Introduction
Infertility is experienced by 10–15% of couples. It
has previously been estimated that half of these cases are due to
male infertility (1,2), a serious form of which is idiopathic
azoospermia (IA). Genetic analysis of patients with IA have
revealed a variety of causes, predominantly chromosome aberrations
or variants of functional genes associated with spermatogenesis
(3,4). Understanding the genetic variants may
contribute to the diagnosis and management of infertility in men,
and is crucial to prevent the passing of genetic defects to
offspring in future generations via in vitro fertilization
procedures.
Androgen receptor (AR) is an important steroid
hormone receptor that is critical during male sexual
differentiation and for the maintenance of normal spermatogenesis
(5–7). AR belongs to a family of nuclear
transcription factors that mediate the action of steroid hormones
(8). There are 4 domains in the AR
protein structure, including the N-terminal transactivation domain,
DNA-binding domain, hinge region and carboxyl ligand-binding domain
(9,10). Binding of androgens, including
testosterone and 5α-dihydrotestosterone, to the ligand-binding
domain of AR results in nuclear translocation, where it acts as a
transcriptional regulator (10).
Previously, a number of AR mutations or
polymorphisms that cause or are associated with a spectrum of
hereditary disorders, including complete and partial androgen
insensitivity syndrome (CAIS and PAIS, respectively), were
identified (11,12). In order to identify variants of the
AR gene in patients with IA, the present study sequenced each exon
of AR in 776 patients diagnosed with IA and 709 proven fertile men.
The results may be important for the accurate diagnosis of IA and
useful for genetic counseling.
Materials and methods
Ethical approval
The present study was approved by the ethics
committee of Peking University Shenzhen Hospital (Shenzhen, China;
approval no. 20090018). The current study was approved on July 18,
2009, initiated on August 1, 2009 and terminated on December 1,
2014.
Patient samples
The patient inclusion criteria was the same as
described previously (13), with
certain modifications. A total of 1,880 azoospermic patients were
recruited for the present study from the Center of Reproductive
Medicine, Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China) between January 2007 and October
2011. Among the subjects, 776 patients fulfilled the following
criteria for IA diagnosis: i) No sperm detected in the pellets of
semen samples on three different occasions; ii) no obstruction,
inflammation or injury of the reproductive system or pelvic cavity;
and iii) no karyotypic abnormality or Y chromosome microdeletion. A
total of 709 fertile men from the Center of Physical Examination,
Peking University Shenzhen Hospital were recruited as controls.
Each had fathered at least one child without assisted reproductive
techniques, including in vitro fertilization,
intracytoplasmic sperm injection or intracytoplasmic
morphologically selected sperm injection. Following panel
re-sequencing and quality control steps, 776 patients aged 24–46
years (mean, 30.6 years) and 709 fertile men aged 29–51 years
(mean, 35.6 years) were available for further analysis. Informed
written consent was obtained from each subject.
Panel re-sequencing
Panel re-sequencing was performed as described
previously (13). Genomic DNA
samples (5 µg) isolated from peripheral blood samples using
the E.Z.N.A. Blood DNA kits (Omega Bio-Tek, Inc., Norcross, GA,
USA) were sent to the Beijing Genomics Institute (Shenzhen, China)
for exome capture and sequencing. The capture procedure was
performed in solution with a NimbleGen custom array (Roche Applied
Science, Madison, WI, USA) that enriched the exonic sequences of
654 infertility- or subfertility- associated genes. The majority
these genes have been reviewed by Matzuk and Lamb (14). Additionally, the present study
selected other genes that have been previously demonstrated to
cause male reproductive defects in mouse models in studies
published between November 2008 and December 2010 (15). Panel re-sequencing was performed
using the Illumina platform (Illumina, Inc., San Diego, CA, USA)
with 90 bp pair-end reads.
FASTQ sequence files were aligned against the human
reference genome (NCBI build 37.1, hg19) with Short Oligonucleotide
Analysis Package aligner software (version 2.21; www.soap.genomics.org.cn). Duplicated paired-end
reads were removed from the merged data sets. Single nucleotide
variants that were different from the hg19 reference genome were
filtered out if they met any of the following criteria: i) A
Phred-like quality score of ≤20; ii) overall depth of ≤8×;
estimated copy number of ≥2; iii) or genomic distance between two
adjacent variants of <5 bp. In addition, the quality score of
the major and minor allele at heterozygous loci were ≥20. The
variants were then annotated using an in-house functional
prediction tool, and were compared with the dbSNP Build 135
(www.ncbi.nlm.nih.gov/projects/SNP/) and 1000 Genomes
(www.1000genomes.org; as of August 2010).
