171, and three
siRNA duplexes were designed to target rat Tbx20 mRNA
transcripts at nucleotide positions 752–772, 1042–1062, and
1089–1109. Negative control shRNA (NC-shRNA) and siRNA (NC-siRNA)
duplexes consisted of random sequences that do not target any known
mammalian genes. siRNA duplexes were chemically synthesized, and
1.0 optical density (20 µM/l) of NC-siRNA was labeled with
the carboxyfluorescin (FAM) fluorophore (GenePharma Co., Ltd.).
NC-shRNA duplexes were cloned into GFP-tagged vectors.
Transfection of siRNA and shRNA into
mammalian cells
Transfection of shRNA and siRNA duplexes into HEK293
and H9c2 cells was achieved using the FuGENE® HD
Transfection Reagent (Roche Diagnostics, Basel, Switzerland)
according to the manufacturer's instructions. At 24 h
post-transfection with GFP-labeled NC-shRNA or FAM-labeled
NC-siRNA, cells were visualized using an inverted fluorescence
microscope with a digital charged-coupled device imaging system
(IX71/DP70; Olympus Corporation, Tokyo, Japan) in order to
determine transfection efficiency.
Cell proliferation assay
Cell viability was determined using a cell counting
kit (CCK-8; Beyotime Institute of Biotechnology). Cells
(5×103/well) were seeded onto 96-well flat-bottom plates
one day prior to transfection. At 24, 48, 72, and 96 h
post-transfection, 10 µl CCK-8 was added to each well, and
cells were incubated for a further 2 h. Sample absorbance was
proportional to the number of living cells and was measured using a
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at 450 nm. The rate of cell proliferation inhibition was calculated
using the following formula: Rate of cell proliferation
inhibition=[(Average absorbance of the control group-average
absorbance of the experimental group)/average absorbance of the
control group]×100%.
Cells harvested at 96 h post-transfection were
subject to western blot analysis for proliferating cell nuclear
antigen (PCNA) using a mouse anti-PCNA antibody (1:500; catalog no.
sc-53407; Santa Cruz Biotechnology, Inc.) and a goat anti-mouse
IgG-HRP secondary antibody (1:2,000; catalog no. sc-2005; Santa
Cruz Biotechnology, Inc.), which is a reliable assay for the
determination of cell proliferation. This was performed using the
same procedures described previously.
Cell apoptosis assay
In order to detect early cell apoptosis, annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining
(BD Biosciences, Franklin Lakes, NJ, USA) and flow cytometry
analysis were performed according to the manufacturer's
instructions. Briefly, cells (5×105 cells/well) were
seeded onto six-well flat-bottom plates. At 48 h post-transfection,
cells were trypsinized, resuspended in binding buffer and incubated
in 5 µl annexin V-FITC and 5 µl PI for 15 min at 25°C
in the dark, prior to flow cytometry analysis. Early FITC-stained
apoptotic cells were represented in the lower-right quadrant of the
fluorescence-activated cell sorting histogram.
In order to detect late cell apoptosis, the DeadEnd™
Fluorometric TUNEL System (Promega Corporation, Madison, WI, USA)
was used according to the manufacturer's instructions. Briefly,
adherent cells in two-well chamber slides were fixed with 4%
formaldehyde and treated with 0.2% Triton X-100. Following
equilibration at room temperature, cells were incubated in buffer
containing nucleotides and the terminal deoxynucleotidyl
transferase enzyme for 1 h. Cells were then stained with PI for 5
min in the dark and visualized under the microscope. Cells were
considered to be apoptotic if they had TUNEL-positive nuclei and
morphological features of cell death, including cell shrinkage,
fragmentation and regions of dense chromatin condensation. The
apoptotic index was defined as the percentage of TUNEL-positive
cells in each well, from three random fields of view
(magnification, ×20).
Cell cycle analysis
Cell cycle analysis was achieved using PI staining
and flow cytometry (FACSCalibur flow cytometer; BD Biosciences).
Briefly, cells were seeded onto six-well plates and transfected
with siRNA or shRNA using the aforementioned procedures. At 48 or
96 h post-transfection, cells were harvested and fixed by adding
70% ethanol and incubating for 12 h at −20°C. Cells were then
stained with PI in a PBS solution containing RNase (KeyGen
Biotechnology, Co., Ltd.) and analyzed by flow cytometry.
In order to determine the expression levels of
factors involved in regulating cell cycle progression, the mRNA
levels of cyclin B1, P27, P16 and P21
were assessed by RT-qPCR and normalized to β-actin or
Gapdh, as described above. The expression levels of these
genes were determined in cells harvested at 48 or 96 h
post-transfection using the aforementioned procedures.
