Introduction
Chronic kidney disease (CKD) is increasingly
considered to be a worldwide public health problem, and affects
>10% of the adult population when it is defined by the presence
of micro- or macroalbuminuria (1).
In 1990, CKD was ranked the 36th cause of death, while it ranked
19th in 2013 (2). Diverse
physiological and pathological events, such as aging, diabetes,
hypertension, obesity and cardiovascular diseases contribute to CKD
(3). Furthermore, renal fibrosis
manifests as CKD progresses, and in turn contributes to a decreased
glomerular filtration rate and associated complications including
anemia (4) and hyperkalemia
(5), leading to end-stage renal
disease. Renal fibrosis, characterized by tubular atrophy,
glomerulosclerosis and tubulointerstitial fibrosis, leads to
interruption of the normal architecture of the kidney, thus
contributing to the exacerbation of CKD (3,6).
Fibroblasts/myofibroblast activation and accumulation cause the
excessive deposition of abundant extracellular matrix (ECM), which
is the hallmark of renal fibrosis (7). During fibroblast/myofibroblast
activation and accumulation, epithelial-to-mesenchymal transition
(EMT) was considered as an essential step (8). With the loss in cell polarity and
change in cell shape from cuboidal to fibroblastoid, epithelial
marker downregulation and mesenchymal marker upregulation are
prominent in EMT (9). TGF-β
signaling activates various transcription factors, such as snail
family zinc finger 1, epithelial cadherin, and β-catenin, which are
significant in the induction of EMT (10). Thus, regulation of EMT in tubular
epithelial cells could be a strategy to alleviate renal
fibrosis.
The klotho (KL) gene was originally
identified as a suppressor of aging, and is primarily expressed in
the brain and kidney. KL−/− mice
exhibit premature aging phenotypes in multiple organs, such as
neural degeneration, osteoporosis, atherosclerosis and a shortened
life span (11), while KL
over-expression mice have an extended life span (12). Recently, experimental studies have
demonstrated that KL is involved in kidney-associated
disorders (13,14), as demonstrated by the reduced
expression levels in acute and CKD, implying its nephron-protective
role. As KL acts as an endogenous inhibitor of multiple
growth factors, including transforming growth factor-β (TGF-β) and
insulin-like growth factor-1, it is obvious that KL
suppresses renal fibrosis (15).
Thus, understanding the mechanism by which KL expression is
regulated may facilitate with improving the therapeutic strategies
for kidney diseases, including CKD.
Epigenetic alterations, such as DNA methylation,
represent a significant method for gene expression silencing. CpG
islands exist in the KL promoter, which are susceptible to
methylation that is mediated by DNA-cytosine methyltransferase 1
(Dnmt1). Indeed, hypermethylation of CpG islands in the KL
promoter causes a loss of KL expression in various cell
types, and potentially contributes to cancer development (16,17),
such as human breast and cervical cancer, and potentially CKD
(15). While DNA demethylating
agent, 2′-deoxy-5-azacytidine could increase KL expression,
emphasizing the essential role of epigenetic regulation in
KL expression, which highlights the promising future of DNMT
agents for numerous disorders.
Curcumin, a polyphenol pigment extracted from
turmeric, has been reported to eliminate reactive oxygen species,
repress proliferation and exert an anti-inflammatory effect, with
no toxic side-effects being observed upon application (18). Our previous study identified that
curcumin prevented TGF-β inducing PAI-1 and α-SMA expression, and
inhibited phosphorylation of the SMAD protein, which limited the
transdifferentiation from renal tubular epithelial cells to
fibroblast cells in a dose- and time-dependent manner (19), thus may be beneficial in the
treatment of renal fibrosis and kidney disorders. Additionally,
curcumin treatment decreases the expression levels of α-SMA and
TGF-β in the kidney and improves the clinical score for
tubulointerstitial fibrosis. In addition, curcumin downregulates
Dnmt1 expression in AML cell lines, in vitro and in
vivo, and in primary AML cells ex vivo, thus functions
as a Dnmt inhibitor (20).
In the present study, the protection effects of
curcumin were found to be due to its inhibition of CpG island
methylation in KL promoters, thus inducing KL
expression. Notably, with the elevated expression level of
KL, the methylation of KL was reduced and in turn
inhibited TGF-β signaling. Therefore, the present study proposes
that curcumin predominantly protects against renal toxicity via the
regulation of KL methylation and may serve as a novel target
for the treatment of chronic fibrosis.
Materials and methods
Reagents and cell lines
Curcumin [1,7-bis
(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione] was purchased
from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions of
curcumin were prepared in dimethyl sulfoxide (Sigma-Aldrich). The
primary and secondary antibodies used were as follows: KL
rabbit anti-human monoclonal (1:1,000; ab181373; Abcam, Cambridge,
MA, USA), DNMT1 rabbit anti-human monoclonal (1:2,000; #5032; Cell
Signaling Technology, Inc., Danvers, MA, USA), horseradish
peroxidase-conjugated anti-rabbit IgG (1:3,000; #7074; Cell
Signaling Technology). Invitrogen TRIzol reagent was purchased from
Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
The HK-2 human proximal tubule epithelial cell line
was obtained from the American Type Culture Collection (Manassas,
VA, USA), and maintained in Hyclone Dulbecco's modified Eagle's
medium (DMEM; Thermo Fisher Scientific, Inc., Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin (100 U/ml) and 1% streptomycin
(100 µg/ml) (Roche Diagnostics, Indianapolis, IN, USA).
Mice and CsA-induced animal model
A total of 72 female C57BL/6 mice (age, 6–8 weeks;
weight, 19.1±0.7 g) were obtained from Shanghai Laboratory Animal
Center of Chinese Academy of Sciences (Shanghai, China) and all
mouse experiments were approved by the Animal Welfare & Ethics
Committee of Zhejiang University (Hangzhou, China). The mice were
maintained in a specific-pathogen-free animal facility, which was
controlled at 23–25°C with a 12-h light/dark cycle. The mice were
caged 5 mice per cage and were fed with normal chow and sterilized
water. Cyclosporine A (CsA; Novartis, Basel, Switzerland) was
dissolved with olive oil at a concentration of 20 mg/ml. Mice were
treated with the vehicle alone (olive oil; 1 ml/kg/day), CsA alone
(15 mg/kg, s.c.), or CsA + curcumin (15 mg/kg/day, i.p.) for 14
days. An s.c. injection of olive oil (15 ml/kg/day) and i.p.
injection of normal saline (0.9% w/v NaCl) served as controls.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Kidney tissues [5×5×5 mm; obtained from mice under
70 mg/kg intraperitoneal sodium pentobarbital anesthesia
(Sigma-Aldrich) and stored at −80°C], or 1×107 kidney
cells were homogenized with 1 ml TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc., San Diego, CA, USA). Chloroform (200 µl)
was added to 1 ml TRIzol, and following vigorous shaking for 30 sec
and centrifugation at 12,000 × g for 20 min at 4°C, the aqueous
phase was collected. An equal volume of isopropanol was added and
mixed thoroughly, and was maintained at room temperature for 15 min
prior to centrifugation again at 12,000 × g for 20 min at 4°C.