Validation of novel missense variants by Sanger sequencing. To
validate the novel missense variants identified by deep sequencing,
polymerase chain reaction (PCR) amplifications were performed and
the PCR products were sequenced in both directions with a 3730 DNA
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The primers for PCR and Sanger sequencing
validation of the AR gene are presented in Table I.
 | Table IPrimers for polymerase chain reaction
and Sanger sequencing validation of the AR gene. |
Table I
Primers for polymerase chain reaction
and Sanger sequencing validation of the AR gene.
| Primer | Sequence (5′−3′) |
|---|
| AR mut1 | |
| Forward |
CTCCGCTGACCTTAAAGACATCCT |
| Reverse |
CTCGCCTTCTAGCCCTTTGGTGTA |
| AR mut2 | |
| Forward |
CTCCCCATCCCCACGCTCGCATCA |
| Reverse |
ATCCAGGGGCCCATTTCGCTTTTG |
| AR mut3 | |
| Forward |
GGTTTAGCAGGTATTTGGGATGAT |
| Reverse |
GAGTCGGGCTGGTTGTTGTC |
Validation of novel missense variants by
Sanger sequencing
As described in Panel re-sequencing, genomic DNA
samples (5 µg) were isolated from peripheral blood samples
using the E.Z.N.A. Blood DNA kits (Omega Bio-Tek, Inc.). PCR was
performed in a volume of 50 µl containing 1 µM of
each forward and reverse primer, 50 ng DNA, and 25 µl
EmeraldAmp PCR Master mix (Takara Bio Inc., Otsu, Japan). Products
were amplified in a thermocycler (MyCycler; Bio-Rad, Hercules CA,
USA) with the following conditions: 30 cycles of 10 sec at 98°C, 30
sec at 60°C, and 40 sec at 72°C. Amplicons were extracted from gels
and sequenced in both directions with a 3730 DNA Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primers for PCR
and Sanger sequencing validation of the AR gene are presented in
Table I.
Protein alignment
Multiple protein alignments were performed with
MegAlign 7.1.0 (DNASTAR, Inc., Madison, WI, USA). The amino acid
sequences of the androgen receptor in humans, chimpanzees, rhesus
monkeys, cows, rats, mice and chickens was determined. The
identification numbers of androgen receptor protein were as
follows: Human, NP_000035.2; chimpanzee, XP_009437511.1; rhesus,
NP_001028083.1; cow, NP_001231056.1; rat, NP_036634.1; mouse,
NP_038504.1; and chicken, NP_001035179.1).
Evaluation of coding single nucleotide
polymorphisms
Sorting Intolerant From Tolerant (SIFT; sift.jcvi.org/) and PolyPhen 2.0 (genetics.bwh.harvard.edu/pph2/) analysis
were used for the evaluation of coding single nucleotide
polymorphisms (16,17). SIFT is based on the premise that
protein evolution is correlated with protein function. SIFT scores
≤0.05 are predicted by the algorithm to be damaging, whereas scores
>0.05 are considered tolerant. Predictions are based on a
combination of phylogenetic, structural and sequence annotation
information characterizing a substitution and its position in the
protein. PolyPhen scores >0.85 are predicted by the algorithm to
be probably damaging, scores >0.15 are considered possibly
damaging, whereas scores <0.15 are considered benign.
Plasmid construction and site-directed
mutagenesis
A human AR expression plasmid was provided as a gift
from Dr Chawnshang Chang (George H. Whipple Lab for Cancer
Research, Departments of Pathology and Urology, University of
Rochester Medical Center, Rochester, NY, USA). Site-directed
mutagenesis was performed to generate AR expression plasmids
bearing the C290R, S495N or R630W variant, as described previously
(13). DNA sequencing was
performed to confirm the introduced variants. The PCR primers used
for site-directed mutagenesis and plasmid construction are
presented in Table II. Products
were amplified in a thermocycler (Bio-Rad) using an Expand High
Fidelity PCR system (Roche, Basel, Switzerland). The 50 µl
PCR reaction was conducted with 50 ng templates, 1 µM primer
pairs, 200 µM dNTPs and 2 units of DNA polymerase. The
extension reaction was initiated by pre-heating the reaction
mixture to 94°C for 3 min; 16 cycles of 94°C for 1 min, 52°C for 1
min and 68°C for 8 min; followed by incubation at 68°C for 15 min.
