Introduction
Drowning is one of the most common causes of
accidental mortality worldwide, and remains a significant medical
and social issue (1). Secondary
acute lung injury (ALI) is a serious complication of drowning.
Seawater aspiration-induced ALI, which has a rapid progress and
high mortality rate, is a common complication of drowning in
seawater (2). Seawater
aspiration-induced ALI exhibits similar characteristics to stress
situations, including trauma, burns and sepsis-induced ALI, and is
characterized by an acute inflammatory response in the pulmonary
parenchyma and interstitial tissue, severe hypoxemia and pulmonary
edema (3). Previous studies
indicated that inflammatory cell infiltration (4–7) and
permeability of the alveolar wall (8) were increased in a rat model of
seawater aspiration-induced ALI, and apoptosis of alveolar
epithelial cells was also involved in seawater aspiration-induced
ALI (9). Another of our previous
studies investigated the role of estrogen in a rat model of
seawater aspiration-induced ALI. The results demonstrated that
treatment with estrogen was able to protect against early stage
seawater aspiration-induced ALI (10). It is well-known that nuclear factor
(NF)-κB is an important molecule associated with sepsis-induced ALI
(11). Recently, it has been
reported that NF-κB and RhoA/Rho kinase pathways are critical in
the progression of seawater aspiration-induced ALI (12). However, whether the inflammatory
response is important in the pathogenesis and potential molecular
mechanisms of seawater aspiration-induced ALI remains to be
elucidated.
To elucidate the potential molecular mechanisms
underlying seawater aspiration-induced ALI, a gene expression
profile analysis was conducted on lungs obtained from a rat model
using Illumina Solexa sequencing technology. A large number of
genes were compared between the experimental groups. In order to
explore the potential molecular mechanisms underlying seawater
aspiration-induced ALI, a distilled water group was used as a
negative control, and a blank control group was also established.
Gene expression profiles were compared in an effort to identify
unique gene classifiers and to differentiate between the three
groups. The present study is the first, to the best of our
knowledge, to use gene expression profile analysis to explore the
potential molecular mechanisms underlying seawater
aspiration-induced ALI.
Materials and methods
Animals and materials
Adult male Sprague-Dawley rats (weight, 180–220 g;
age, 6 weeks) were obtained from the Animal Center of the Fourth
Military Medical University (Xi'an, China). The rats were housed in
filtered, temperature-controlled units (24°C, 50% humidity), with
ad libitum access to food and water and with diurnal
variation of light. The experiments were approved by the Animal
Care and Use Committee of the Fourth Military Medical University,
and were conducted in accordance with the Declaration of the
National Institutes of Health Guide for Care and Use of Laboratory
Animals (Publication No. 85-23; revised 1985). Seawater
(osmolality, 1,300 mmol/l; pH 8.2; specific gravity, 1.05; NaCl,
26.518 g/l; MgSO4, 3.305 g/l; MgCl2, 2.447
g/l; CaCl2, 1.141 g/l; KCl, 0.725 g/l;
NaHCO3, 0.202 g/l; NaBr, 0.083 g/l) was prepared
according to the major composition of the East China Sea provided
by the Chinese Ocean Bureau (Beijing, China). Distilled water
(osmolality, 0 mmol/l) was prepared according to distilled water
preparation.
Animal modeling and grouping
The seawater aspiration model was produced according
to the method described previously (10), with some modifications. The rats
were anesthetized with 3% pentobarbital sodium (1.5 ml/kg of body
weight; intraperitoneal administration) prior to exposure of the
tracheae.
Sprague-Dawley rats were randomly assigned into
three groups (n=3/group): Blank control group (C group), distilled
water negative control group (D group) and seawater
aspiration-induced ALI group (S group). Seawater or distilled water
(4 ml/kg) was instilled into the rat lungs via the trachea in 4 min
at a steady speed using a 2 ml syringe. In the blank control group
the rats' tracheae were exposed without instillation.
RNA isolation and lung
histopathology
The rats were sacrificed by aortic transection 6 h
after tracheal exposure. Rats were anesthetized with 3%
pentobarbital sodium (1.5 ml/kg of body weight; intraperitoneal
administration) prior to aortic transection. A section from the
right lower lobe of the lung was removed from each rat, immediately
frozen and stored in liquid nitrogen until further use. RNA
extraction, and quantification and quality assessment of the
extracted RNA was performed as previously described (13). Another section from the right lower
lobe of the lung was fixed with 4% paraformaldehyde for 24 h. The
tissues were then embedded in paraffin and cut into 5 μm
sections. Hematoxylin-eosin staining was performed according to a
standard protocol (14).
Histopathological quantification
Three samples from each group underwent
quantification of lung histopathology. The sections were stained
with hematoxylin-eosin and were analyzed independently by two
pathologists who were blind to the groups the animals belonged to.
After viewing ~10 fields per section at low and high magnification
with a light microscope, each section was given a lung injury
score, which ranged between 0 and 4, based on edema, neutrophil
infiltration, hemorrhage, bronchiole epithelial desquamation and
hyaline membrane formation, according to a previously described
standard assessment (15). The
scores (0 to 4) represent severity of lung injury: 0, no injury
(normal appearing lung); 1, modest injury (limited congestion and
edema, but no interstitial neutrophil infiltration with occasional
red blood cells and neutrophils in the alveolar spaces); 2,
intermediate injury (mild congestion, edema, and interstitial
neutrophil infiltration with occasional red blood cells and
neutrophils in the alveolar spaces); 3, widespread injury (more
prominent congestion and edema, with neutrophils partially filling
the alveolar spaces but with consolidation); and 4, widespread
injury (most prominent, marked congestion and edema, with
neutrophilic infiltrate nearly filling the alveolar spaces or with
frank lung consolidation) (16).
Lung wet/dry ratios
To quantify the magnitude of pulmonary edema, the
wet/dry weight ratios of the lung samples from the upper and middle
lobes of the right lung from each rat were determined. Tissue
samples were obtained 6 h after instillation, were immediately
weighed, and were then subjected to desiccation in an oven at 70°C
until a stable dry weight was achieved after 72 h. The wet/dry
weight ratios were calculated, in order to quantify the magnitude
of pulmonary edema (14).
Preparation of bronchoalveolar lavage
fluid (BALF)
Rats were sacrificed 6 h after instillation, then
the left lungs (n=3) were excised from the rats, and BALF was
collected using intratracheal injections of 2 ml ice-cold
phosphate-buffered saline. More than 90% of the BALF was recycled.
Subsequently, BALF was centrifuged (12,000 × g, 10 min, 4°C), and
protein concentrations in BALF were determined according to Lowry's
method (17), with bovine serum
albumin as a standard.
Sequencing analysis
One biological replicate was tested from each group.
To minimize the impact of interindividual variability on the
sequencing data, each biological replicate consisted of pooled lung
RNA samples prepared from three rats per group: C group sample (a
pooled sample from three blank control samples), D group sample (a
pooled sample from three distilled water negative control samples),
and S group sample (a pooled sample from three seawater
aspiration-induced ALI samples).