Ethanol was used to prepare the RNA. The total RNA was
reverse-transcribed using an RT-qPCR kit (Invitrogen; Thermo Fisher
Scientific, Inc., San Diego, CA, USA). The polymerase used was Taq
DNA polymerase (Invitrogen; Thermo Fisher Scientific, Inc.), and
the DNA ladder and ethidium bromide used were also from Invitrogen
(Thermo Fisher Scientific, Inc.). qPCR was conducted on an ABI
Prism 7900HT (Applied Biosystems; Thermo Fisher Scientific, Inc.)
using the SuperScript III Platinum SYBR Green One-step qRT-PCR
system from Invitrogen (Thermo Fisher Scientific, Inc), which
included SuperScript III, Platinum Taq, reaction mix and dNTPs. The
cycling conditions were as follows: Initial denaturation at 95°C
for 5 min, 35 cycles of 95°C for 15 sec and 60°C for 60 sec, then
72°C for 5 min. The primers were as follows: Forward, 5′-CCCTA
CCCAG CACCT TCAAA-3′ and reverse, 5′-GTGGC CGATG TTTCC AGTCT-3′ for
Col I; forward, 5′-GGGCA CTACC ATGTA CCCAG-3′ and reverse, 5′-TGAAG
GCGCT GATCC ACAAA A-3′ for α-SMA; forward, 5′-GGCCA CCAAC TTCGG
AGTAA-3′ and reverse, 5′-TGTTC CATGA CCCCA TGAGC-3′ for PAI-1;
forward, 5′-AGTGG CCGAG AGAGT TTTGG-3′ and reverse, 5′-GGTGT AATGA
CTCAC CGCCA-3′ for KL; forwrd, 5′-GCTGT TCCTT GTAGG CGAGT-3′
and reverse, 5′-GGGGA CTCAA ACCTT GCGTA-3′ for Dnmt1. GAPDH served
as an internal control to normalize for differences in the quantity
of total RNA in each sample, and expression was calculated using
the 2−ΔΔCt method (21). A total of 3 independent experiments
were performed.
Histological staining
Kidneys harvested from the control, CsA and CsA +
curcumin groups were fixed for 3–12 h in 10% formalin, followed by
incubation at room temperature for 30 min in 30% sucrose (Sinopharm
Chemical Reagent Co., Ltd., Shanghai, China) in phosphate-buffered
saline (PBS; Gibco; Thermo Fisher Scientific, Inc.), and flash
frozen in cold isopentane (Sinopharm Chemical Reagent Co., Ltd.). A
cryostat (CM3050S; Leica Microsystems, Inc., Buffalo Grove, IL,
USA) was used to prepare 10-µm sections for staining.
Alternatively, MAs were embedded in paraffin by Tissue-Tek VIP
(Sakula Finetek, Flemingweg, Netherlands) automated instrument and
sectioned at 6 µm for analysis. The sections were stained
using hematoxylin and eosin (Beyotime Institute of Biotechnology,
Nantong, China). The sections were assessed using a light
microscope (DM6 B, Leica Microsystems, Inc.).
Cell proliferation assay
HK-2 cells were seeded into 96-well plates at a
density of 1×103 cells per well. Cell viability was
quantified at 24 h in the absence or presence of curcumin using a
Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), measuring absorbance at 450 nm according to the
manufacturer's protocols.
Cell transfection
HK-2 cells were plated in 24-well plates
(2×105 cells/well) and were transfected with 30 pmol
KL siRNA (5′-GGU UGG AAU AAA CUU GUCA-dtdt-3′) or control
siRNA (5′-UUC UCC GAA CGU GUC ACGU-dtdt-3′), using 2 µl
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Cells were used for
further experiment after 48 hours.
Western blot analysis
Total protein was extracted using a Protein
Extraction kit according to the manufacturer's instructions (EMD
Millipore, Billerica, MA, USA) and western blotting was performed
as previously described (22).
Methylation-specific PCR (MSP) and
bisulfite sequencing
Genomic DNA was extracted from the cell lines and
bisulfite modification of the DNA was performed using the EZ DNA
Methylation kit (Zymo Research Corp, Irvine, CA, USA) according to
the manufacturer's instructions. Methylation-specific primers were
used to amplify a 161-bp region of KL promoter and the
primers were as follows: Forward, AGCTG GGAGA AACAG GTGCC and
reverse, TGGCA ATAAT TACCT GCGAG. The PCR products were purified
and subcloned using pGEM-T Easy Vector Systems (Promega
Corporation, Madison, WI, USA) for subsequent sequencing analysis.
Methylation levels at each position were calculated from the
quantified data of bisulfite sequencing according to the formula
(height of C peak) / [(height of C peak) × (height of T peak)].
CpG MSP reporter assay
Various forward primers, in the presence or absence
of a methyl group to one specific cytosine residue, were used in
combination with the common RV4 reverse primer (Promega
Corporation). PCR products from three independent reactions were
extracted, and co-transfected with Renilla luciferase into
HK-2 cells using Lipofectamine 2000. Luciferase activity was
measured 48 h later using the Dual-Luciferase® Reporter Assay
system (Promega Corporation) according to the manufacturer's
instructions. Data were normalized to the level of Renilla
expression for each well. Unmethylated control luciferase activity
was defined as 100%.
Statistical analysis
All of the measurements were collected in triplicate
for each independent preparation. The results were statistically
analyzed using Student's t-test or one-way analysis of
variance with SPSS software, version 20 (IBM SPSS, Armonk, NY,
USA). P<0.05 were considered to indicate a statistically
significant difference.