Statistical analysis
The data are expressed as the mean ± standard
deviation and differences between the means were evaluated using
analysis of variance and the Student's t-test with SPSS
software (version 16.0; SPSS Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
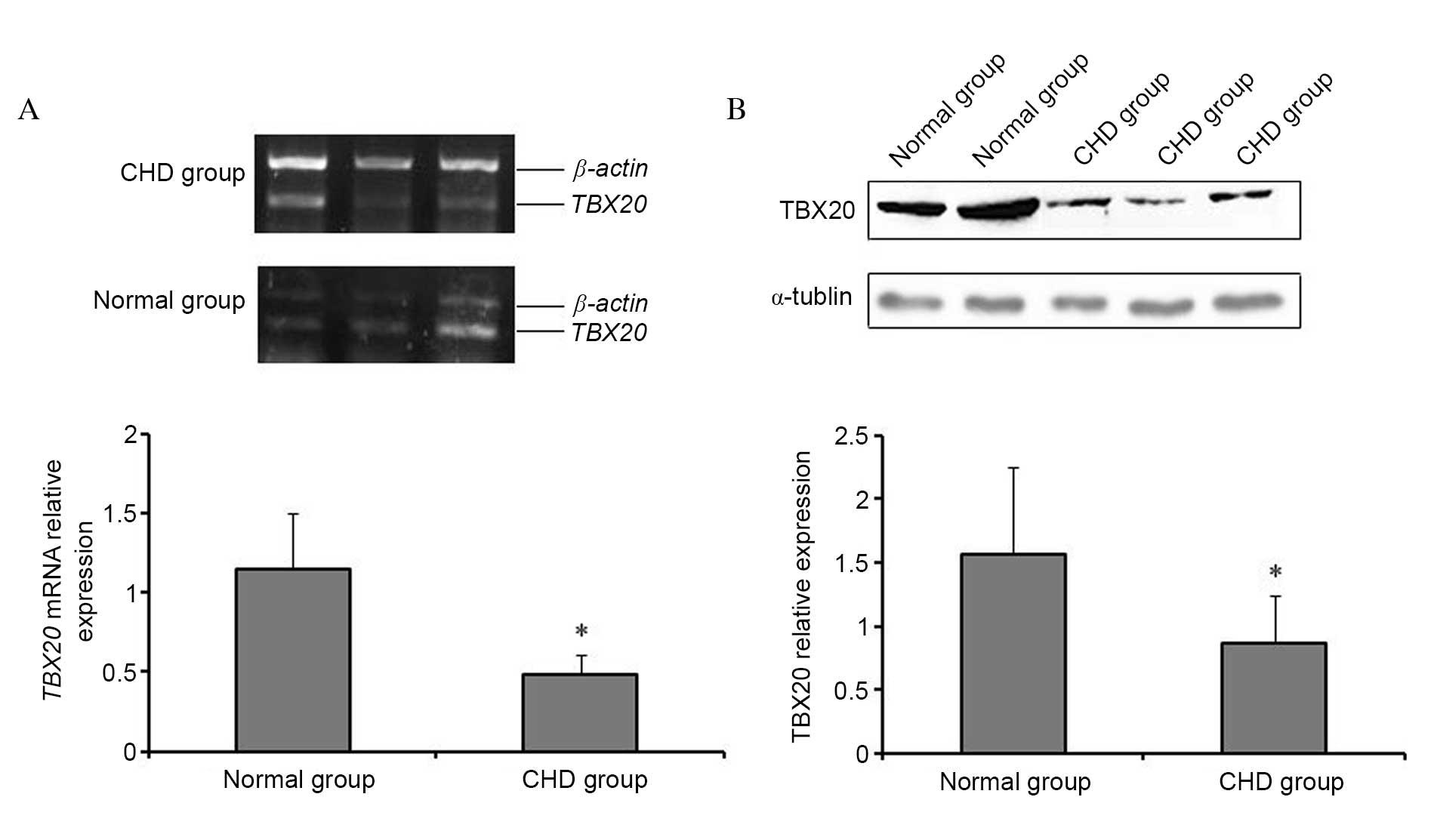
TBX20 expression is decreased in CHD
cardiac tissues
The mRNA expression levels of TBX20 were
significantly reduced in cardiac tissues from CHD patients compared
to cardiac tissues from normal controls (P=0.023; Fig. 1A), which was confirmed by western
blotting analysis (P=0.031; Fig.
1B).
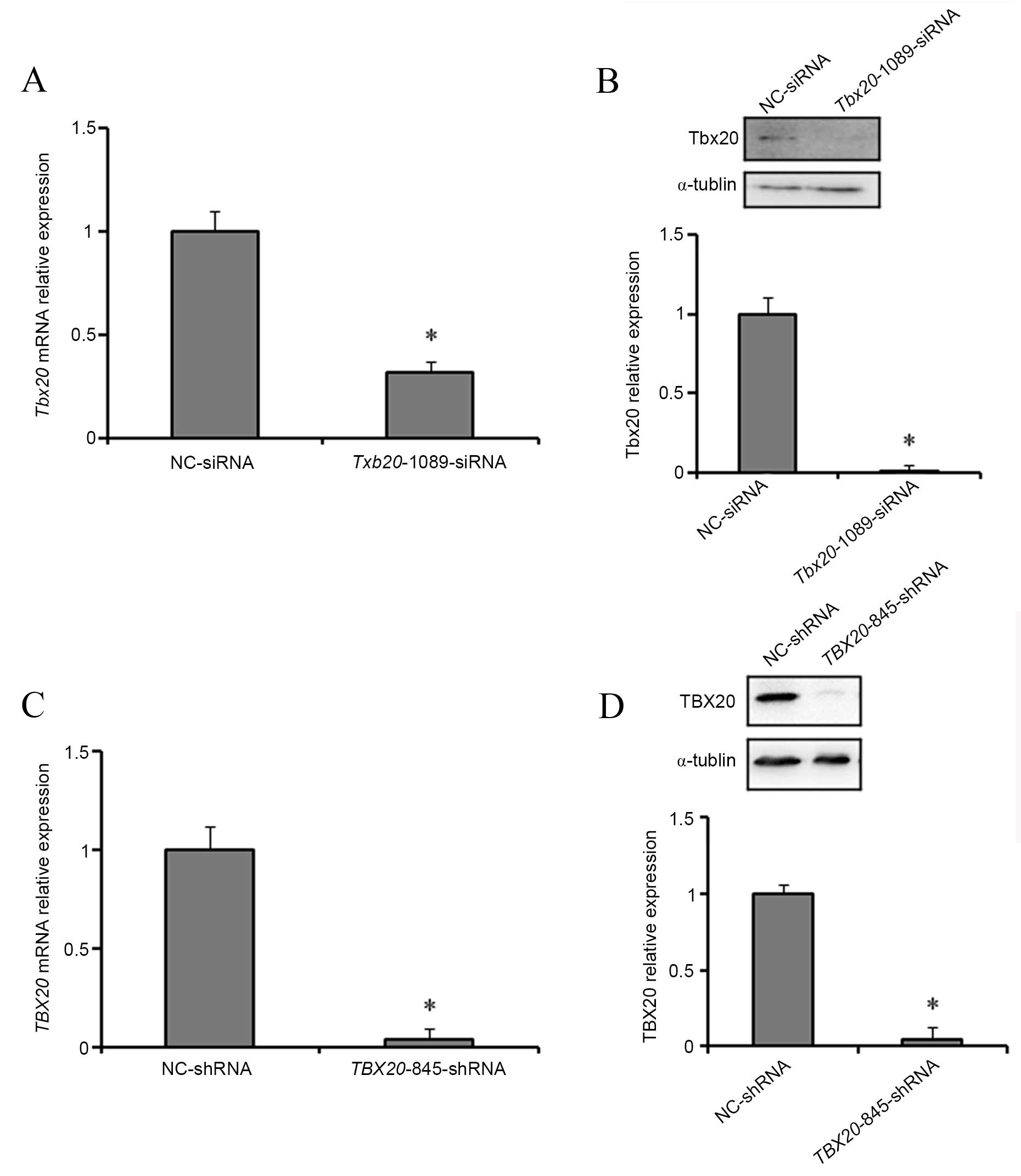
TBX20 expression is reduced following
transfection of H9c2 and HEK293 cells with TBX20 siRNA and shRNA
duplexes, respectively
At 24 h post-transfection with
fluorescently-labelled TBX20 siRNA and shRNA duplexes, the
transfection efficiency was determined using fluorescence
microscopy by comparing optical microscope images of identical
fields of view. Following confirmation of a high transfection
efficiency for TBX20 siRNA and shRNA duplexes in H9c2 (80%)
and HEK293 (90%) cells, respectively, the mRNA and protein
expression levels of TBX20 were determined by RT-qPCR and
western blotting analysis. At 48 h post-transfection with
Tbx20-1089-siRNA, H9c2 cells exhibited a significant
decrease (P=0.021 and P=0.011) in the expression levels of
TBX20 mRNA and protein compared with normal controls
(Fig. 2A and B). Consistent with
these observations, at 96 h following transfection of HEK293 cells
with TBX20-845-shRNA, the expression levels of TBX20
mRNA and protein were significantly reduced (P=0.018 and P=0.012,
respectively; Fig. 2C and D).