The Illumina standard kit was used for mRNA-Seq
sample preparation, according to the manufacturer's protocol
(Illumina, San Diego, CA, USA). Sequencing was performed on a
Genome Analyzer (GAII; Illumina) using a 50-bp read length. The
sequence reads were processed and contrasted to the Ensembl
(http://asia.ensembl.org/info/data/ftp/index.html)
rat reference genome using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml). The
relative abundance of genes was measured using Cufflinks
(http://cole-trapnell-lab.github.io/cufflinks/)
compared with normalized RNA-Seq fragments. The unit of measurement
is fragments per kilobase of exon per million fragments mapped
(FPKM). The confidence intervals for the FPKM estimates were
calculated according to a Bayesian inference method. The
differentially expressed genes were visualized using the CummeRbund
program (http://compbio.mit.edu/cummeRbund/).
Gene products that were ≥2-fold differentially
expressed were further analyzed, as described in the results
section. Differentially expressed genes were classified according
to their gene ontology (GO; http://www.geneontology.org/), using goatools
(https://github.com/tanghaibao/goatools). Cellular
pathway association was analyzed according to the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) using the KEGG Orthology
Based Annotation System (http://kobas.cbi.pku.edu.cn/home.do). The sequencing
data have been deposited in the National Center for Biotechnology
Information short read archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under
the accession number SRP052721.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT was performed on total RNA (200 ng) in a total
volume of 13 μl containing 1 μl 50 μM random
hexamers, 1 μl 10 mM dNTP mix, DEPC water. RNA-primer mix
was incubated at 65°C for 5 min, then incubated on ice for >1
min. 5X SuperScript IV Buffer (4 μl; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 mM dithiothreitol (1
μl), RNase inhibitor (1 μl) and 200 U/μl
SuperScript Reverse Transcriptase (1 μl; Thermo Fisher
Scientific, Inc.) were added to the reaction. The samples were
incubated at 23°C for 10 min, 50°C for 10 min, and 80°C for 10 min.
qPCR reactions were performed using the QuantiTect SYBR Green PCR
kit (Qiagen, Inc., Valencia, CA, USA) to detect the mRNA expression
levels of the following genes: C-X-C motif chemokine ligand
(CXCL)2, CXCL13, C-C motif chemokine ligand (CCL)20, CCL3,
interleukin (IL)-6, IL-1 receptor type 2 (IL1R2) and tumor necrosis
factor superfamily member 10 (TNFSF10). Each value obtained is the
mean of three independent biological replicates. The following
primers were used: CXCL2, forward 5′-CAGCCTCGAGTCTCACCTGT-3′;
reverse 5′-CACTCTTTGGTCCAGAGCCAT-3′. CXCL13, forward
5′-GCTCACCTTGGAACACCTAC-3′, reverse 5′-ACACCTAGGTTGTATGCCCA-3′;
CCL20, forward 5′-AAAAGATCCGTGTGCGCTGA-3′, reverse
5′-CTCTGCTCCCTTATTCCCGTC-3′; CCL3, forward
5′-CGTGGAATTTGCCGTCCATAG-3′, reverse 5′-GTCCTGTCCTGCTCAAGGAT-3′;
IL-6, forward 5′-CCAGATAGCTGCTCAGAAGGG-3′, reverse
5′-GTCTTTGCAAAAGAGAGGCCC-3′; IL1R2, forward
5′-CTATCTGGGGGACGCTGAAA-3′, reverse 5′-TTCAACGCCCTACAAGGAGAC-3′;
TNFSF10, forward 5′-CCGGAATGGTTGGAAGTGGTT-3′, reverse
5′-GGTATGGCGGAGTTAGCACG-3′; and L32, forward
5′-GATCTTGGGCTTCACCAGAGG-3′, reverse 5′-CTTGAGACACGCAGCTCCAC-3′ The
cycling condition used were incubation at 95°C for 20 sec, followed
by 40 cycles of denaturation at 95°C for 3 sec, followed by
annealing, extension and data acquisition at 60°C for 30 sec using
the ABI 7500 Fast Real-Time PCR system (Thermo Fisher Scientific,
Inc.). The internal reference was L32 and quantification was
performed using the standard curve method.
Statistical analysis
Statistical differences between groups were
determined by one-way analysis of variance followed by Tukey's test
for qPCR. For non-continuous histological injury score data, the
Kruskal-Wallis test was used. Analysis was performed using SPSS
software (version 13.0; SPSS, Inc., Chicago, IL, USA). Results are
presented as the mean ± standard error of the mean. P<0.05 was
considered to indicate a statistically significant difference.
Results
Histopathological alterations and lung
injury scores in seawater aspiration-induced ALI
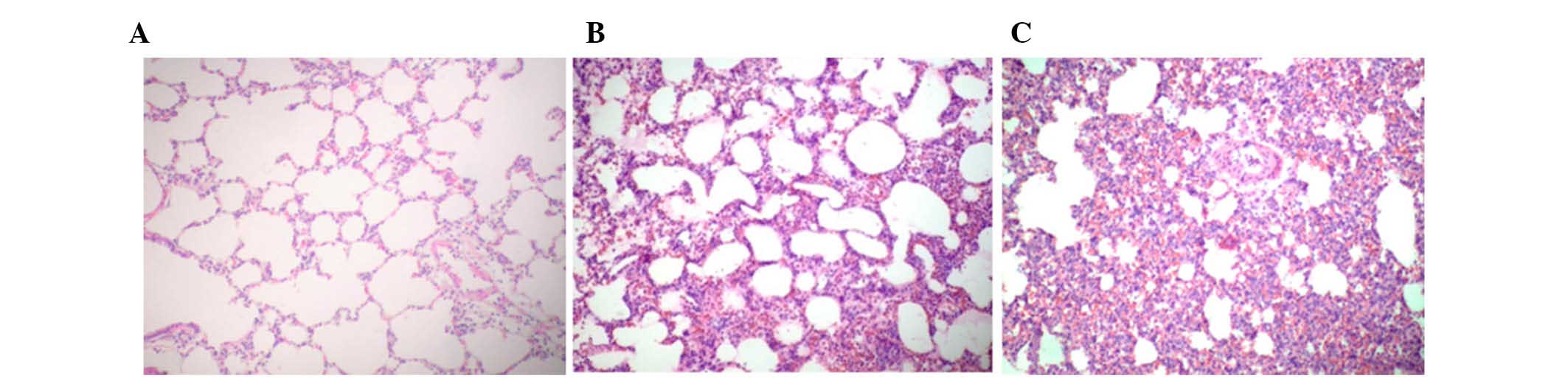
Histopathology images are presented in Fig. 1. The results demonstrated that
seawater aspiration induced pulmonary edema, infiltration of
inflammatory cells into the lung tissues and alveoli, and alveolar
collapse (S group; Fig. 1C). The
distilled water negative control group exhibited similar lung
injury (D group; Fig. 1B).