Results
Curcumin exerts protective effects
against CsA-induced renal fibrosis
The mechanism of CsA-induced renal fibrosis has been
demonstrated to be correlated with EMT (23); thus, to investigate the possible
mechanism of the protective effects induced by curcumin against
renal fibrosis, the expression of PAI-1 and α-SMA (considered as
crucial proteins in the TGF-β signaling pathway) (24–26)
were analyzed in the present study. The expression level of Col I
served as a readout for the severity of renal toxicity. The HK-2
human proximal tubule epithelial cell line was treated with
curcumin, and the mRNA expression levels of α-SMA, PAI-1 and Col I
were determined by RT-qPCR. The results indicated that the
expression levels of these three genes became elevated from 2 h and
continued to increase for 22 h in the presence of CsA (8
µM). However, the increase in expression was less in the CsA
(8 µM) + curcumin group (10 µM; Fig. 1A–C). In addition, curcumin
significantly inhibited the proliferation of HK-2 cells in a
concentration-dependent manner (Fig.
1D), suggesting that curcumin administered in a culture medium
(DMEM supplemented with FBS and penicillin and streptomycin) could
downregulate the expression of genes associated with TGF-β
signaling. Furthermore, cell apoptosis was detected and no
significant change was identified upon curcumin treatment.
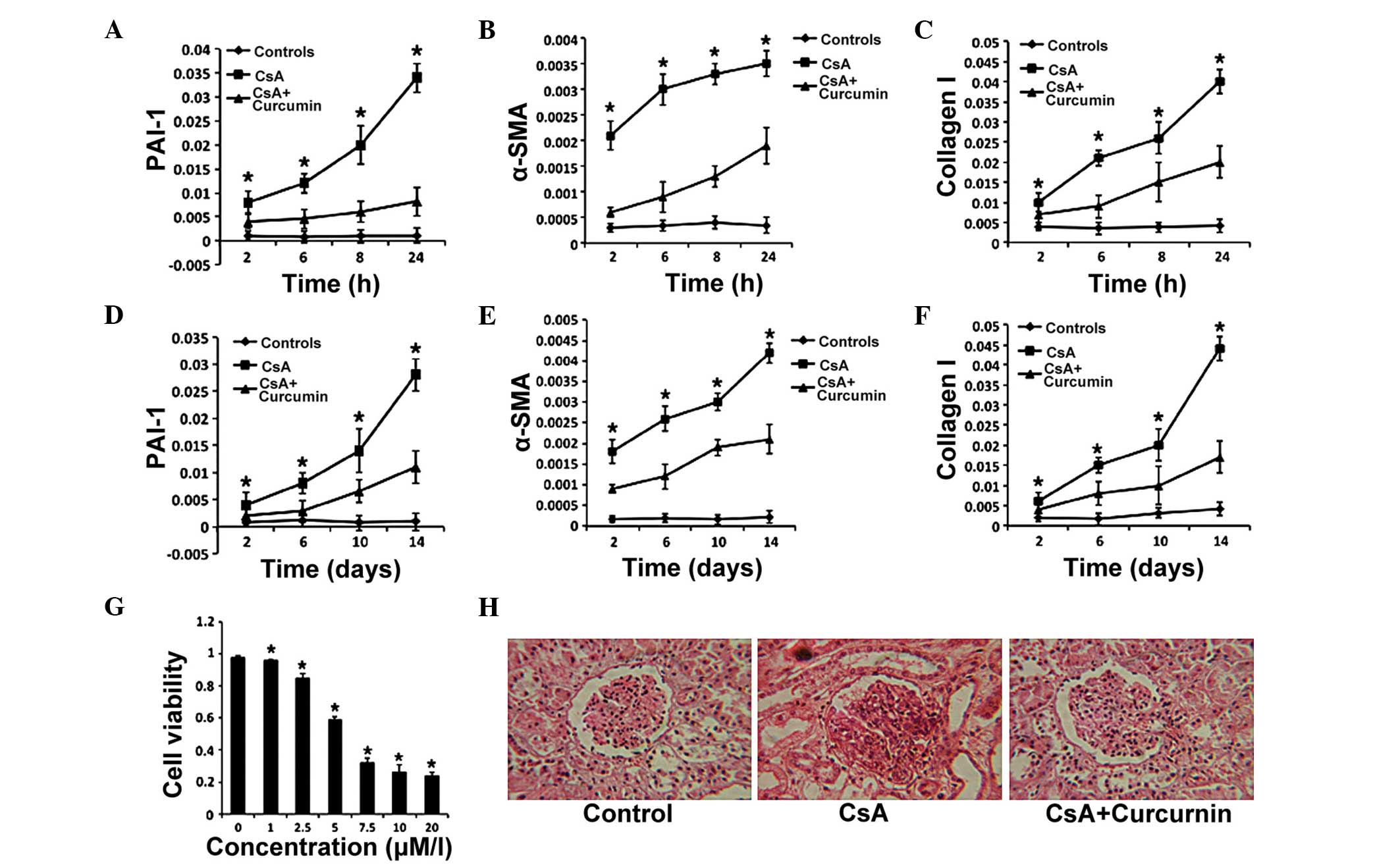 | Figure 1Curcumin exerts protective effects on
CsA-induced renal fibrosis. (A–C) The expression levels of PAI-1,
α-SMA and Col I were detected in HK-2 cells by reverse
transcription-quantitative polymerase chain reaction at the
indicated time-points in the control, CsA and CsA + curcumin
groups. (D) HK-2 cell proliferation in the presence of curcumin.
(E–G) The expression levels of PAI-1, α-SMA and Col I were measured
in the kidney tissues of mice in the control, CsA and CsA +
curcumin groups. (H) Histological staining of kidney tissues from
mice following 2 weeks of the indicated treatments (magnification,
×100). In the CsA group, glomerular fibrosis was severe and the
structure was destroyed. The error bars represent the mean ±
standard error. *P<0.05, (A–C and E–G) CsA vs. CsA +
curcumin, (D) vs. 0 µM/l. CsA, cyclosporine A; PAI-1,
plasminogen activator inhibitor-1; α-SMA, α-smooth muscle actin;
Col I, collagen I. |
To further confirm the effects of curcumin on renal
fibrosis, CsA-induced renal fibrosis model was established using
mice. Renal tissue samples were harvested from the 6 mice and
evaluated the expression of the same genes in the tissue samples
after 2 weeks. The expression of α-SMA, PAI-1 and Col I were
elevated from 2 days for the next 12 days in the CsA alone and the
CsA + curcumin groups. The findings demonstrated that in the CsA +
curcumin group, the increasing expression levels were lower than in
the CsA group (Fig. 1E–G).