These results indicate that Tbx20-1089-siRNA and
TBX20-845-shRNA duplexes inhibit TBX20 expression in
H9c2 and HEK293 cells, respectively.
TBX20 inhibition represses cell
proliferation in H9c2 and HEK293 cells
To investigate the association between TBX20
inhibition and H9c2 and HEK293 cell proliferation, cell viability
was assessed using a CCK-8 assay following transfection of cells
with TBX20 siRNA and shRNA duplexes, respectively. Compared
with NC-siRNA-transfected controls, a significant time-dependent
decrease in cell proliferation rates were observed at 72 and 96 h
(48.3±0.036 and 51.8±0.110%, respectively) following transfection
of H9c2 cells with Tbx20-1089-siRNA (P<0.01; Fig. 3A). A significant time-dependent
reduction in cell proliferation rates were also observed in HEK293
cells at 72, 96 and 120 h (57.3±0.049, 42.3±0.034 and 43.3±0.020%,
respectively) following transfection with TBX20-845-shRNA
(P<0.01; Fig. 3B).
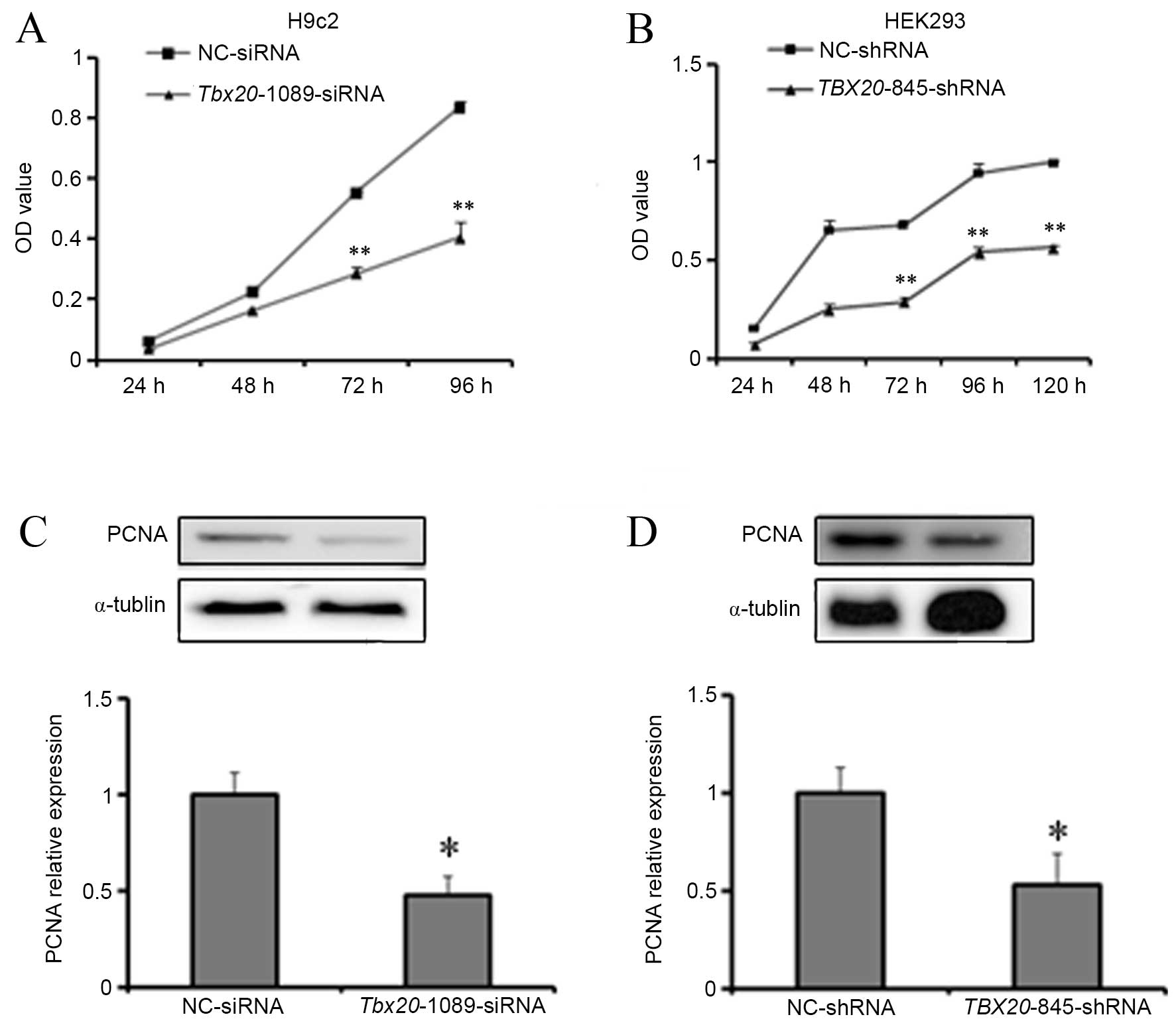 | Figure 3TBX20 inhibition suppresses
H9c2 and HEK293 cell proliferation. Cell counting kit-8 assay
analysis of (A) H9c2 cells at 24, 48, 72 and 96 h post-transfection
with Tbx20-1089-siRNA and (B) HEK293 cells at 24, 48, 72, 96
and 120 h post-transfection with TBX20-845-shRNA. Western
blotting analysis of PCNA expression in (C) H9c2 cells 96 h
post-transfection with Tbx20-1089-siRNA and NC-siRNA
controls, and (D) HEK293 cells 96 h post-transfection with
TBX20-845-shRNA and NC-shRNA controls. *P<0.05
and **P<0.01 vs. controls. TBX20, T-box 20; siRNA,
small-interfering RNA; shRNA, short-hairpin RNA; NC, normal
control; PCNA, proliferating cell nuclear antigen. |
The putative repressive effect of TBX20
inhibition on cell proliferation was then investigated by western
blotting analysis for PCNA, which is only expressed in
proliferating cells (20). A
significant reduction in PCNA protein expression levels was
observed in H9c2 cells at 96 h post-transfection with
Tbx20-1089-siRNA (P=0.017; Fig.