However, alterations in the S group were more severe compared with
in the D group. Normal histopathology was observed in the lung
samples of the blank control group (C group; Fig. 1A). The assigned lung injury scores
of each group with regards to edema, neutrophil infiltration,
hemorrhage, bronchiole epithelial desquamation and hyaline membrane
formation are shown in Table
I.
 | Table IAssigned lung injury scores in the
seawater aspiration-induced acute lung injury group and the control
groups. |
Table I
Assigned lung injury scores in the
seawater aspiration-induced acute lung injury group and the control
groups.
| Group | Edema | Neutrophil
infiltration | Hemorrhage | Bronchiole epithelial
desquamation | Hyaline membrane |
|---|
| Blank control | 0.1±0.16 | 0.2±0.26 | 0.1±0.15 | 0.2±0. 23 | 0.0±0.00 |
| Distilled water
negative control | 3.0±0.15a,b | 3.2±0.17a,b | 2.2±0.25a,b | 2.7±0.28a,b | 0.2±0.24 |
| Seawater | 3.7±0.24a | 3.6±0.25a | 3.1±0.37a | 3.5±0.36a | 0.3±0.36 |
Lung wet/dry ratios in seawater
aspiration-induced ALI
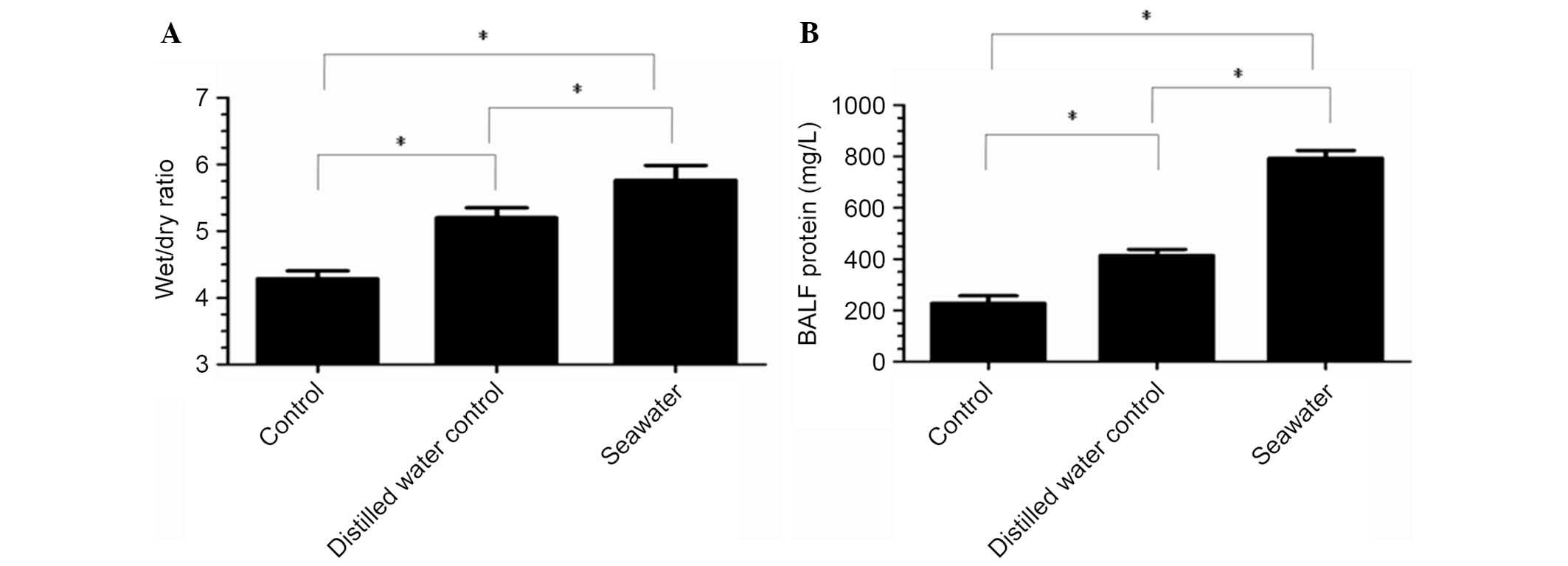
To investigate the severity of pulmonary edema in
the S group, lung wet/dry ratios were measured (Fig. 2A). The results indicated that
seawater aspiration significantly increased the lung wet/dry ratios
(5.77±0.22) compared with the C group (4.29±0.12; P<0.05; n=3)
and the D group (5.2±0.15; P<0.05; n=3).
Total protein concentrations in BALF in
seawater aspiration-induced ALI
Total protein concentrations in BALF were increased
in the S group (793.67±30.37) compared with the C group
(228.33±28.95; P<0.05; n=3) and the D group (414.33±23.53;
P<0.05; n=3) (Fig. 2B).
Gene expression analysis
A total of 29,516 genes were attained from each
group by RNA-Seq short sequence read analysis, which were aligned
to the Ensembl rat reference genome. A total of 277 and 250 genes
in the S group were revealed to be up and downregulated
respectively, as compared with the C group. In addition, there were
152 and 130 genes that were up and downregulated in the S group
respectively, as compared with the D group. The top 10 up and
downregulated genes are listed in Tables II and III.
 | Table IITop 10 up and downregulated genes in
the seawater aspiration-induced acute lung injury group compared
with the blank control group. |
Table II
Top 10 up and downregulated genes in
the seawater aspiration-induced acute lung injury group compared
with the blank control group.
| No. | Gene | S (FPKM) | C (FPKM) | Fold changea | P-value |
|---|
| Up | | | | | |
| 1 | Cxcl2 | 158.856 | 0.935096 | 169.89 | <0.01 |
| 2 | F1M1Q7_RAT | 615.196 | 4.13873 | 148.63 | <0.01 |
| 3 | Orm1 | 104.343 | 0.718 | 145.32 | 8.49E-12 |
| 4 | Irg1 | 75.7689 | 0.635845 | 119.16 | <0.01 |
| 5 | B1H259_RAT | 5.58517 | 0.091918 | 60.76253 | 5.75E-05 |
| 6 | Cxcl6 | 110.903 | 2.12643 | 52.15451 | 7.89E-08 |
| 7 | D3ZDX3_RAT | 19.8754 | 0.389918 | 50.9733 | 0.000122 |
| 8 | Csf3 | 9.44276 | 0.186485 | 50.6356 | 0.000126 |
| 9 | Fcrls | 117.251 | 2.67019 | 43.91129 | <0.01 |
| 10 | Tnn | 6.79358 | 0.169375 | 40.10972 | 2.41E-06 |
| Down | | | | | |
| 1 | F1LVW9_RAT | 0.561806 | 14.1462 | 25.17989 | 0.002012 |
| 2 | Rnase17 | 1.16126 | 21.15 | 18.21292 | 0.038443 |
| 3 | D3Z9Q1_RAT | 2.15125 | 37.4138 | 17.39161 | 1.06E-09 |
| 4 | Vipr1 | 12.7842 | 196.93 | 15.40418 | 1.21E-07 |
| 5 | Ear11 | 4.0495 | 61.6461 | 15.2232 | 4.98E-10 |
| 6 | Slc6a2 | 0.61605 | 7.91125 | 12.84193 | 3.46E-06 |
| 7 | Nova2 | 0.458669 | 5.88786 | 12.83686 | 5.59E-05 |
| 8 | Csrp3 | 0.649324 | 8.33526 | 12.83686 | 0.000771 |
| 9 | F1M5M6_RAT | 8.74595 | 111.67 | 12.76826 | 2.96E-07 |
| 10 | Cyp1a1 | 2.47813 | 29.744 | 12.00256 | 1.07E-11 |
 | Table IIITop 10 up and downregulated genes in
the seawater aspiration-induced acute lung injury group compared
with the distilled water negative control group. |
Table III
Top 10 up and downregulated genes in
the seawater aspiration-induced acute lung injury group compared
with the distilled water negative control group.