DNA sequencing was conducted with a 3730 DNA Analyzer (Applied
Biosystems) to confirm the introduced variants.
 | Table IIPrimers for polymerase chain reaction
used for site-directed mutagenesis and plasmid construction. |
Table II
Primers for polymerase chain reaction
used for site-directed mutagenesis and plasmid construction.
| Primer | Sequence (5′−3′) |
|---|
| hAR mut1 (T-C) | |
| Forward | TGCCCCATTGGCCGAA |
|
cGCAAAGGTTCTCTGCT |
| Reverse | AGCAGAGAACCTTTGC |
|
gTTCGGCCAATGGGGCA |
| hAR mut2 (G-A) | |
| Forward | CTGGCGGGCCAGGAAA |
|
aCGACTTCACCGCACCT |
| Reverse | AGGTGCGGTGAAGTCG |
|
tTTTCCTGGCCCGCCAG |
| hAR mut3 (C-T) | |
| Forward | ATGACTCTGGGAGCC |
|
tGGAAGCTGAAGAAACTT |
| Reverse |
AAGTTTCTTCAGCTTCC |
| aGGCTCCCAGAGTCAT |
Luciferase assay
Luciferase analysis was performed as described
previously, with certain modifications (18). HeLa and TM4 cells (American Type
Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) and 100 µg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C, 95% humidity and 5%
CO2 atmosphere. Cells were seeded in 24-well tissue
culture plates for 24 h prior to transfection. Equivalent
concentrations (100 ng) of wild-type (WT) or mutant AR expression
plasmids were cotransfected with mouse mammary tumor virus (pMMTV)
long terminal repeat plasmids (a gift from Dr Chawnshang Chang)
into HeLa and TM4 cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Cells were treated with or without 100 nM
testosterone (Sigma-Aldrich) after 6 h of transfection and
harvested 24 h after treatment. Firefly and Renilla
luciferase expression was assessed using a Dual-Luciferase Reporter
Assay System (cat no. E1910; Promega Corporation, Madison, WI,
USA). Renilla luciferase activity was normalized to that of
firefly luciferase. The Modulus/9200-003 luminometer (Turner
Biosystems, Sunnydale, CA, USA) was used in this study. Following
normalization for transfection efficiency, induction factors were
calculated as the ratio of the mean luciferase value for
testosterone-stimulated versus non-testosterone-stimulated (ethanol
vehicle-treated) samples.
Ultrasound examinations
All ultrasonographic measurements were conducted by
the same ultrasonographer. Patients with novel AR variants were
examined with scrotal ultrasonography to test the size of testes.
All examinations were performed using ultrasound scanner (EUB-7500,
Hitachi Medical Corporation, Tokyo, Japan) equipped with a linear
array transducer (6.5 MHz; Model EUP-L65; Hitachi Medical
Corporation).
Statistical analysis
All experiments were repeated at least three times.
Data of the luciferase assay are expressed as the mean ± standard
deviation. SPSS version 17.0 statistical software (SPSS, Inc.,
Chicago, IL, USA) was used for statistical analysis. Analysis of
variance (ANOVA) and Dunnett's t-test were used to compare the
difference in the means between the variants and WT. P<0.05 was
considered to indicate a statistically significant difference. Data
were log-transformed before ANOVA to satisfy the equal variances
assumption.