Furthermore, the renal fibrosis was markedly reduced in the CsA +
curcumin group (Fig. 1H). Taken
together, these results indicate that curcumin attenuated TGF-β
signaling and exerts protective effects against renal fibrosis.
Inhibition of TGF-β signaling by curcumin
is KL-dependent in renal tubule epithelial cells
TGF-β is the master regulatory gene for EMT and is
required for activation of diverse EMT biomarkers, such as α-SMA
and β-catenin, which are significant in fibrosis (10). KL has been shown to suppress
TGF-β signaling via binding to the type II TGF-β receptor, thus
inhibiting the EMT (27).
Therefore, the present study investigated whether curcumin inhibits
TGF-β signaling via inducing KL expression, and used the
expression of PAI-1, α-SMA and Col I as readouts for TGF-β
signaling. To support our hypothesis, HK-2-Si KL (HK-2-Si)
cells, with the KL gene knocked down, were used. The HK-2-Si
cells were incubated with curcumin and the expression levels of
α-SMA, PAI-1 and Col I were evaluated. The expression level of
KL was significantly downregulated at the mRNA and protein
levels (Fig. 2A and B),
demonstrating the key role of curcumin in regulating the expression
of KL. Knockdown of KL almost completely obviated the
downregulation in PAI-1, α-SMA and Col I in mRNA expression levels
(Fig. 2C–E), illustrating the
essential role of KL in curcumin-mediated inhibition of
TGF-β signaling. Thus, it was demonstrated that regulation of
KL expression is crucial for the inhibitory affects on TGF-β
signaling (a master regulator of fibrosis), thus indicating that
regulating the expression of KL may be important for
protection against CsA-induced renal fibrosis.
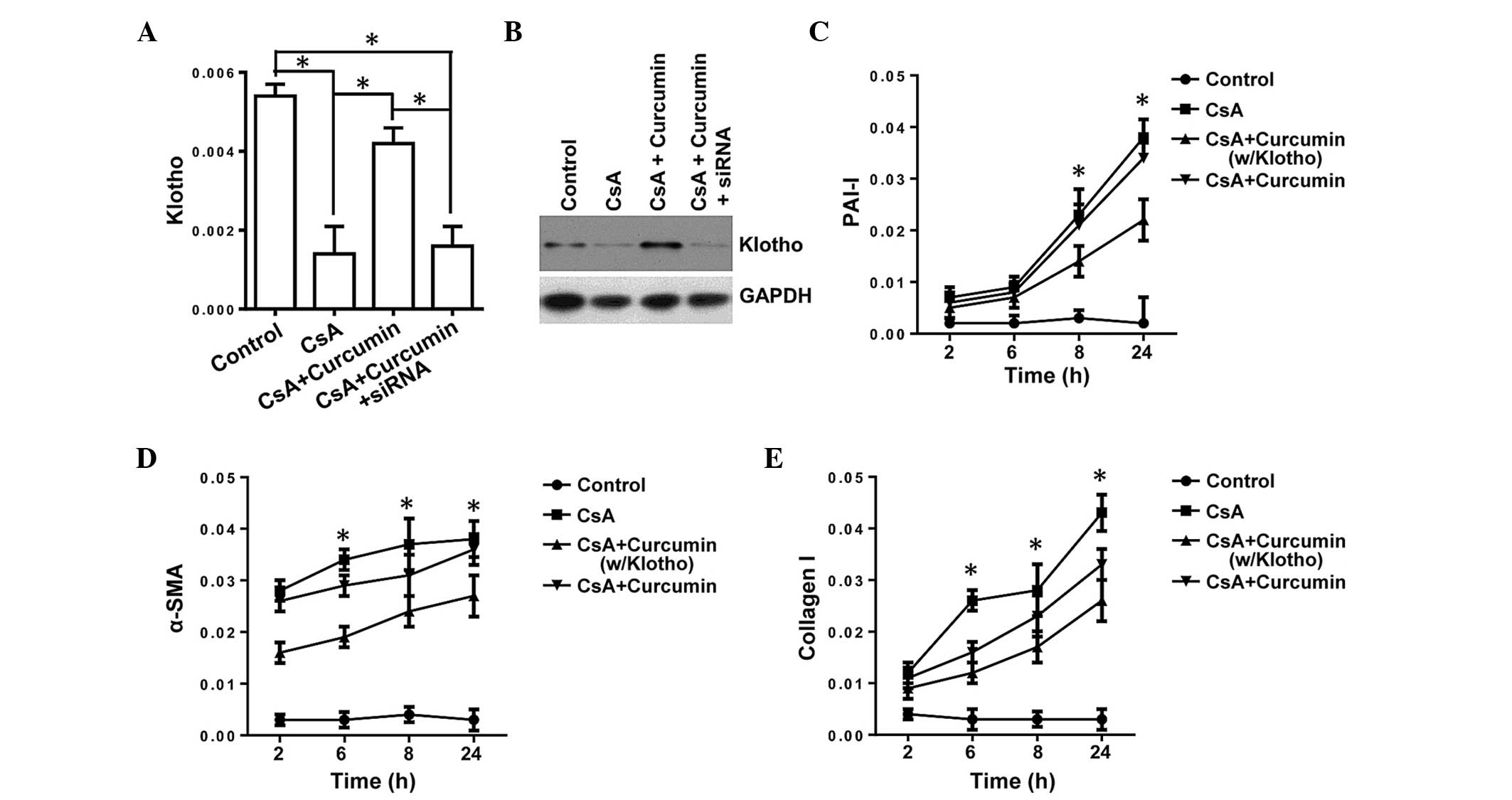 | Figure 2Inhibition of transforming growth
factor-β by curcumin is KL-dependent in tubule epithelial
cells. (A and B) siRNA was used to transfect HK-2 cells (HK-2-Si),
and the mRNA and protein expression levels of KL were detected by