| No. | Gene | S (FPKM) | D (FPKM) | Fold changea | P-value |
|---|
| Up | | | | | |
| 1 | F1M1Q7_RAT | 615.196 | 2.90533 | 211.7473 | <0.01 |
| 2 | F1M663_RAT | 358.937 | 9.77567 | 36.71735 | 8.44E-15 |
| 3 | Il6 | 26.1417 | 1.0887 | 24.01194 | 1.34E-09 |
| 4 | D4A7S8_RAT | 79.1362 | 3.4686 | 22.81499 | 1.69E-06 |
| 5 | Irg1 | 75.7689 | 3.44331 | 22.0047 | <0.01 |
| 6 | F1LWN4_RAT | 112.483 | 5.1964 | 21.64617 | 9.04E-08 |
| 7 | Fcrls | 117.251 | 5.88037 | 19.9394 | <0.01 |
| 8 | Slamf9 | 183.298 | 10.7154 | 17.10599 | 4.44E-16 |
| 9 | LOC100363638 | 38.4476 | 2.44753 | 15.70868 | 0.000528 |
| 10 | Atp6v0a4 | 5.02422 | 0.339452 | 14.80097 | 4.02E-06 |
| Down | | | | | |
| 1 | Cyp1a1 | 2.47813 | 51.4444 | 20.75936 | 2.22E-16 |
| 2 | F1LVW9_RAT | 0.561806 | 9.76356 | 17.37884 | 0.000146 |
| 3 | F1MAE7_RAT | 25.1881 | 393.779 | 15.63362 | 2.43E-13 |
| 4 | Wnk2 | 0.279078 | 4.32902 | 15.51186 | 8.09E-08 |
| 5 | Vipr1 | 12.7842 | 164.067 | 12.83365 | 9.70E-07 |
| 6 |
ENSRNOG00000030444 | 1.37746 | 15.7838 | 11.45861 | 3.90E-07 |
| 7 | Ear11 | 4.0495 | 43.4886 | 10.73924 | 8.88E-08 |