Results
Identification of AR variant in patients
with IA
To examine whether AR genetic defects were
associated with IA, the present study screened for AR exonic
variants in 776 patients with IA and 709 fertile men using massive
parallel sequencing technology. As demonstrated in Table III, seven missense variants and
two synonymous variants were detected in AR. Two of these missense
variants (F827L and M887V) have been previously reported in the
ExAC database. Another variant at amino acid 290 (C290W) has been
previously described in the ExAC database, however, this variant
was different from the variant identified in the present study
(C290R). The three patient-specific missense variants (C290R, S495N
and R630W) have not been previously reported in the dbSNP135
database, 1000 Genome Project dataset or ExAC database. These three
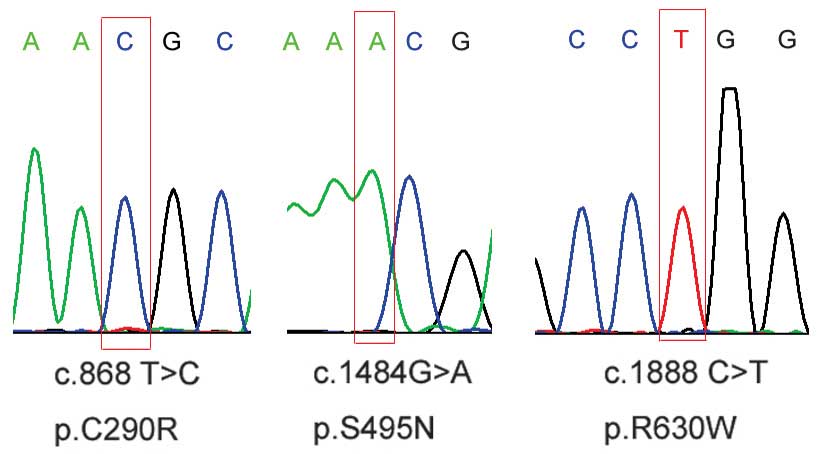
novel patient-specific missense variants were further confirmed by
Sanger sequencing (Fig. 1).
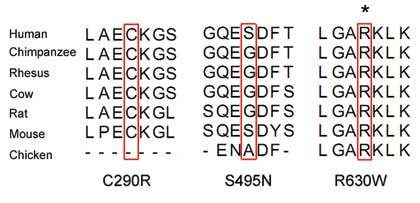
Alignment of the amino acid sequence of AR to its orthologs in
different species demonstrated that the R630W variant affected a
highly conserved amino acid (Fig.
2). Based on this conservation behavior, the variant R630W was
predicted to be possibly damaging to the protein, according to SIFT
and PolyPhen-2.0 analysis (Table
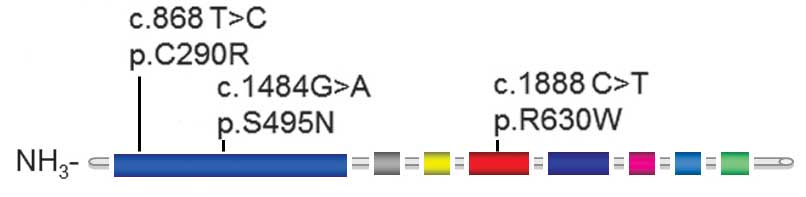
IV). The locations of these three missense variants within the
AR protein are demonstrated in Fig.
3.
 | Table IIIAndrogen receptor variants and single
nucleotide polymorphisms identified in patients with idiopathic
azoospermia (IA; n=776) and fertile controls (n=709). |
Table III
Androgen receptor variants and single
nucleotide polymorphisms identified in patients with idiopathic
azoospermia (IA; n=776) and fertile controls (n=709).
| Variant no. | Sequence variant | Amino acid
change | IA patients, n | Fertile controls,
n | Reported | Sample ID |
|---|
| Missense | | | | | | |
| 1 | c.868 T>C | p.C290R | 1 | 0 | No | w320 |
| 2 | c.1484 G>A | p.S495N | 1 | 0 | No | w691 |
| 3 | c.1888 C>T | p.R630W | 1 | 0 | No | w530 |
| 4 | c.569 C>T | p.T190I | 0 | 1 | No | 262 |
| 5 | c.616 A>G | p.S206G | 0 | 1 | No | 285 |
| 6 | c.2481 C>A | p.F827L | 0 | 1 | Yes | 81 |
| 7 | c.2659 A>G | p.M887V | 0 | 1 | Yes | 57 |
| Synonymous | | | | | | |
| 8 | c.639 G>A | None | 0 | 1 | Yes | 177 |
| 9 | c.1149 C>T | None | 2 | 0 | No | w325,w671 |
 | Table IVList of missense variants predicted
to be functionally significant by SIFT and PolyPhen 2.0
programs. |
Table IV
List of missense variants predicted
to be functionally significant by SIFT and PolyPhen 2.0
programs.