RT-qPCR and western blotting, respectively. (C–E) The expression
levels of PAI-1, α-SMA and Collagen I in HK-2-Si cells or HK-2
cells (w/KL) by RT-qPCR. The error bars represent the mean ±
standard error. *P<0.05, (A) HK-2-Si vs. HK-2 cells,
(C–E) CsA +curcumin (w/Klotho) vs. CsA + curcumin. CsA,
cyclosporine A; PAI-1, plasminogen activator inhibitor-1; α-SMA,
α-smooth muscle actin; si, small interfering; KL, klotho; w/Klotho,
with klotho; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
Promoter methylation suppresses KL gene
expression in renal tubule epithelial cells
Hypermethylation of CpG islands rich in KL
promoter causes a reduction in KL expression in various
types of cells. Therefore, the present study hypothesized that
curcumin inhibits methylation of the KL promoter in HK-2
cells. The KL promoter region −275 to −115 base pairs, which
is particularly CpG-rich, was amplified and bisulfite sequencing
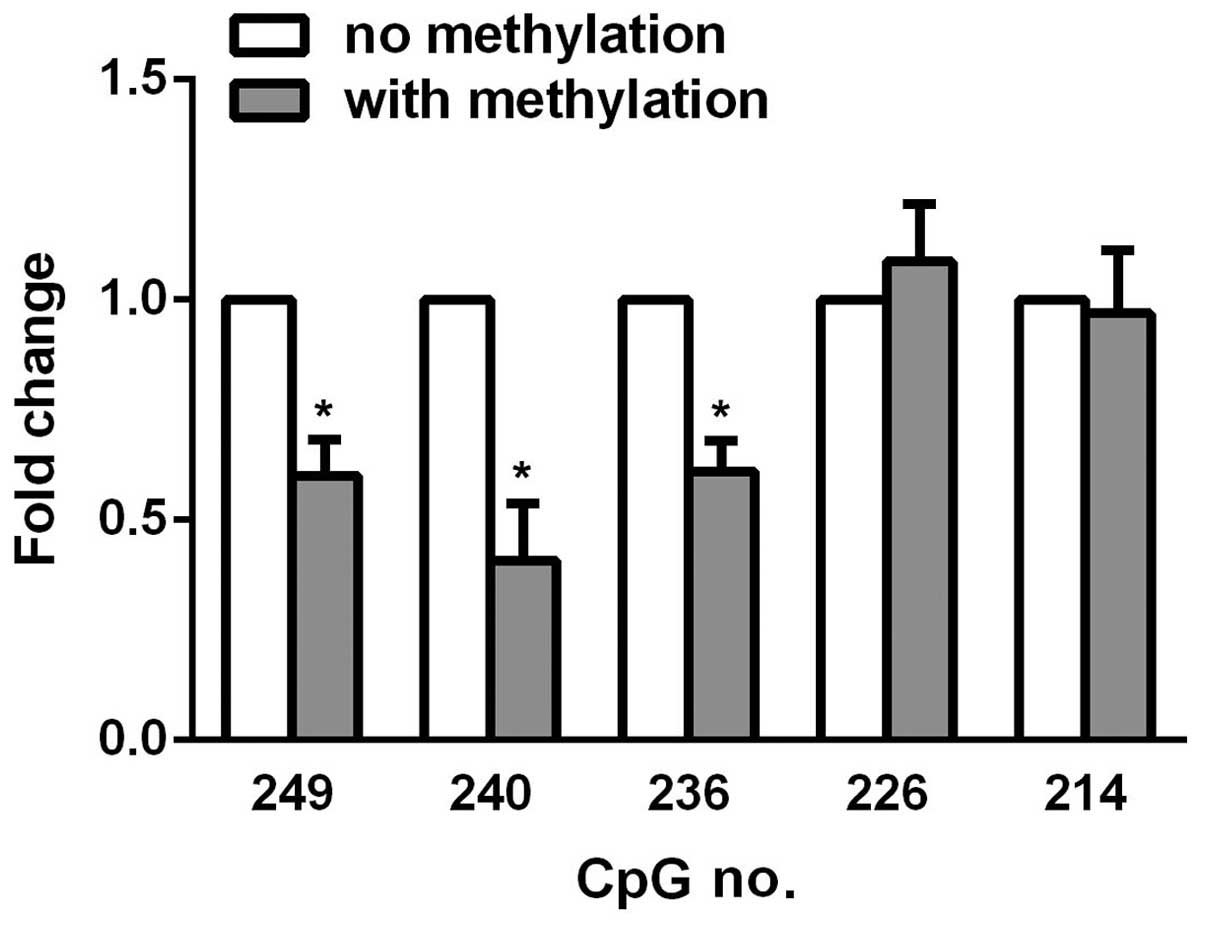
was performed for statistical analysis. As shown in Table I, CsA promoted CpG methylation at
various sites, while curcumin reversed the hypermethylation effects
at the majority of the sites, particularly at site 249, 240, and
236, suggesting that these sites are pivotal to KL
expression in HK-2 renal tubule epithelial cells. To further
emphasize the important role of these sites, PCR was used to
introduce methyl groups to individual C-terminal residues, and the
promoter was amplified using forward primers differing only by
containing a methyl group. The reverse primer was common to all PCR
reactions amplifying from the 3′-end of the firefly luciferase
gene. The linear DNA was subsequently co-transfected with
Renilla luciferase into the HK-2 cells. After 48 h,
luciferase activity was measured, and in vitro methylation
of CpG 249, 240, and 236 was observed to cause decreased luciferase
activity upon methylation (Fig.