| 8 | Nr1d1 | 3.18831 | 32.3391 | 10.14298 | 0.000344 |
| 9 | LOC100365881 | 1.44117 | 13.8846 | 9.634234 | 8.34E-06 |
| 10 | Zbtb16 | 6.00713 | 54.2278 | 9.027272 | 4.31E-11 |
Functional clusters analysis
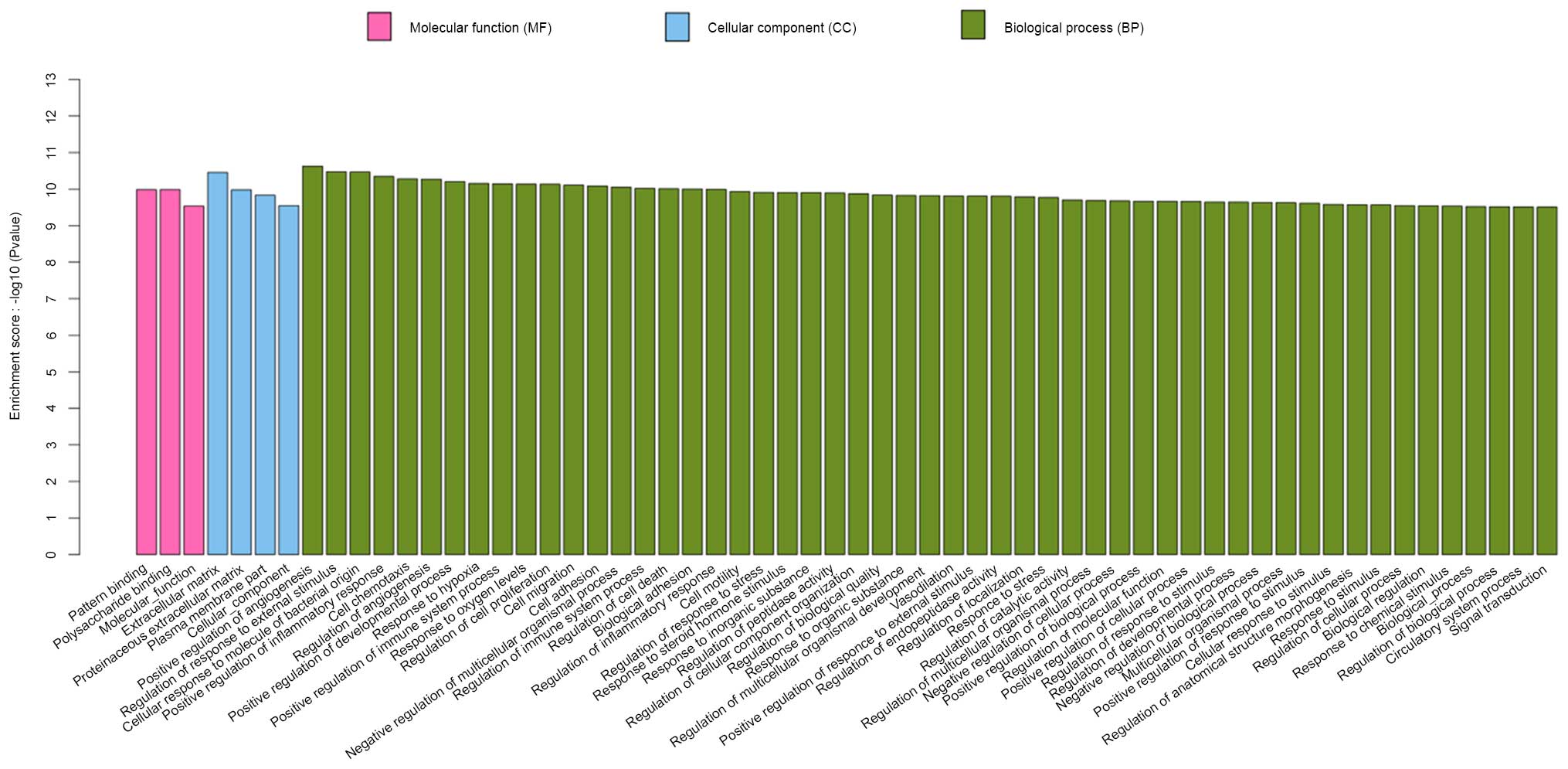
Functional enrichment analysis of the GO and KEGG
pathways was carried out with a significance threshold of 0.05 for
the adjusted P-value. Identified genes in the S group were
associated with 245 specific enriched GO terms. A total of 60 of
the most significant GO terms are presented in Fig. 3, of which three were associated
with molecular function, four were associated with cellular
component, and 53 were associated with biological process. As shown
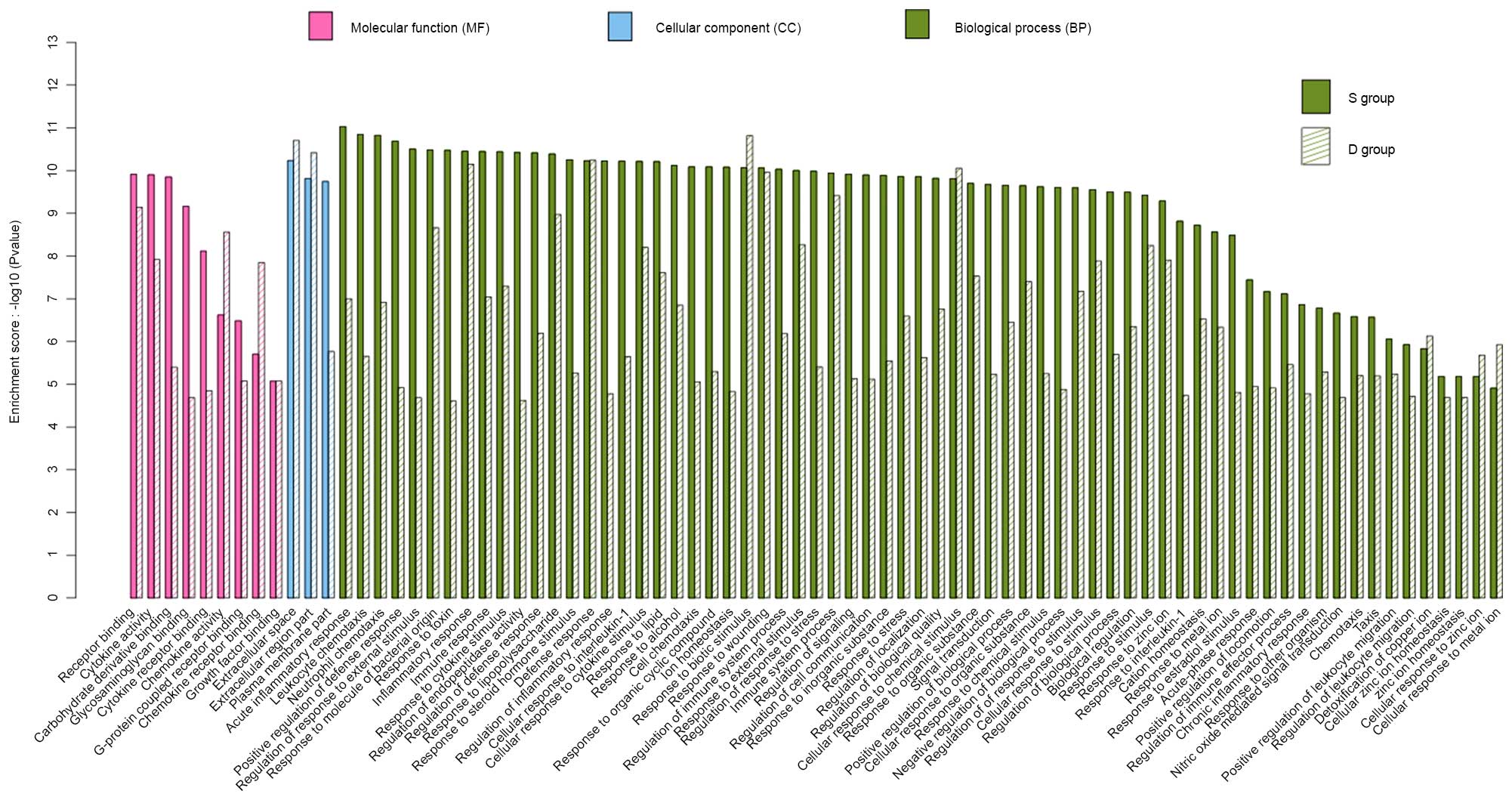
in Fig. 4, there were 80 enriched
GO terms in the S and D groups. All major genes were associated
with biological process, cellular component and molecular function.
These genes were, for the most part, associated with neutrophil
chemotaxis, immune and defense responses, and cytokine activity.
Changes to these GO terms were more obvious in the S group compared
with in the D group.