3C). In addition, a significant reduction in PCNA protein
expression levels was observed in HEK293 cells at 96 h
post-transfection with TBX20-845-shRNA (P=0.022; Fig. 3D). These results are consistent
with those obtained from the CCK-8 assays, which suggests that
TBX20 inhibition suppresses the proliferation of H9c2 and
HEK293 cells.
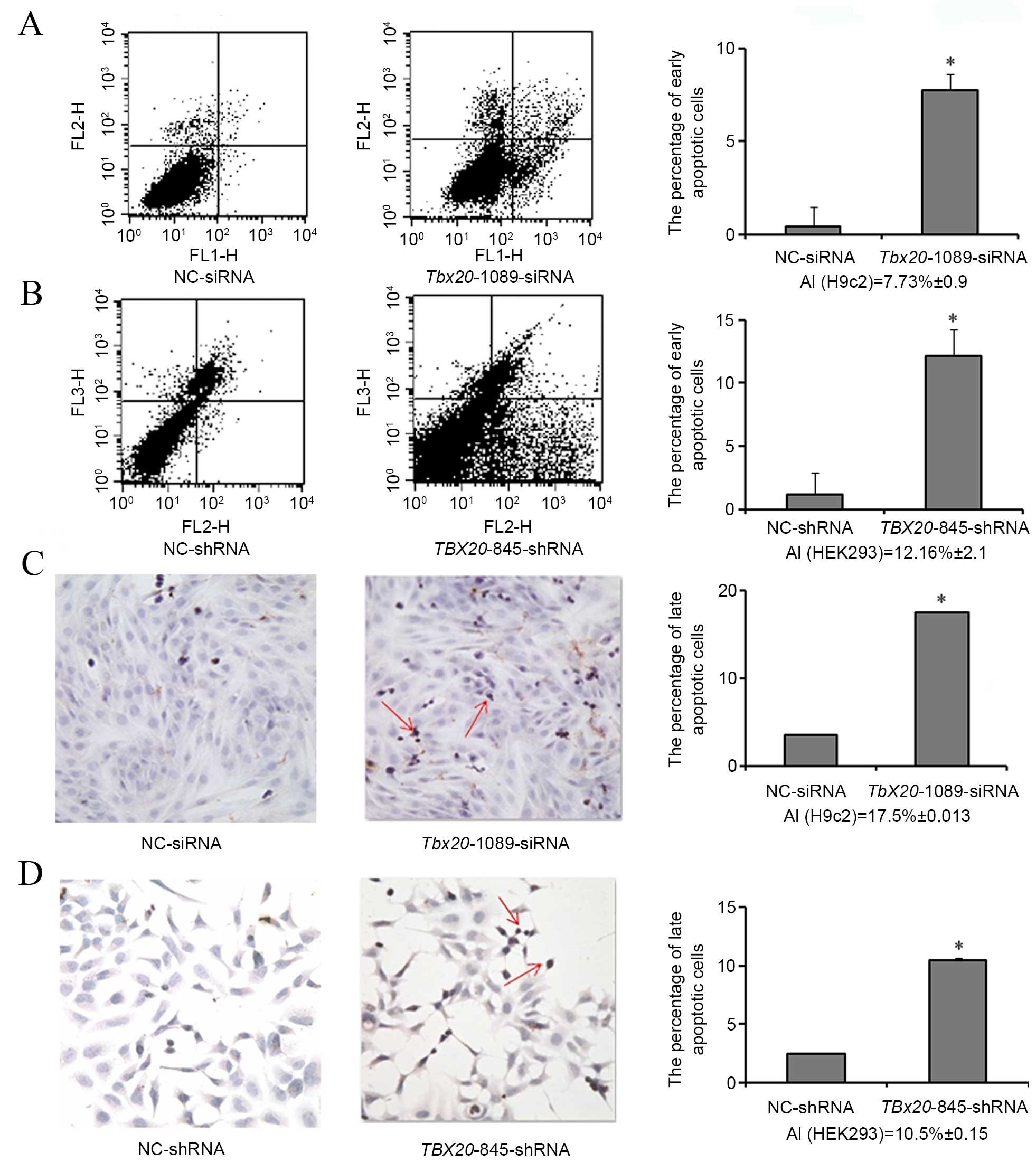
TBX20 inhibition induces cell apoptosis
in H9c2 and HEK293 cells
Annexin V-FITC/PI staining was performed to detect
early cell apoptosis. Compared with negative controls, the
percentage of early apoptotic cells was observed to increase
significantly in H9c2 cells transfected with
Tbx20-1089-siRNA (P=0.027; 7.73±0.9%), and in HEK293 cells
transfected with TBX20-845-shRNA (P=0.034; 12.16±2.1%;
Fig. 4A and B).
TUNEL staining was then performed in order to
determine late cell apoptosis. As demonstrated in Fig. 4C and D, a significant increase in
the percentage of late apoptotic cells was observed in H9c2 cells
transfected with Tbx20-1089-siRNA (P=0.028; 17.5±0.013%) and
in HEK293 cells transfected with TBX20-845-shRNA (P=0.024;
10.5±0.15%). These results suggest that TBX20 inhibition may
induce apoptosis of H9c2 and HEK293 cells.
TBX20 inhibition leads to cell cycle
arrest in G2 phase of H9c2 and HEK293 cells
To investigate the effects of TBX20
inhibition on cell cycle progression, PI staining and flow
cytometry analyses were conducted to examine the cell cycle phases
of H9c2 and HEK293 cells transfected with TBX20 siRNA and
shRNA duplexes respectively. As demonstrated in Table II and Fig. 5A and B, the percentage of cells in
the G2/M phase were significantly increased in
Tbx20-1089-siRNA-transfected H9c2 cells (P=0.036;
6.38±0.78%) and TBX20-845-shRNA-transfected HEK293 cells
(P=0.025; 7.86±1.56%) compared with negative controls.