3), which highlighted the hypermethylation effects of curcumin
on HK-2 cells.
 | Table ICpG methylation levels of
klotho promoter. |
Table I
CpG methylation levels of
klotho promoter.
| HK-2 | CpG methylation
level at nucleotide position (%)
|
|---|
| 249 | 246 | 242 | 240 | 236 | 233 | 226 | 214 | 208 |
|---|
| Control | 18 | 41 | 67 | 0 | 20 | 34 | 63 | 0 | 0 |
| CsA | 53 | 68 | 71 | 13 | 74 | 42 | 69 | 0 | 39 |
| CsA/CMN | 26 | 44 | 69 | 0 | 38 | 38 | 66 | 0 | 0 |
Curcumin suppresses CpG island
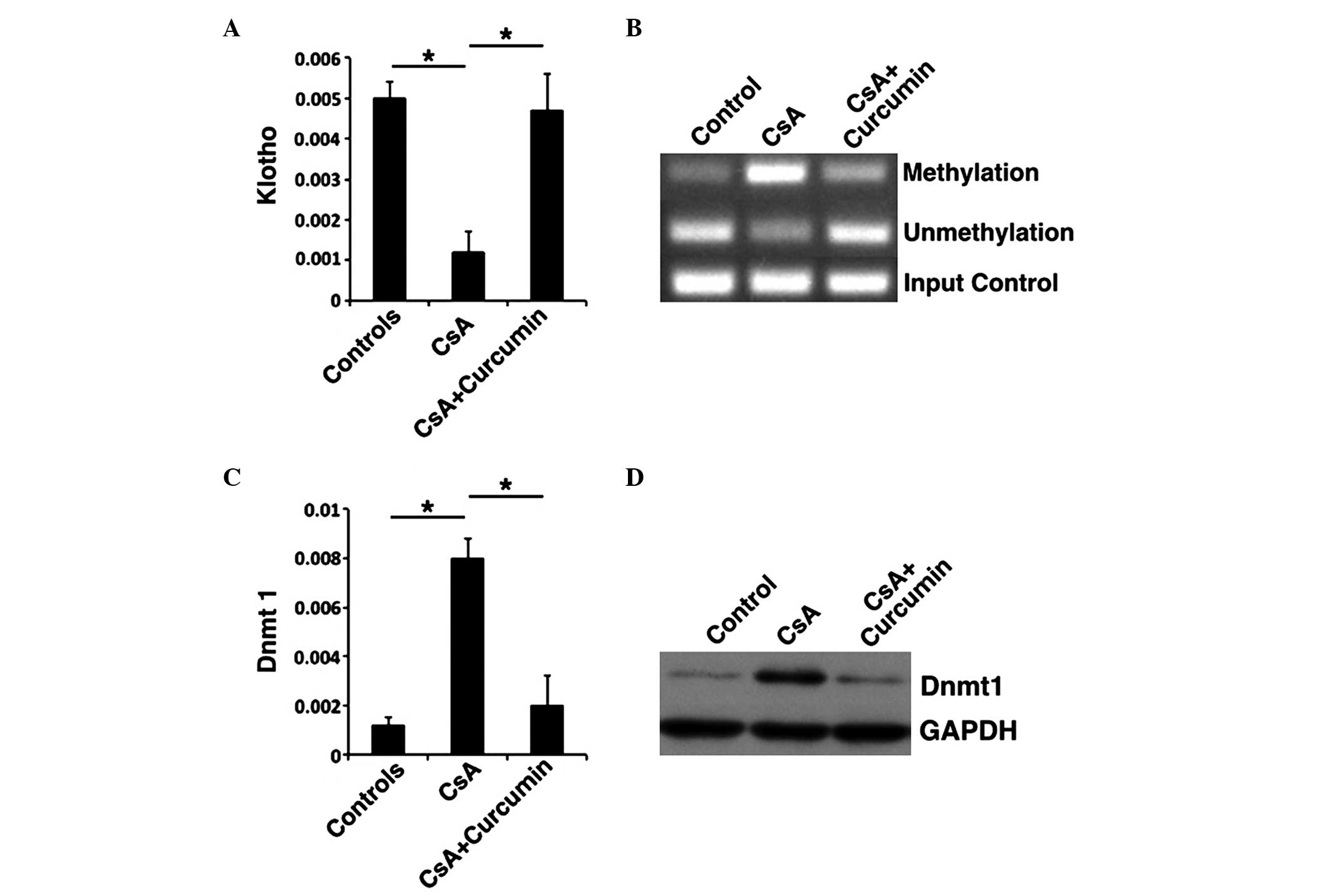
hypermethylation via Dnmt1 inhibition
Previous studies have demonstrated that KL
expression was negatively correlated with the expression of PAI-1,
α-SMA and SMAD2/3 (19).
Therefore, whether the expression of KL altered following
the addition of curcumin to the culture medium was evaluated in
HK-2 cells. KL expression levels were elevated in the CsA +
curcumin group, whereas they were inhibited in the CsA group
(Fig. 4A and B). Previous studies
proposed that KL expression was primarily dominated by Dnmt1
(28). To further investigate the
mechanism of KL expression in the presence of curcumin, MSP
was performed to evaluate the methylation of KL, and RT-qPCR
and western blot were conducted to analyze the expression of Dnmt1.
Methylation of KL was inhibited in the presence of curcumin
when compared with the CsA group (Fig.
4B). In addition, the Dnmt1 expression correlated with
increased methylation, with methylation being downregulated in the
CsA + curcumin group (Fig. 4C and
D). These results indicate that curcumin may inhibit the
methylation of KL and, as result, downregulate the
expression of Dnmt1, regulate specific genes in the TGF-β signaling
pathway and induce a protective effect against renal fibrosis.
Discussion
In the present study, the results demonstrated that
curcumin reduced CsA-induced renal fibrosis effectively by
suppressing the methylation of KL, thus regulating the
expression of KL in tubular epithelial cells. Additionally,
TGF-β signaling was observed to be inhibited due to the suppressed
methylation of KL in the presence of curcumin during renal
fibrosis process, as KL knockdown obviated the majority of
the effects of curcumin on TGF-β signaling, which was demonstrated
by changes in PAI-1 and α-SMA expression. Thus, the present study
provides an insight into the underlying mechanism of curcumin in
renal fibrosis, and supports the feasibility of administrating
curcumin for the therapeutic treatment of renal fibrosis.
CsA may induce oxidative stress injury by increasing
the production of reactive oxygen species and decreasing the
activities of antioxidant enzymes, such as mitochondrial
antioxidant manganese superoxide dismutase and glutathione
(29), and exerts a marked
nephrotoxic effect, as shown in the present study by considerable
renal fibrosis (Fig. 1H). The
in vivo and in vitro results display significant
levels of expression of various ECM components, such as PAI-1,
α-SMA and Col I, all of which may be the downstream biological
events of the TGF-β signaling pathway. Previous studies have
indicated that curcumin influences cytokine secretion and
inflammation in CKD patients (30,31);
the current data indicated that curcumin exerts nephroprotective
effects, at least partially, via anti-TGF-β signaling.
In kidney injury or CKD, the expression of KL
decreases while the expression of PAI-1 increases (32), and our previous study demonstrated
that curcumin suppresses TGF-β activity via inhibiting SMAD2/3
signaling (19), leading us to
hypothesize that curcumin induces KL expression in tubular
epithelial cells, thus producing nephroprotective effects.