KEGG analysis demonstrated that several gene
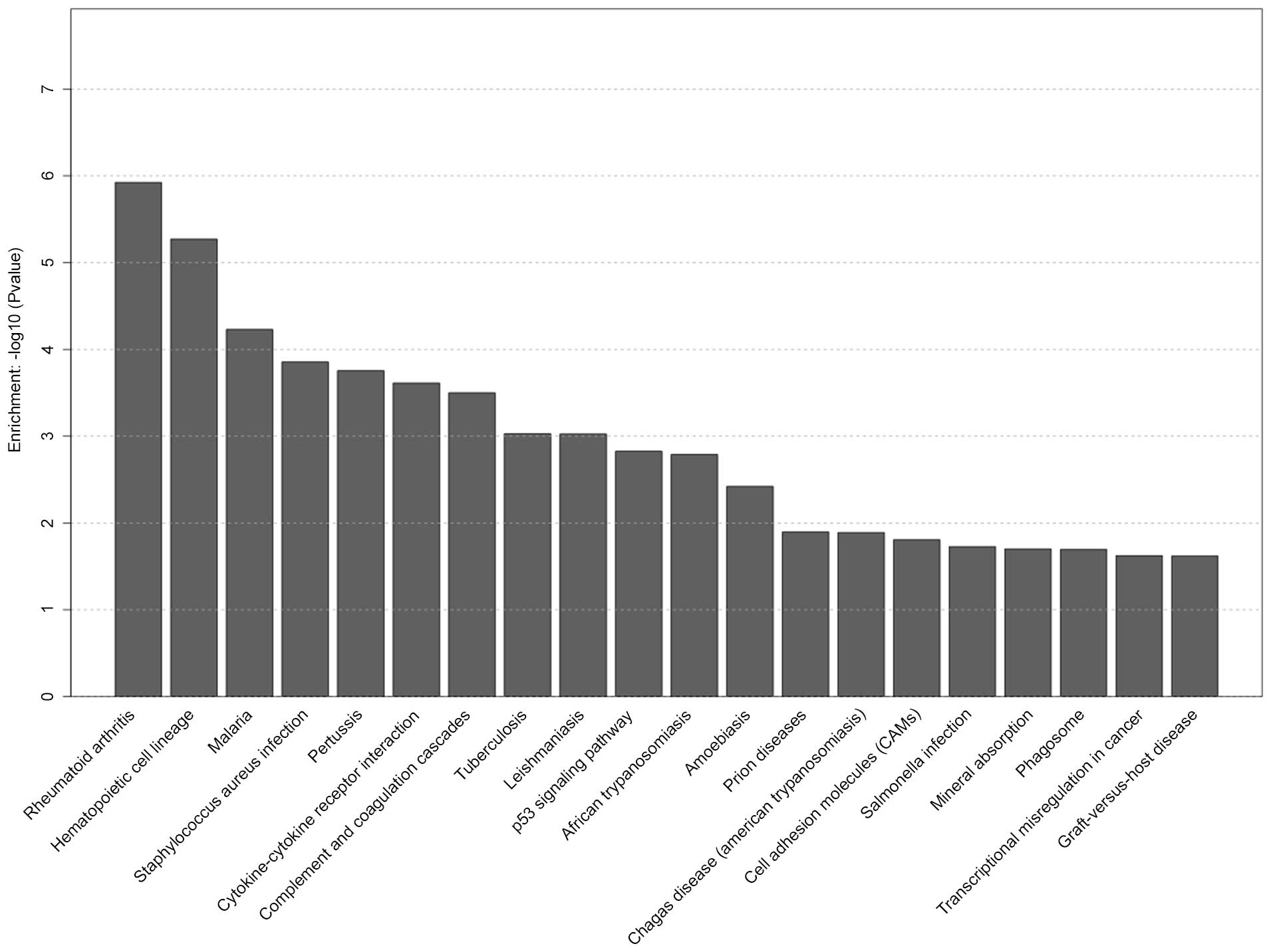
pathways were significantly affected in the S group. The top 20
selected pathways in the S group are presented in Fig. 5, including hematopoietic cell
lineage, cytokine-cytokine receptor interaction, p53 signaling
pathway, cell adhesion molecule and phagosome pathway. Selected
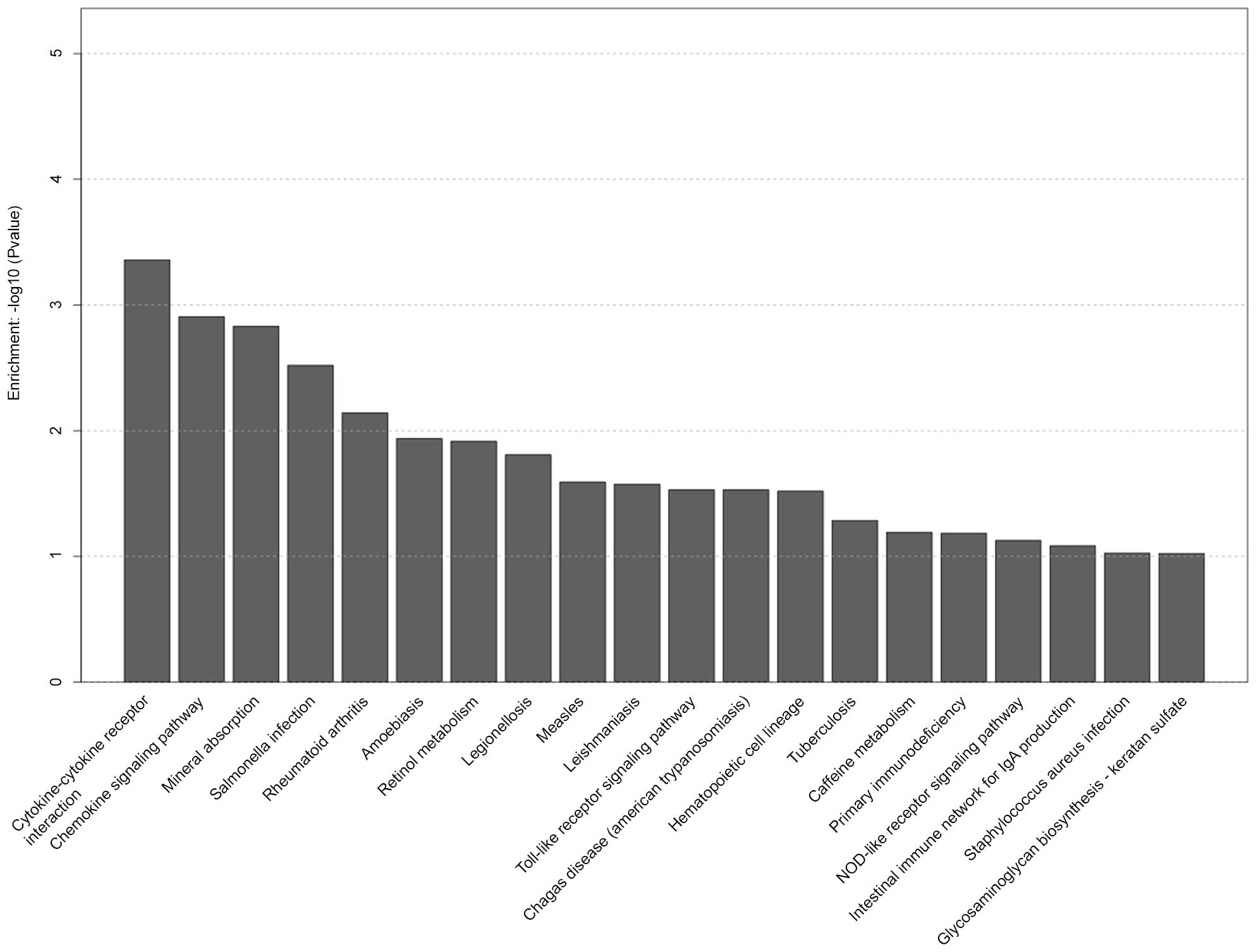
pathways in the D group are shown in Fig. 6, including cytokine-cytokine
receptor interaction, chemokine signaling pathway, toll-like
receptor signaling pathway and hematopoietic cell lineage.