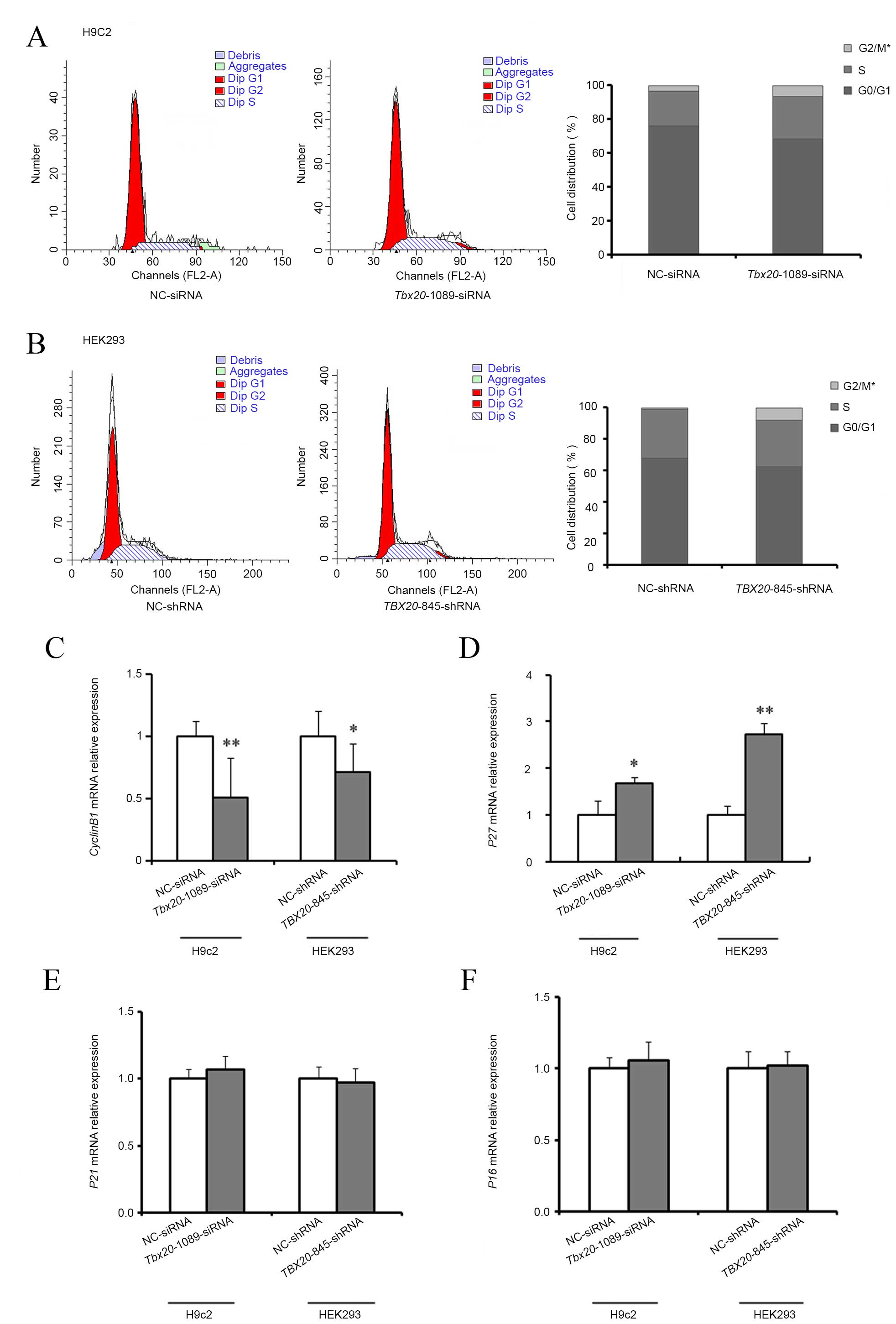 | Figure 5TBX20 inhibition arrests the
cell cycle at G2 phase in H9c2 and HEK293 cells. Cell cycle
distribution analysis of (A) H9c2 following transfection with
Tbx20-1089-siRNA and NC-siRNA, and (B) HEK293 cells
following transfection with TBX20-845-shRNA and NC-shRNA.
The mRNA expression levels of (C) cyclin B1, (D) P27,
(E) P21, and (F) P16 in H9c2 cells following
transfection of Tbx20-1089-siRNA and NC-siRNA controls, and
HEK293 cells following transfection of TBX20-845-shRNA and
NC-shRNA controls. *P<0.05 and **P<0.01
vs. controls. TBX20, T-box 20; siRNA, small-interfering RNA; shRNA,
short-hairpin RNA; NC, normal control; P27, cyclin-dependent kinase
inhibitor (CDKI) 1B; P21, CDKI1A; P16, CDKI2A. |
 | Table IIPercentage of H9c2 and HEK293 cells
in different cell cycle phases following silencing of TBX20
expression. |
Table II
Percentage of H9c2 and HEK293 cells
in different cell cycle phases following silencing of TBX20
expression.
| Group | G0/G1 phase
(%) | S phase (%) | G2/M phase (%) |
|---|
| H9c2-NC-siRNA | 75.99±1.33 | 20.78±1.44 | 3.22±0.99 |
|
H9c2-Tbx20-1089-siRNA | 67.69±0.83 | 24.95±1.03 | 6.38±0.78a |
|
HEK293-NC-shRNA | 67.81±0.24 | 31.11±1.54 | 1.08±1.30 |
|
HEK293-TBX20-845-shRNA | 62.33±1.92 | 29.82±3.47 | 7.86±1.56a |
To investigate the mechanisms of cell cycle arrest
following TBX20 inhibition, RT-qPCR analysis was used to
detect the expression of a number of important cell cycle
regulators including, cyclin B1, P27, P21 and
P16. Compared with negative controls, at 48 h
post-transfection of H9c2 cells with Tbx20-1089-siRNA,
Tbx20 inhibition resulted in a significant reduction in the
expression of cyclin B1 mRNA expression levels (P=0.003;
Fig. 5C), with a concomitant
significant increase in the expression levels of p27
(P=0.015; Fig. 5D), and no
considerable alterations in p16 and p21 expression
levels (P=0.23; Fig. 5E and F).
Similarly, at 96 h following transfection of HEK293 cells with
TBX20-845-shRNA, the mRNA expression levels of cyclin
B1 were significantly reduced (P=0.026; Fig. 5C), P27 expression levels
were significantly increased (P=0.006; Fig. 5D), and no notable alterations in
P16 and P21 expression levels were observed when
compared with negative controls (P=0.38; Fig. 5E and F).