Consistent with our hypothesis, curcumin treatment promoted the
expression of KL, and inhibited TGF-β signaling in the
present study. Furthermore, the specific knockdown of KL
attenuated the inhibitory effect of curcumin on TGF-β signaling,
clearly demonstrating the essential role of KL in
curcumin-mediated inhibition of TGF-β signaling. In addition, it is
well accepted that KL protects organs from aging in rodents,
as shown by KL−/− mice, which
exhibited premature aging phenotypes in various organs, including
the kidney, and shortened life spans (11). A previous study demonstrated more
serious tubulointerstitial fibrosis in 24-month-old mice, when
compared with 2-month-old mice, as well as reduced KL
expression in the kidneys (33).
These studies highlighted the nephroprotective role of
KL.
Previous studies have found that curcumin may
inhibit cytokine expression and reduce inflammation, as well as
inhibit the secretion of TGF-β (34). These studies suggested that
curcumin may be beneficial during fibrosis, and as multiple
pathways, including inflammation, TGF-β and KL have been
demonstrated to be involved in kidney fibrosis the current study
investigated whether curcumin alleviates fibrosis through a
KL-mediated pathway, while it is possible that other
pathways are involved. However, the KL-mediated
anti-fibrosis effect is essential, at least during renal fibrosis,
as knockdown of KL almost completely obviated the beneficial
effects of curcumin. An additional finding which supports the
hypothesis that the curcumin effect is KL-dependent is that
CsA-induced nephropathy is associated with decreased expression of
KL (26,35). In the present study, increased
expression levels of KL were observed in HK-2 cells after
treatment with curcumin, which was potentially via the suppression
of KL promoter hypermethylation. CpG methylation in the
KL promoter appears to be key in the maintenance of tissue
homeostasis, as aberrant methylation of CpG sites is frequently
observed in malignant transformation of tissues, such as those of
the cervix and mammary glands (16,17).
DNMT inhibitor, 2′-deoxy-5-azacytidine may suppress CpG methylation
in the KL promoter and induce the expression of KL in
various human cervical cancer cell lines, such as siHA and SNU-1299
(16). The current study showed
that curcumin inhibited hypermethylation in the KL promoter
region (−275 to −115) in HK-2 cells, indicating the underlying
mechanism of hypermethylation in suppressing KL gene
expression. This region was selected as it is CpG-rich (19 CpG
sites in 161 bp), and curcumin was found to suppress methylation
the most markedly at sites 249, 240 and 236. Additionally, a CpG
methylation PCR reporter assay was used to further confirm that the
methylation of just one of these sites is adequate to inhibit
KL expression by half. However, the effects of CpG
methylation at different sites are varied, and methylation at
certain sites has no effect on expression and even promotes
KL expression. The comprehensive and specific effects of CpG
methylation on KL expression require further research.
DNMTs are the key enzymes for the regulation of DNA
methylation, and during kidney injury, oxidative stress affects
DNMT expression, such as Dnmt1, -3a, and -3b, and subsequently
modulates the epigenetic regulation of gene expression (28). Dnmt1 is the most abundant Dnmt, and
is considered to be the key maintenance methyltransferase in
mammals (36). Dnmt1 has been
suggested to contribute to CpG methylation in the KL
promoter, and suppresses KL expression (28). Furthermore, the present study
indicates Dnmt1 as a candidate for CpG methylation of the KL
promoter, however it is not necessarily the only one. Whether
multiple DNMTs, or specific DNMTs in specific cell types,
contribute to methylation requires further investigation.
Elucidating this will facilitate with the screening for small
molecular inhibitors for these DNMTs, and aid the development of
therapeutic approaches for kidney diseases and other associated
diseases.
In conclusion, the present study indicates that
curcumin exerts its protective effects against renal fibrosis in
vitro and in vivo via regulating the methylation of
KL. This may provide an additional mechanism for resistance
to renal fibrosis and provide a novel molecular target for therapy
during the pathogenesis of chronic CsA-induced nephropathy.
Acknowledgments
The present study was supported by research grants
from the Science and Technology Foundation of Zhejiang Province,
China (grant no. LY12H05005).
References
|
1
|
James MT, Hemmelgarn BR and Tonelli M:
Early recognition and prevention of chronic kidney disease. Lancet.
375:1296–1309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global, regional and national age-sex
specific all-cause and cause-specific mortality for 240 causes of
death, 1990–2013: A systematic analysis for the Global burden of
disease study 2013. Lancet. 385:117–171. 2015. View Article : Google Scholar
|
|
3
|
Levey AS and Coresh J: Chronic kidney
disease. Lancet. 379:165–180. 2012. View Article : Google Scholar
|
|
4
|
Garrido P, Ribeiro S, Fernandes J, Vala H,
Bronze-da-Rocha E, Rocha-Pereira P, Belo L, Costa E, Santos-Silva A
and Reis F: Iron-hepcidin dysmetabolism, anemia and renal hypoxia,
inflammation and fibrosis in the remnant kidney rat model. PLoS
One. 10:e01240482015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sinha AD and Agarwal R: Chronic renal
disease progression: treatment strategies and potassium intake.
Semin Nephrol. 33:290–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jha V, Garcia-Garcia G, Iseki K, Li Z,
Naicker S, Plattner B, Saran R, Wang AY and Yang CW: Chronic kidney
disease: Global dimension and perspectives. Lancet. 382:260–272.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Falke LL, Gholizadeh S, Goldschmeding R,
Kok RJ and Nguyen TQ: Diverse origins of the
myofibroblast-implications for kidney fibrosis. Nat Rev Nephrol.
11:233–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kriz W, Kaissling B and Le Hir M:
Epithelial-mesenchymal transition (EMT) in kidney fibrosis: Fact or
fantasy? J Clin Invest. 121:468–474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi
H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et
al: Mutation of the mouse klotho gene leads to a syndrome
resembling ageing. Nature. 390:45–51. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurosu H, Yamamoto M, Clark JD, Pastor JV,
Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M,
Kawaguchi H, et al: Suppression of aging in mice by the hormone
Klotho. Science. 309:1829–1833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sugiura H, Yoshida T, Shiohira S, Kohei J,
Mitobe M, Kurosu H, Kuro-o M, Nitta K and Tsuchiya K: Reduced
Klotho expression level in kidney aggravates renal interstitial
fibrosis. Am J Physiol Renal Physiol. 302:F1252–F1264. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu MC, Kuro-o M and Moe OW: Renal and
extrarenal actions of Klotho. Semin Nephrol. 33:118–129. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Azuma M, Koyama D, Kikuchi J, Yoshizawa H,
Thasinas D, Shiizaki K, Kuro-o M, Furukawa Y and Kusano E: Promoter
methylation confers kidney-specific expression of the Klotho gene.