The cytokine-cytokine receptor interaction pathway
was one of the most important pathways in the S and D groups. The
cytokine-cytokine receptor interaction pathway in S group is shown
in Fig. 7. mRNA sequencing
demonstrated that certain genes were upregulated, including CXCL2,
CXCL13, CCL20, CCL3, IL-6, IL-10, IL-23A, oncostatin M (OSM),
colony stimulating factor (CSF), CSF3 receptor, TNF receptor
superfamily member 11a (TNFSF11A) and TNFRSF9. However, other genes
were downregulated, including KIT and TNFSF10. As shown in Fig. 8, the expression of certain genes
associated with the cytokine-cytokine receptor interaction pathway
were also altered in the D group, including CXCL2, CCL20, CCL3, OSM
receptor and IL1R2, which were upregulated, and CXCL9 and TNFSF10,
which were downregulated.
qPCR
qPCR is used to measure gene expression levels, and
is often used to confirm the results of sequencing studies. Seven
cytokine-cytokine receptor interaction pathway-related genes
(CXCL2, CXCL13, CCL20, CCL3, IL6, IL1R2 and TNFSF10) were selected
for qPCR. The results are shown in Table IV. The results of the gene
expression profiles determined by sequencing were consistent with
the results of qPCR.
 | Table IVqPCR validation of cytokine-cytokine
receptor interaction pathway-related gene expression data. |
Table IV
qPCR validation of cytokine-cytokine
receptor interaction pathway-related gene expression data.
| Gene_ID | Gene | Description | Seawater
| Distilled water
|
|---|
Sequencing
| qPCR
| Sequencing
| qPCR
|
|---|
| Fold changea | P-value | Fold changea | P-value | Fold changea | P-value | Fold changea | P-value |
|---|
|
ENSRNOG00000002792 | CXCL2 | C-X-C motif
chemokine 2 | 169.88 | <0.001 | 152.10 | <0.01 | 12.41 | <0.001 | 12.15 | <0.01 |
|
ENSRNOG00000024899 | CXCL13 | C-X-C motif
chemokine 13 |
12.14 | <0.001 |
11.32 | <0.01 |
3.59 | <0.001 |
4.07 | <0.01 |
|
ENSRNOG00000015992 | CCL20 | C-C motif chemokine
20 |
5.60 | <0.001 |
6.10 | <0.01 |
6.85 | <0.001 |
7.24 | <0.01 |
|
ENSRNOG00000011205 | CCL3 | C-C motif chemokine
3 |
33.02 | <0.001 |
30.14 | <0.01 |
4.27 | <0.001 |
4.58 | <0.01 |
|
ENSRNOG00000010278 | IL-6 | Interleukin-6 |
27.09 | <0.001 |
25.69 | <0.01 |
1.13 | 0.86 |
1.10 | >0.01 |
|
ENSRNOG00000014378 | IL1R2 | Interleukin-1
receptor type 2 |
21.22 | <0.001 |
24.2 | <0.01 |
21.87 | <0.001 | 22.97 | <0.01 |
|
ENSRNOG00000013269 | TNFSF10 | Tumor necrosis
factor ligand superfamily member 10 |
0.13 | <0.001 |
0.15 | <0.01 |
0.26 | <0.001 |
0.31 | <0.01 |
Discussion
One of the serious complications of drowning is
secondary acute lung injury (ALI), which is characterized by
relentless deterioration and mortality (18,19).
Previous studies have focused on seawater aspiration-induced ALI
(4–10); however, few have involved pathway
and GO analyses of seawater aspiration-induced ALI. The present
study is the first, to the best of our knowledge, to develop a
genetic signature for seawater aspiration-induced ALI with gene
expression profiles.
The histopathological analysis demonstrated that
seawater aspiration-induced ALI was able to induce pulmonary edema,
infiltration of inflammatory cells into the lung tissues, and
alveolar collapse. These alterations were more serious in the S
group compared with in the D group (20). To investigate the pathogenesis of
seawater aspiration-induced ALI a gene expression profile analysis
was conducted using KEGG and GO analyses. The results identified
differential gene expression patterns among the S group, the D
group and the C group.
GO and KEGG pathway analyses provided a global view
on the function of the identified genes. The results of the GO
analysis demonstrated that there were 80 enriched GO terms in the S
and D groups, including biological process, cellular component and
molecular function. Neutrophil chemotaxis, immune and defense
responses, and cytokine activity were the most significantly
enriched GO terms. These GO terms were more enriched in the S group
compared with in the D group. These enriched GO terms indicated
that regulation of response to external stimulus and inflammatory
responses may be the main biological reactions and molecular
functions in the S and D groups (21); however, these reactions were more
intense in the S group.
The present study also revealed that hematopoietic
cell lineage, cytokine-cytokine receptor interaction, p53 signaling
pathway, cell adhesion molecule and phagosome pathway were
significantly altered in the S group, whereas cytokine-cytokine
receptor interaction, chemokine signaling pathway, toll-like
receptor signaling pathway and hematopoietic cell lineage were
significantly altered in the D group. Among these, we focused on
the cytokine-cytokine receptor interaction pathway as one of the
essential pathways of pathogenesis in the S and D groups. It has
previously been reported that inflammation and cytokines have a key
role in the pathogenesis of seawater aspiration-induced ALI
(3,21), as is consistent with the present
study. Furthermore, the expression of genes associated with the
cytokine-cytokine receptor interaction pathway were altered in the
S group, including CXCL2, CXCL13, CCL20, CCL3, IL6, IL1R2 and
TNFSF10. With regards to the inflammatory response, lung tissue in
the S and D groups expressed higher levels of chemotactic and
inflammatory factors, and lower levels of members of the tumor
necrosis factor ligand superfamily compared with in the C group.
However, these gene expression changes were markedly more obvious
in the S group compared with in the D group as detected by qPCR. In
particular, several studies have reported an association between
TNFSF10 and neutrophilic inflammation. It has been demonstrated
that the absence of TNFSF10 [alias TNF-related apoptosis-inducing
ligand (TRAIL)] results in enhancement of neutrophilic inflammation
in ALI and sepsis-induced organ injury, and exogenous TRAIL can
induce apoptosis of neutrophils in tissues (22,23).
Therefore, the downregulation of TNFSF10 may substantially
contribute to the development of neutrophilic inflammation in
seawater aspiration-induced ALI; however, the detailed mechanism
remains unclear.
Notably, the proinflammatory cytokine IL-6 was
markedly increased in the S group, but not in the D group compared
with the C group. It has previously been demonstrated that IL-6
mRNA expression is increased in response to high glucose
concentrations in human osteoblastic cells (24). In addition, under hyperosmotic
stress, IL-6 was increased in an ex vitro human conjunctival
epithelial cell model of dry-eye (25). Therefore, the differential
expression of IL-6 in the S group compared with in the D group may
be due to hyperosmotic stress.
In conclusion, the differential gene expression
patterns detected in the present study suggested that activation of
the cytokine-cytokine receptor interaction pathway has an important
role in the pathogenesis of seawater aspiration-induced ALI.
Downregulation of TNFSF10 may enhance inflammation, whereas the
differential expression of IL-6 may be due to variations in
hyperosmotic stress between the S and D groups. Therefore, IL-6 may
be considered a biomarker to distinguish the seawater
aspiration-induced ALI group from the distilled water negative
control group. However, further research is required to elucidate
the mechanistic relationship and the biological implications of the
overexpression or downregulation of cytokine-cytokine receptor
interaction-related genes in seawater aspiration-induced ALI.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81270124).
References
|
1
|
van Beeck EF, Branche CM, Szpilman D,
Modell JH and Bierens JJ: A new definition of drowning: Towards
documentation and prevention of a global public health problem.