| Nucleotide
change | Amino acid
change | SIFTa
|
PolyPhen-2.0b
|
|---|
| Score | Prediction | Score | Prediction |
|---|
| c.868 T>C | p.C290R | 0.340 | Tolerated | 0.995 | Possibly
damaging |
| c.1484G>A | p.S495N | 0.480 | Tolerated | 0.066 | Benign |
| c.1888 C>T | p.R630W | 0.000 | Damaging | 1.000 | Probably
damaging |
Effects of AR variants on AR
function
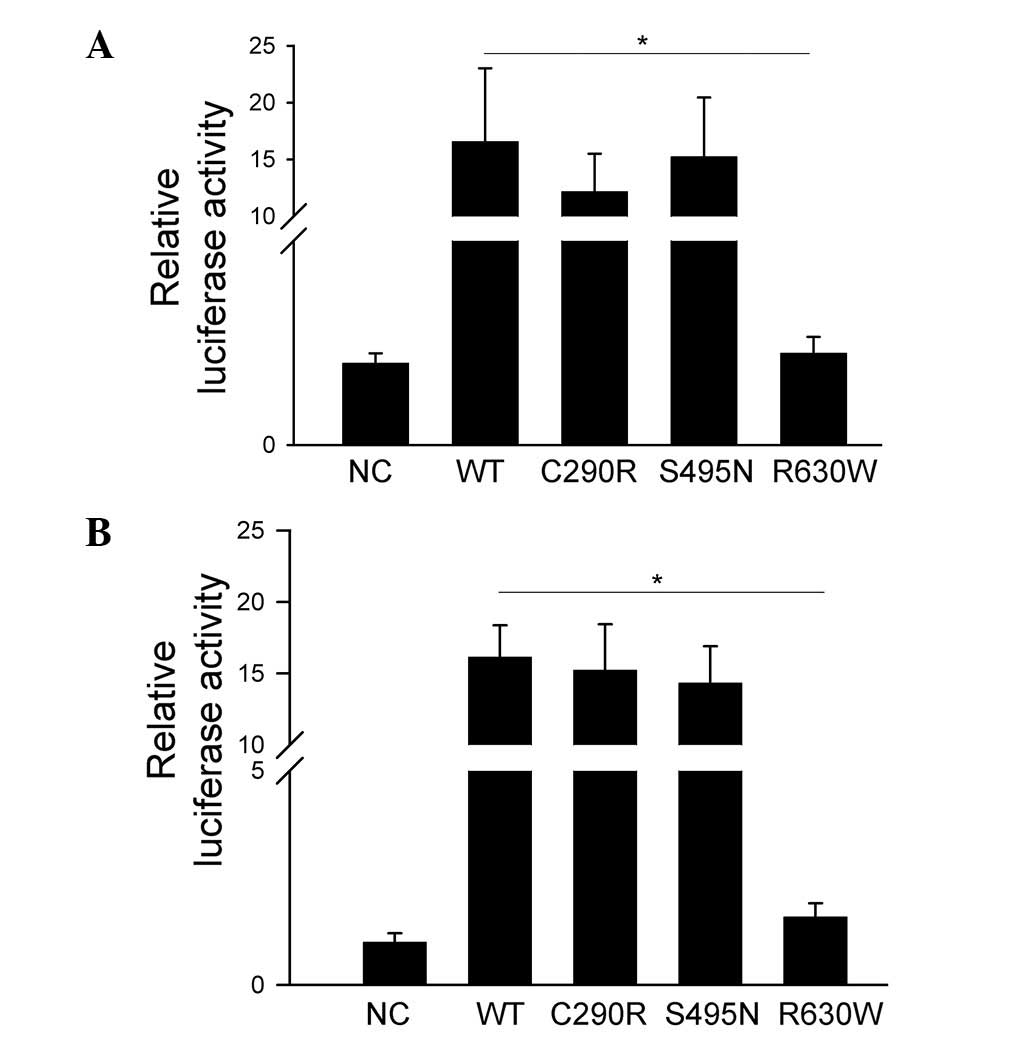
To evaluate whether the identified patient-specific
missense variants of AR inhibit its regulatory function, luciferase
reporter constructs containing the androgen responsive elements
(MMTV promoter) were transfected into HeLa and TM4 cell lines. AR
WT, C290R and S495N, but not R630W variants, significantly
increased MMTV promoter activity compared with empty vector in both
cell types (P<0.05; Fig. 4).