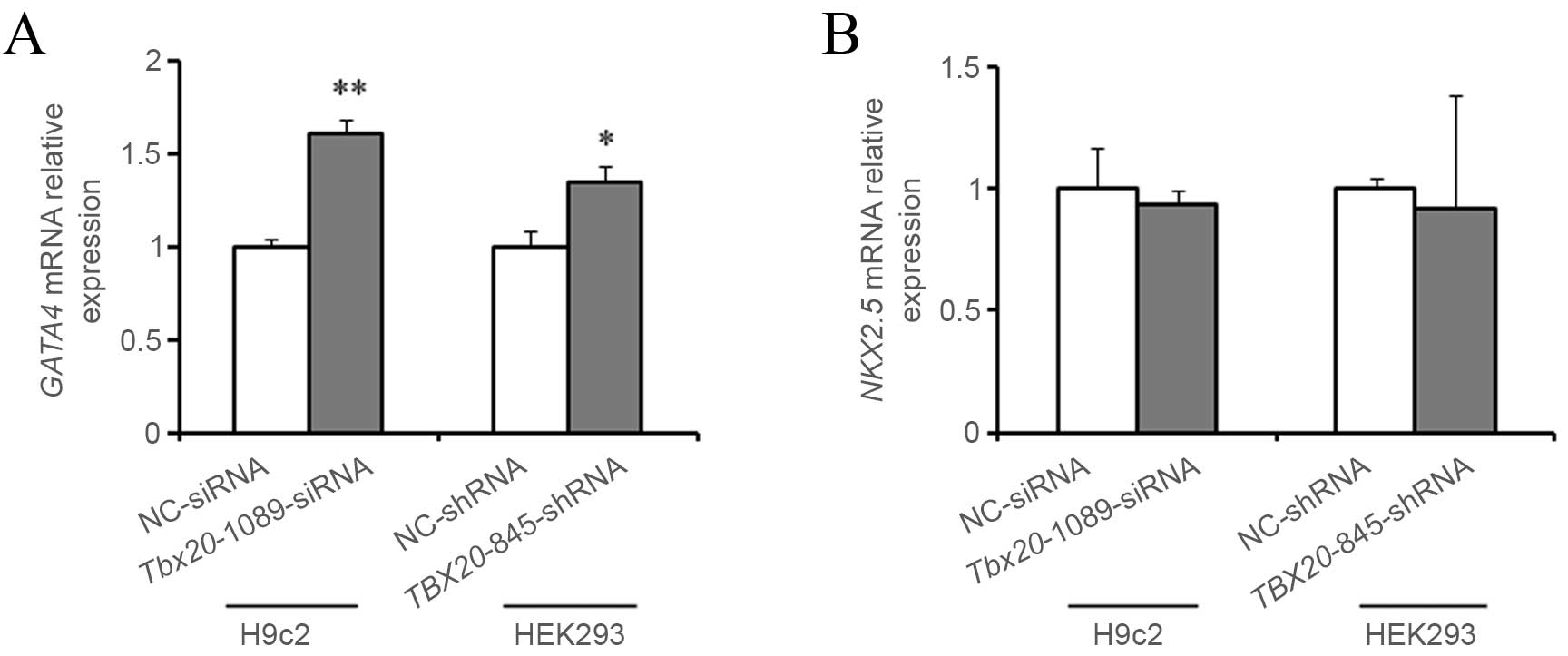
TBX20 inhibition upregulates GATA4 mRNA
expression in H9c2 and HEK293 cells
In order to investigate the role of TBX20 in
heart development, the mRNA expression levels of GATA4 and
NKX2-5 were determined in H9c2 and HEK293 cells following
transfection with TBX20-siRNA and shRNA duplexes
respectively. As demonstrated in Fig.
6A and B, the mRNA expression levels of Gata4 were
significantly increased (P=0.001) within H9c2 cells 48 h following
transfection with Tbx20-1089-siRNA compared with negative
controls, whereas no significant alterations in Nkx2-5
expression levels were observed. Similarly, the expression levels
of GATA4 mRNA were significantly increased (P=0.012) in
HEK293 cells 96 h following transfection with
TBX20-845-shRNA and no notable alterations in NKX2-5
expression levels were observed, when compared with controls
(P=0.09; Fig. 6A and B).
Discussion
During the process of human heart morphogenesis,
both cardiomyocyte proliferation and enlargement contribute to
postnatal heart growth (15).
Notably, targeted disruption of cardiomyocyte proliferation at
mid-gestation, leads to hypoplastic ventricular walls and impaired
trabeculation (16). Therefore,
normal cardiomyocyte proliferation is necessary to support the
growth and development of the postnatal human heart. The T-box
family of transcription factors serve critical functional roles in
embryonic development and organogenesis including, cell type
specification, tissue patterning and morphogenesis (21). In particular, the endocardium,
myocardium and epicardium of the developing heart express TBX20,
which suggests that TBX20 has numerous roles in cardiac development
(22). The results of the present
study suggest that TBX20 is a key mediator of cell proliferation,
particularly cell cycle progression.
TBX20 is a dose-sensitive regulator. In
zebrafish and mouse models, knockout or knockdown of Tbx20
is associated with abnormal heart cyclization, right ventricular
dysplasia, severe damage of the outflow tract and disordered
chamber differentiation, which suggests that maintaining normal
Tbx20 expression is critical for normal heart development
(7–10,23).
In the present study, the mRNA and protein
expression levels of TBX20 in CHD patients were significantly lower
than normal controls, which is consistent with previous animal
studies (7–10,23).
Therefore, it was hypothesized that this low level of TBX20
expression may be insufficient to maintain normal heart development
in CHD patients and therefore be responsible for heart
malformations. In contrast, Hammer et al (19) reported that TBX20 expression
was increased in patients with tetralogy of fallot. However, this
may due to the study population and sample size, as this was a
German study performed on 13 patients and 6 healthy controls.
During the heart development process, the number of
cardiomyocytes increases due to mitosis and heart volume increases
to support the rising hemodynamic load. Therefore, the ordered
proliferation of cardiomyocytes is essential for normal heart
development (15). A number of
studies have confirmed that a reduction in the proliferation rate
of fetal rat cardiomyocytes results in thinning of the myocardial
compact layer and derangement of the heart trabeculae, which leads
to cardiac septal defects as well as other structural deformities
(7,16,22).
The results of the present study demonstrate that
TBX20 participates in cardiomyocyte proliferation, which is
consistent with previous mouse studies (7,22).
Additionally, the results provide evidence of a possible mechanism
by which TBX20 may regulate cardiomyocyte proliferation.
Cyclin B1 is the primary activator of cyclin-dependent kinase 1
(CDK1). Through complex formation with CDK1, cyclin B1 controls the
G2/M transition during cell cycle progression (24,25).
P27 is a member of the kinase-inhibiting protein 1 family
and controls G2/M transition by repressing CDK1 (26,27).