Bull World Health Organ. 83:853–856. 2005.PubMed/NCBI
|
|
2
|
Salomez F and Vincent JL: Drowning: A
review of epidemiology, pathophysiology, treatment and prevention.
Resuscitation. 63:261–268. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xinmin D, Yunyou D, Chaosheng P, Huasong
F, Pingkun Z, Jiguang M, Zhiqian X and Qinzhi X: Dexamethasone
treatment attenuates early seawater instillation-induced acute lung
injury in rabbits. Pharmacol Res. 53:372–379. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma L, Li Y, Zhao Y, Wang Q, Nan Y, Mu D,
Li W, Sun R, Jin F and Liu X: 3,5,4′-tri-O-acetylresveratrol
ameliorates seawater exposure-induced lung injury by upregulating
connexin 43 expression in lung. Mediators Inflamm. 2013:1821322013.
View Article : Google Scholar
|
|
5
|
Zhang Y, Zhang B, Xu DQ, Li WP, Xu M, Li
JH, Xie XY, Fan QX, Liu W, Mu DG, et al: Tanshinone IIA attenuates
seawater aspiration-induced lung injury by inhibiting macrophage
migration inhibitory factor. Biol Pharm Bull. 34:1052–1057. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Zhao Y, Li B, Wang Q, Liu X, Chen X,
Nan Y, Liang L, Chang R, Liang L, et al:
3,5,4′-Tri-O-acetylresveratrol attenuates seawater
aspiration-induced lung injury by inhibiting activation of nuclear
factor-kappa B and hypoxia-inducible factor-1α. Respir Physiol
Neurobiol. 185:608–614. 2013. View Article : Google Scholar
|
|
7
|
Liu W, Dong M, Bo L, Li C, Liu Q, Li Y, Ma
L, Xie Y, Fu E, Mu D, et al: Epigallocatechin-3-gallate ameliorates
seawater aspiration-induced acute lung injury via regulating
inflammatory cytokines and inhibiting JAK/STAT1 pathway in rats.
Mediators Inflamm. 2014:6125932014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Xu M, Fan Q, Xie X, Zhang Y, Mu D,
Zhao P, Zhang B, Cao F, Wang Y, et al: Tanshinone IIA ameliorates
seawater exposure-induced lung injury by inhibiting aquaporins
(AQP) 1 and AQP5 expression in lung. Respir Physiol Neurobiol.
176:39–49. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han F, Luo Y, Li Y, Liu Z, Xu D, Jin F and
Li Z: Seawater induces apoptosis in alveolar epithelial cells via
the Fas/FasL-mediated pathway. Respir Physiol Neurobiol. 182:71–80.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan Q, Zhao P, Li J, Xie X, Xu M, Zhang Y,
Mu D, Li W, Sun R, Liu W, et al: 17β-Estradiol administration
attenuates seawater aspiration-induced acute lung injury in rats.
Pulm Pharmacol Ther. 24:673–681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hiroshima Y, Hsu K, Tedla N, Chung YM,
Chow S, Herbert C and Geczy CL: S100A8 induces IL-10 and protects
against acute lung injury. J Immunol. 192:2800–2811. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M, Dong M, Liu W, Wang L, Luo Y, Li
Z and Jin F: 1α, 25-dihydroxyvitamin D3 ameliorates seawater
aspiration-induced acute lung injury via NF-κB and RhoA/Rho kinase
pathways. PLoS One. 9:e1045072014. View Article : Google Scholar
|
|
13
|
Nosotti M, Falleni M, Palleschi A,
Pellegrini C, Alessi F, Bosari S and Santambrogio L: Quantitative
real-time polymerase chain reaction detection of lymph node lung
cancer micrometastasis using carcinoembryonic antigen marker.
Chest. 128:1539–1544. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Zhang B, Chen XJ, Xu DQ, Wang YX,
Dong HY, Ma SR, Sun RH, Hui YP and Li ZC: Osthole protects
lipopolysaccharide-induced acute lung injury in mice by preventing
downregulation of angiotensin-converting enzyme 2. Eur J Pharm Sci.
48:819–824. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou ZH, Sun B, Lin K and Zhu LW:
Prevention of rabbit acute lung injury by surfactant, inhaled
nitric oxide, and pressure support ventilation. Am J Respir Crit
Care Med. 161:581–588. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen XJ, Zhang B, Hou SJ, Shi Y, Xu DQ,
Wang YX, Liu ML, Dong HY, Sun RH, Bao ND, et al: Osthole improves
acute lung injury in mice by upregulating Nrf-2/thioredoxin 1.
Respir Physiol Neurobiol. 188:214–222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scharfman A, Hayem A, Lebas J, Laine A and
Sablonnière B: Comparison of three methods for protein measurement:
application to bronchoalveolar lavage fluids. Bull Eur Physiopathol
Respir. 16:785–789. 1980.PubMed/NCBI
|
|
18
|
Saidel-Odes LR and Almog Y: Near-drowning
in the Dead Sea: A retrospective observational analysis of 69
patients. Isr Med Assoc J. 5:856–858. 2003.PubMed/NCBI
|
|
19
|
Gregorakos L, Markou N, Psalida V,
Kanakaki M, Alexopoulou A, Sotiriou E, Damianos A and Myrianthefs
P: Near-drowning: Clinical course of lung injury in adults. Lung.
187:93–97. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rui M, Duan YY, Wang HL, Zhang XH and Wang
Y: Differences between seawater- and freshwater-induced lung
injuries. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 21:416–420.
2009.In Chinese. PubMed/NCBI
|
|
21
|
Miyazato T, Ishikawa T, Michiue T and
Maeda H: Molecular pathology of pulmonary surfactants and cytokines
in drowning compared with other asphyxiation and fatal hypothermia.
Int J Legal Med. 126:581–587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McGrath EE, Marriott HM, Lawrie A, Francis
SE, Sabroe I, Renshaw SA, Dockrell DH and Whyte MK: TNF-related
apoptosis-inducing ligand (TRAIL) regulates inflammatory neutrophil
apoptosis and enhances resolution of inflammation. J Leukoc Biol.
90:855–865. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beyer K, Poetschke C, Partecke LI, von
Bernstorff W, Maier S, Broeker BM and Heidecke CD: TRAIL induces
neutrophil apoptosis and dampens sepsis-induced organ injury in
murine colon ascendens stent peritonitis. PLoS One. 9:e974512014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garcia-Hernández A, Arzate H,
Gil-Chavarria I, Rojo R and Moreno-Fierros L: High glucose
concentrations alter the biomineralization process in human
osteoblastic cells. Bone. 50:276–288. 2012. View Article : Google Scholar
|
|
25
|
Kim JH, Kang SS, Kim ES, Kim JY, Kim MJ
and Tchah H: Osmoprotective effects of supplemental epidermal
growth factor in an ex vivo multilayered human conjunctival model
under hyperosmotic stress. Graefes. Arch Clin Exp Ophthalmol.
251:1945–1953. 2013. View Article : Google Scholar
|