FASEB J. 26:4264–4274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee J, Jeong DJ, Kim J, Lee S, Park JH,
Chang B, Jung SI, Yi L, Han Y, Yang Y, et al: The anti-aging gene
Klotho is a novel target for epigenetic silencing in human cervical
carcinoma. Mol Cancer. 9:1092010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rubinek T, Shulman M, Israeli S, Bose S,
Avraham A, Zundelevich A, Evron E, Gal-Yam EN, Kaufman B and Wolf
I: Epigenetic silencing of the tumor suppressor klotho in human
breast cancer. Breast Cancer Res Treat. 133:649–657. 2012.
View Article : Google Scholar
|
|
18
|
Srivastava RM, Singh S, Dubey SK, Misra K
and Khar A: Immunomodulatory and therapeutic activity of curcumin.
Int Immunopharmacol. 11:331–341. 2011. View Article : Google Scholar
|
|
19
|
Hu Y, Liang H, Du Y, Zhu Y and Wang X:
Curcumin inhibits transforming growth factor-beta activity via
inhibition of Smad signaling in HK-2 cells. Am J Nephrol.
31:332–341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu J, Peng Y, Wu LC, Xie Z, Deng Y, Hughes
T, He S, Mo X, Chiu M, Wang QE, et al: Curcumin down-regulates DNA
meth-yltransferase 1 and plays an anti-leukemic role in acute
myeloid leukemia. PLoS One. 8:e559342013. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Cai W, Du A, Feng K, Zhao X, Qian L,
Ostrom RS and Xu C: Adenylyl cyclase 6 activation negatively
regulates TLR4 signaling through lipid raft-mediated endocytosis. J
Immunol. 191:6093–6100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hazzan M, Hertig A, Buob D, Copin MC, Noël
C, Rondeau E and Dubois-Xu YC: Epithelial-to-mesenchymal transition
predicts cyclosporine nephrotoxicity in renal transplant
recipients. J Am Soc Nephrol. 22:1375–1381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Damiano S, Scanni R, Ciarcia R, Florio S
and Capasso G: Regulation of sodium transporters in the kidney
during cyclosporine treatment. J Nephrol. 23(Suppl 16): S191–S198.
2010.
|
|
25
|
Neria F, Castilla MA, Sanchez RF, Gonzalez
Pacheco FR, Deudero JJ, Calabia O, Tejedor A, Manzarbeitia F, Ortiz
A and Caramelo C: Inhibition of JAK2 protects renal endothelial and
epithelial cells from oxidative stress and cyclosporin A toxicity.
Kidney Int. 75:227–234. 2009. View Article : Google Scholar
|
|
26
|
Yoon HE, Ghee JY, Piao S, Song JH, Han DH,
Kim S, Ohashi N, Kobori H, Kuro-o M and Yang CW: Angiotensin II
blockade upregulates the expression of Klotho, the anti-ageing
gene, in an experimental model of chronic cyclosporine nephropathy.
Nephrol Dial Transplant. 26:800–813. 2011. View Article : Google Scholar :
|
|
27
|
Doi S, Zou Y, Togao O, Pastor JV, John GB,
Wang L, Shiizaki K, Gotschall R, Schiavi S, Yorioka N, et al:
Klotho inhibits transforming growth factor-beta1 (TGF-beta1)
signaling and suppresses renal fibrosis and cancer metastasis in
mice. J Biol Chem. 286:8655–8665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun CY, Chang SC and Wu MS: Suppression of
Klotho expression by protein-bound uremic toxins is associated with
increased DNA methyltransferase expression and DNA
hypermethylation. Kidney Int. 81:640–650. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khan M, Shobha JC, Mohan IK, Rao Naidu MU,
Prayag A and Kutala VK: Spirulina attenuates cyclosporine-induced
nephrotoxicity in rats. J Appl Toxicol. 26:444–451. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shing CM, Adams MJ, Fassett RG and Coombes
JS: Nutritional compounds influence tissue factor expression and
inflammation of chronic kidney disease patients in vitro.
Nutrition. 27:967–972. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moreillon JJ, Bowden RG, Deike E, Griggs
J, Wilson R, Shelmadine B, Cooke M and Beaujean A: The use of an
anti-inflammatory supplement in patients with chronic kidney
disease. J Complement Integr Med. 10:143–152. 2013. View Article : Google Scholar
|
|
32
|
Yamada K, Doi S, Nakashima A, Kawaoka K,
Ueno T, Doi T, Yokoyama Y, Arihiro K, Kohno N and Masaki T:
Expression of age-related factors during the development of renal
damage in patients with IgA nephropathy. Clin Exp Nephrol.
19:830–837. 2015. View Article : Google Scholar
|
|
33
|
Lim JH, Kim EN, Kim MY, Chung S, Shin SJ,
Kim HW, Yang CW, Kim YS, Chang YS, Park CW and Choi BS:
Age-associated molecular changes in the kidney in aged mice. Oxid
Med Cell Longev. 2012:1713832012. View Article : Google Scholar
|
|
34
|
Kliem C, Merling A, Giaisi M, Köhler R,
Krammer PH and Li-Weber M: Curcumin suppresses T cell activation by
blocking Ca2+ mobilization and nuclear factor of activated T cells
(NFAT) activation. J Biol Chem. 287:10200–10209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han DH, Piao SG, Song JH, Ghee JY, Hwang
HS, Choi BS, Kim J and Yang CW: Effect of sirolimus on calcineurin
inhibitor-induced nephrotoxicity using renal expression of KLOTHO,
an antiaging gene. Transplantation. 90:135–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mohan KN and Chaillet JR: Cell and
molecular biology of DNA methyltransferase 1. Int Rev Cell Mol
Biol. 306:1–42. 2013. View Article : Google Scholar : PubMed/NCBI
|