In the present study, transfection of siRNA and shRNA duplexes
targeting TBX20 in rat myocardial cells and human embryonic kidney
cells respectively, was associated with a significant reduction in
the expression levels of cyclin B1 mRNA and a significant
increase in P27 mRNA expression levels. Through the
inactivation of CDK1, this decrease in cyclin B1 and
increase in P27 expression was hypothesized to have lead to
cell cycle arrest in G2, thereby blocking mitotic division and
inhibiting cell proliferation. However, it is unclear whether
TBX20 regulates cyclin B1 and P27 through
direct or indirect mechanisms. Future research is necessary to
clarify this further.
In addition to an adequate number of cardiomyocytes,
normal heart development requires correct cardiomyocyte
differentiation and maturation. GATA4 and NKX2-5 can be detected at
an early stage of heart development, and regulate the
differentiation and maturation of cardiomyocytes by interacting
with myocyte enhancer factor 2, serum response factor, and atrial
natriuretic factor (28–33). GATA4 and NKX2-5 are
dosage-sensitive regulators of cardiac morphogenesis, and
insufficient or excessive expression may result in a hypoplastic
heart or abnormal cardiac hypertrophy (34–40).
In the present study, TBX20 inhibition upregulated GATA4
mRNA expression levels in rat myocardial cells, and had no effects
on NKX2-5 mRNA expression, which suggested that TBX20
may participate in cardiomyocyte differentiation and maturation.
Combined with the decreased expression of TBX20 in cardiac
tissue samples from CHD patients, this may partially explain
abnormal cardiac hypertrophy observed in some CHD patients. The
functions of TBX20 and GATA4 have been studied extensively in early
cardiac cells (8,9,15,35,41);
however, the results of the present study demonstrate that TBX20
may additionally regulate the expression of GATA4 in human kidney
cells. This may be due to the presence of analogous signaling
pathways for heart and kidney development. The present results
therefore provide novel evidence to suggest that TBX20 and GATA4
may serve a functional role in human kidney development, which
should be investigated further using in vivo
methodologies.
In conclusion, the results of the present study
identified reduced TBX20 expression in cardiac tissues samples, and
silencing of TBX20 in H9c2 and HEK293 cells significantly inhibited
cell proliferation and induced cell apoptosis and G2/M cell cycle
arrest. A reduction in TBX20 expression was associated with
a significant decrease in cyclin B1 expression and a
significant increase in P27 expression, which may have
resulted in the observed cell cycle arrest of rat myocardial and
human embryonic kidney cells in G2 phase. These results suggest
that TBX20 may serve a functional role in cardiomyocyte
proliferation by regulating cyclin B1 and P27
expression during heart morphogenesis. Furthermore, increased
expression of GATA4 was observed following inhibition of
TBX20 in the same cell lines, which may affect the maturation and
differentiation of cardiomyocytes in vivo and lead to
cardiac hypertrophy observed in CHD patients. We hypothesize that
the inhibition of TBX20 expression alters normal development of the
heart and leads to the occurrence of CHDs, and that a role is
played by TBX20 in heart development.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81070131) and the
Program for Liaoning Excellent Talents in University (grant no.
LJQ2012069).
References
|
1
|
Hoffman JI and Kaplan S: The incidence of
congenital heart disease. J Am Coll Cardiol. 39:1890–1900. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van der Linde D, Konings EE, Slager MA,
Witsenburg M, Helbing WA, Takkenberg JJ and Roos-Hesselink JW:
Birth prevalence of congenital heart disease worldwide: A
systematic review and meta-analysis. J Am Coll Cardiol.
58:2241–2247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thienpont B, Mertens L, de Ravel T,
Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR
and Devriendt K: Submicroscopic chromosomal imbalances detected by
array-CGH are a frequent cause of congenital heart defects in
selected patients. Eur Heart J. 28:2778–2784. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Corrigan N, Brazil DP and McAuliffe F:
Fetal cardiac effects of maternal hyperglycemia during pregnancy.
Birth Defects Res A Clin Mol Teratol. 85:523–530. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong LG, Qiu GR, Jiang H, Xu XY, Zhu HY
and Sun KL: Analysis of single nucleotide polymorphisms and
haplotypes in HOXC gene cluster within susceptible region 12q13 of
simple congenital heart disease. Zhonghua Yi Xue Yi Chuan Xue Za
Zhi. 22:497–501. 2005.PubMed/NCBI
|
|
6
|
Srivastava D: Making or breaking the
heart: From lineage determination to morphogenesis. Cell.
126:1037–1048. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai CL, Zhou W, Yang L, Bu L, Qyang Y,
Zhang X, Li X, Rosenfeld MG, Chen J and Evans S: T-box genes
coordinate regional rates of proliferation and regional
specification during cardiogenesis. Development. 132:2475–2487.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh MK, Christoffels VM, Dias JM, Trowe
MO, Petry M, Schuster-Gossler K, Bürger A, Ericson J and Kispert A:
Tbx20 is essential for cardiac chamber differentiation and
repression of Tbx2. Development. 132:2697–2707. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stennard FA, Costa MW, Lai D, Biben C,
Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI,
Dunwoodie SL, et al: Murine T-box transcription factor Tbx20 acts
as a repressor during heart development, and is essential for adult
heart integrity, function and adaptation. Development.
132:2451–2462. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeuchi JK, Mileikovskaia M,
Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M,
Georges R, Davidson L, Mo R, et al: Tbx20 dose-dependently
regulates transcription factor networks required for mouse heart
and motoneuron development. Development. 132:2463–2474. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qian L, Mohapatra B, Akasaka T, Liu J,
Ocorr K, Towbin JA and Bodmer R: Transcription factor
neuromancer/TBX20 is required for cardiac function in Drosophila
with implications for human heart disease. Proc Natl Acad Sci USA.
105:19833–19838. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kirk EP, Sunde M, Costa MW, Rankin SA,
Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, et al:
Mutations in cardiac T-box factor gene TBX20 are associated with
diverse cardiac pathologies, including defects of septation and
valvulogenesis and cardiomyopathy. Am J Hum Genet. 81:280–291.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu C, Shen A, Li X, Jiao W, Zhang X and
Li Z: T-box transcription factor TBX20 mutations in Chinese
patients with congenital heart disease. Eur J Med Genet.