These results indicate that C290R and S495N did not affect the
transcriptional regulation activity of AR. However, the
transcriptional regulation activity of AR was inhibited by the
R630W variant.
Clinical variable and hormone
analysis
Scrotal color Doppler ultrasonography of the three
patients with novel AR variants revealed small testes in the
scrotal sac, with a homogeneous echotexture and wide
hypoechogenicity. No solid or cystic lesions were observed.
Table V summarizes the clinical
and hormone data of these three patients, none of which had a
family history of male infertility, CAIS or PAIS.
 | Table VClinical and hormone profile of
patients with idiopathic azoospermia with novel androgen receptor
missense variants. |
Table V
Clinical and hormone profile of
patients with idiopathic azoospermia with novel androgen receptor
missense variants.
| Sample ID | Age, years | FSH, IU/la | LH, IU/lb | Estradiol,
pg/mlc | Testosterone,
ng/mld |
|---|
| w320 | 25 | 27.19 | 14.51 | 29.64 | 4.23 |
| w530 | 30 | 8.12 | 11.46 | 66.61 | 8.75 |
| w691 | 24 | 25.18 | 8.40 | 20.59 | 3.15 |
Discussion
Accumulating evidence indicates that AR is a
ligand-dependent transcription factor that regulates the expression
of androgen-responsive genes (19). Androgens and AR are essential in
male spermatogenesis and fertility (20,21).
Although the AR gene has >700 reported mutants
and polymorphisms, only 5 mutants located in exon 1 have been
observed in azoospermia patients with varying degrees of impaired
spermatogenesis or mild AIS (22).
In agreement with the present study, all previous subjects
presented normal external genitalia, complicating the clinical
diagnosis. Additionally, a previous study observed that male mice
with total AR knockout exhibited female-typical external
appearance, which was similar to human AIS or testicular
feminization variant mice (23).
The current study sequenced the coding sequence of
AR in a large group of patients with IA. The R630W variant was
observed in 1 of the 776 patients with IA, but was not detected in
any of the 709 fertile men sequenced or in other individuals
previously reported in the public databases. The R630W variant was
localized in the conserved DNA-binding domain of AR, which is in
accordance with the abolished transcriptional activity of AR caused
by this mutation. Computerized analysis using PolyPhen-2.0 and SIFT
software classified the R630W variant as damaging, predicting it to
have a deleterious effect on the protein structure. Additionally,
local alignment analysis of the amino acid sequences of AR
demonstrated that the affected arginine residue was highly
conserved in multiple species, including chickens. The evolutionary
preservation of the entire region around this residue across
multiple species indicates that variants in this region may affect
the normal functions of the AR protein.
When evaluating the pathogenic effect of AR variants
in patients with fertility issues, an important question to address
is whether the variant affects transcriptional regulatory function
of AR. The present study demonstrated that the R630W variant
affected the transcriptional regulatory function of AR at the MMTV
promoter.
The hormonal profile of the patient carrying the
R630W mutation (w530) was different to the other two patients with
novel AR variants. Compared with patients w320 and w691, patient
w530 exhibited low follicle stimulating hormone, and high estradiol
and testosterone. Thus, the R630W mutation potentially reduces the
transcriptional activity of AR, which leads to an imbalance of the
hormonal profile.
In conclusion, the current study identified seven
missense variants and two synonymous novel variants of AR using
massive parallel sequencing technology. Functional analysis
confirmed that the R630W variant suppressed the normal
transcriptional regulatory function of AR. These results suggested
that AR was important in human spermatogenesis. This study also
demonstrated that systematic analysis of the genetic mutations in
large cohorts of patients complemented by subsequent functional
assays may provide novel insights into the cause of IA in
humans.
Acknowledgments
The current study was supported by grants from the
National Natural Science Foundation of China (grant nos. 31271244
and 81200465), Guangdong Natural Science Foundation of China (grant
no. 2014A030313785), Shenzhen Foundation of Science and Technology
(grant nos. GJHZ20140414170821192 and JCYJ20140414170821337), the
'Three Outstanding Projects' of Shenzhen, the Project of Shenzhen
Engineering Center and the Key Laboratory Project of Shenzhen
Second People's Hospital.
References
|
1
|
Jarvi K, Lo K, Fischer A, Grantmyre J,
Zini A, Chow V and Mak V: CUA Guideline: The workup of azoospermic
males. Can Urol Assoc J. 4:163–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hirsh A: Male subfertility. BMJ.