51:580–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Posch MG, Gramlich M, Sunde M, Schmitt KR,
Lee SH, Richter S, Kersten A, Perrot A, Panek AN, Al Khatib IH, et
al: A gain-of-function TBX20 mutation causes congenital atrial
septal defects, patent foramen ovale and cardiac valve defects. J
Med Genet. 47:230–235. 2010. View Article : Google Scholar :
|
|
15
|
Pan Y, Geng R, Zhou N, Zheng GF, Zhao H,
Wang J, Zhao CM, Qiu XB, Yang YQ and Liu XY: TBX20 loss-of-function
mutation contributes to double outlet right ventricle. Int J Mol
Med. 35:1058–1066. 2015.PubMed/NCBI
|
|
16
|
Chen J, Sun F, Fu J and Zhang H:
Association of TBX20 gene polymorphism with congenital heart
disease in Han Chinese neonates. Pediatr Cardiol. 36:737–742. 2015.
View Article : Google Scholar
|
|
17
|
Shen T, Aneas I, Sakabe N, Dirschinger RJ,
Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al: Tbx20
regulates a genetic program essential to adult mouse cardiomyocyte
function. J Clin Invest. 121:4640–4654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Hammer S, Toenjes M, Lange M, Fischer JJ,
Dunkel I, Mebus S, Grimm CH, Hetzer R, Berger F and Sperling S:
Characterization of TBX20 in human hearts and its regulation by
TFAP2. J Cell Biochem. 104:1022–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kubben FJ, Peeters-Haesevoets A, Engels
LG, Baeten CG, Schutte B, Arends JW, Stockbrügger RW and Blijham
GH: Proliferating cell nuclear antigen (PCNA): A new marker to
study human colonic cell proliferation. Gut. 35:530–535. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stennard FA and Harvey RP: T-box
transcription factors and their roles in regulatory hierarchies in
the developing heart. Development. 132:4897–4910. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chakraborty S and Yutzey KE: Tbx20
regulation of cardiac cell proliferation and lineage specialization
during embryonic and fetal development in vivo. Dev Biol.
363:234–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shelton EL and Yutzey KE: Tbx20 regulation
of endocardial cushion cell proliferation and extracellular matrix
gene expression. Dev Biol. 302:376–388. 2007. View Article : Google Scholar
|
|
24
|
Tang L, Zhang Y, Pan H, Luo Q, Zhu XM,
Dong MY, Leung PC, Sheng JZ and Huang HF: Involvement of cyclin B1
in progesterone-mediated cell growth inhibition, G2/M cell cycle
arrest and apoptosis in human endometrial cell. Reprod Biol
Endocrinol. 7:1442009. View Article : Google Scholar
|
|
25
|
Paruthiyil S, Cvoro A, Tagliaferri M,
Cohen I, Shtivelman E and Leitman DC: Estrogen receptor β causes a
G2 cell cycle arrest by inhibiting CDK1 activity through the
regulation of cyclin B1, GADD45A and BTG2. Breast Cancer Res Treat.
129:777–784. 2011. View Article : Google Scholar
|
|
26
|
Yadav V, Sultana S, Yadav J and Saini N:
Gatifloxacin induces S and G2-phase cell cycle arrest in pancreatic
cancer cells via p21/p27/p53. PloS One. 7:e477962012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mitrea DM, Yoon MK, Ou L and Kriwacki RW:
Disorder-function relationships for the cell cycle regulatory
proteins p21 and p27. Biol Chem. 393:259–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese
RJ, Markham BE and Izumo S: The cardiac tissue-restricted homeobox
protein Csx/Nkx2.5 physically associates with the zinc finger
protein GATA4 and cooperatively activates atrial natriuretic factor
gene expression. Mol Cell Biol. 18:3120–3129. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sepulveda JL, Vlahopoulos S, Iyer D,
Belaguli N and Schwartz RJ: Combinatorial expression of GATA4,
Nkx2-5, and serum response factor directs early cardiac gene
activity. J Biol Chem. 277:25775–25782. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vincentz JW, Barnes RM, Firulli BA, Conway
SJ and Firulli AB: Cooperative interaction of Nkx2.5 and Mef2c
transcription factors during heart development. Dev Dyn.
237:3809–3819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Snyder M, Huang XY and Zhang JJ: Stat3
directly controls the expression of Tbx5, Nkx2.5, and GATA4 and is
essential for cardiomyocyte differentiation of P19CL6 cells. J Biol
Chem. 285:23639–23646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schlesinger J, Schueler M, Grunert M,
Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I and
Sperling SR: The cardiac transcription network modulated by Gata4,
Mef2a, Nkx2.5, Srf, histone modifications and microRNAs. PLoS
Genet. 7:e10013132011. View Article : Google Scholar
|
|
33
|
Chen Y and Cao X: NFAT directly regulates
Nkx2-5 transcription during cardiac cell differentiation. Biol
Cell. 101:335–349. 2009. View Article : Google Scholar
|
|
34
|
Zhao R, Watt AJ, Battle MA, Li J, Bondow
BJ and Duncan SA: Loss of both GATA4 and GATA6 blocks cardiac
myocyte differentiation and results in acardia in mice. Dev Biol.
317:614–619. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pu WT, Ishiwata T, Juraszek AL, Ma Q and
Izumo S: GATA4 is a dosage-sensitive regulator of cardiac
morphogenesis. Dev Biol. 275:235–244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oka T, Maillet M, Watt AJ, Schwartz RJ,
Aronow BJ, Duncan SA and Molkentin JD: Cardiac-specific deletion of
Gata4 reveals its requirement for hypertrophy, compensation, and
myocyte viability. Circ Res. 98:837–845. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guner-Ataman B, Paffett-Lugassy N, Adams
MS, Nevis KR, Jahangiri L, Obregon P, Kikuchi K, Poss KD, Burns CE
and Burns CG: Zebrafish second heart field development relies on
progenitor specification in anterior lateral plate mesoderm and
nkx2.5 function. Development. 140:1353–1363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao L, Ju D, Gao Q, Zheng X and Yang G:
Over-expression of Nkx2.5 and/or cardiac α-actin inhibit the
contraction ability of ADSCs-derived cardiomyocytes. Mol Biol Rep.
39:2585–2595. 2012. View Article : Google Scholar
|
|
39
|
Liu H, Harris TM, Kim HH and Childs G:
Cardiac myocyte differentiation: The Nkx2.5 and Cripto target genes
in P19 clone 6 cells. Funct Integr Genomics. 5:218–239. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamak A, Temsah R, Maharsy W, Caron S,
Paradis P, Aries A and Nemer M: Cyclin D2 rescues size and function
of GATA4 haplo-insufficient hearts. Am J Physiol Heart Circ
Physiol. 303:H1057–H1066. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Laforest B and Nemer M: GATA5 interacts
with GATA4 and GATA6 in outflow tract development. Dev Biol.
358:368–378. 2011. View Article : Google Scholar : PubMed/NCBI
|