327:669–672. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alechine E and Corach D: High-throughput
screening for spermatogenesis candidate genes in the AZFc region of
the Y chromosome by multiplex real time PCR followed by high
resolution melting analysis. PLoS One. 9:e972272014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu M, Qin Y, Qu J, Lu C, Wang Y, Wu W,
Song L, Wang S, Chen F, Shen H, et al: Evaluation of five candidate
genes from GWAS for association with oligozoospermia in a Han
Chinese population. PLoS One. 8:e803742013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heemers HV and Tindall DJ: Androgen
receptor (AR) coregulators: A diversity of functions converging on
and regulating the AR transcriptional complex. Endocr Rev.
28:778–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heinlein CA and Chang C: Androgen receptor
(AR) coregulators: An overview. Endocr Rev. 23:175–200. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Patrão MT, Silva EJ and Avellar MC:
Androgens and the male reproductive tract: An overview of classical
roles and current perspectives. Arq Bras Endocrinol Metabol.
53:934–945. 2009. View Article : Google Scholar
|
|
8
|
Li CY, Earley RL, Huang SP and Hsu Y:
Fighting experience alters brain androgen receptor expression
dependent on testosterone status. Proc Biol Sci. 281:2014.15322014.
View Article : Google Scholar
|
|
9
|
Ferlin A, Bartoloni L, Rizzo G, Roverato
A, Garolla A and Foresta C: Androgen receptor gene CAG and GGC
repeat lengths in idiopathic male infertility. Mol Hum Reprod.
10:417–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gottlieb B, Lombroso R, Beitel LK and
Trifiro MA: Molecular pathology of the androgen receptor in male
(in)fertility. Reprod Biomed Online. 10:42–48. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bermúdez de la Vega JA, Fernández-Cancio
M, Bernal S and Audí L: Complete androgen insensitivity syndrome
associated with male gender identity or female precocious puberty
in the same family. Sex Dev. 9:75–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shao J, Hou J, Li B, Li D, Zhang N and
Wang X: Different types of androgen receptor mutations in patients
with complete androgen insensitivity syndrome. Intractable Rare Dis
Res. 4:54–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mou L, Wang Y, Li H, Huang Y, Jiang T,
Huang W, Li Z, Chen J, Xie J, Liu Y, et al: A dominant-negative
mutation of HSF2 associated with idiopathic azoospermia. Hum Genet.
132:159–165. 2013. View Article : Google Scholar
|
|
14
|
Matzuk MM and Lamb DJ: The biology of
infertility: Research advances and clinical challenges. Nature Med.
14:1197–1213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Huang Y, Li H, Hu J, Liu X, Jiang T,
Sun G, Tang A, Sun X, Qian W, et al: Excess of rare variants in
genes that are key epigenetic regulators of spermatogenesis in the
patients with non-obstructive azoospermia. Sci Rep. 5:87852015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ng PC and Henikoff S: Predicting
deleterious amino acid substitutions. Genome Res. 11:863–874. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ramensky V, Bork P and Sunyaev S: Human
non-synonymous SNPs: server and survey. Nucleic Acids Res.
30:3894–3900. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mou L, Zhang Q, Wang Y, Zhang Q, Sun L, Li
C, Huang W, Yuan Y, Duan Y, Diao R, et al: Identification of Ube2b
as a novel target of androgen receptor in mouse sertoli cells. Biol
Reprod. 89:322013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Van der Steen T and Tindall DJ: Are
androgen receptor variants a substitute for the full-length
receptor? Nat Rev Urol. 12:137–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aquila S and De Amicis F: Steroid
receptors and their ligands: Effects on male gamete functions. Exp
Cell Res. 328:303–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smith LB and Walker WH: The regulation of
spermatogenesis by androgens. Semin Cell Dev Biol. 30:2–13. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gottlieb B, Beitel LK, Nadarajah A,
Paliouras M and Trifiro M: The androgen receptor gene mutations
database: 2012 update. Hum Mutat. 33:887–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H,
Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, et al: Generation
and characterization of androgen receptor knockout (ARKO) mice: An
in vivo model for the study of androgen functions in selective
tissues. Proc Natl Acad Sci USA. 99:13498–13503. 2002. View Article : Google Scholar : PubMed/NCBI
|