Introduction
The intervertebral disc lies between adjacent
cartilaginous endplates in the spine and is composed of a gel-like
core, termed the nucleus pulposus (NP), and the surrounding annulus
fibrosus (1). In the NP,
proteoglycan is the primary aggrecan, while collagen II is the
primary collagen (2). Decreased
levels of aggrecan and collagen II in the NP result in the
initiation of intervertebral disc degeneration (3,4).
Tumor necrosis factor-α (TNF-α) and interleukin 1-β (IL-lβ) are the
pro-inflammatory cytokines principally associated with the
progression of intervertebral disc degeneration and have been
previously demonstrated to increase the degradation of the
extracellular matrix (ECM), including aggrecan and collagen II
(5–7). Matrix metalloproteinases (MMPs),
disintegrins and metalloproteinases with thrombospondin motifs
(ADAMTSs) and cyclooxygenase 2 (Cox2) are the major catabolic
enzymes associated with ECM degradation induced by pro-inflammatory
factors in the intervertebral disc (6–10).
Syndecan 4 (SDC4), a cell surface heparin sulfate proteoglycan,
serves a major role in matrix catabolism through the activation of
MMP3, ADAMTS4 and ADAMTS5 in the intervertebral disc (6,7,10).
Mitogen-activated protein kinase (MAPK) inhibition
can decrease the upregulation of MMP3, ADAMTS4 and ADAMTS5
expression induced by TNF-α and IL-1β (6,7,11).
In addition, nuclear factor κ-light-chain-enhancer of activated B
cells (NF-κB) and SDC4 are involved in the signaling pathways in NP
cells that mediate TNF-α and IL-1β induced aggrecan degradation by
MMP3, ADAMTS4 and ADAMTS5 (6,7,11).
Similarly, the induction of MMP13 and Cox2 expression by TNF-α and
IL-1β is regulated through the MAPK and NF-κB signaling pathways
(12,13). Specificity protein-1 (Sp1) is an
important transcription factor that contains a zinc finger
DNA-binding domain, and binds to GC-rich regions in the promoters
of genes to activate transcription (14). It is also involved in the
expression of ECM-associated genes in various cells, including
renal tubular epithelial cells and melanoma cells (15,16).
Inhibition of Sp1 and Sp1 knockdown has been previously
demonstrated to partially inhibit ADAMTS4 and Cox2 induction by
IL-1β or Toll-like receptor 4 in chondrocytes (17,18).
However, the effect of the Sp1 transcription factor on the
expression of catabolic enzymes induced by inflammatory cytokines
in NP cells remains to be determined.
The aim of the current study was to examine the role
of the Sp1 transcription factor in the regulation of TNF-α-induced
catabolic factor, including MMP3 and ADAMTS4, gene expression in NP
cells.
Materials and methods
Plasmid constructs and reagents
Sp1 short hairpin RNA (shRNA) and plasmid (p)LKO.1
shRNA were provided by Dr Hyoung-Pyo Kim of Yonsei University
College of Medicine (Seoul, Korea) (19). psPAX2, pMD2.G and pRL-TK plasmids
were provided by Dr Dong Xiao of Nanfang Medical University
(Guangzhou, China) (20). To assay
the promoter activity, the 5′-flanking region of the MMP3, ADAMTS4
and ADAMTS5 genes was inserted into the firefly lucif-erase
reporter vector, pGL3-Basic (Promega Corporation, Madison, WI,
USA), which contained no eukaryotic promoter or enhancer element.
To construct the promoter, genomic DNA was isolated from 293T
cells, which were provided by Dr Dong Xiao of Nanfang Medical
University (20), using a Gentra
genomic DNA isolation kit (Qiagen, Inc., Valencia, CA, USA).
Polymerase chain reaction (PCR) was used to amplify the 2.3 kb
MMP-3 promoter (MMP3-Luc, F 5′-GGCGCCTCGAGTTGACATTTGCTATGAGC-3′; R
5′-CAAGCTTGTCTTGCCTGCCTCCTTGTAGGT-3′); 2.1 kb ADAMTS4 promoter
(ADAMTS4-Luc, F 5GCGTGCTAGCCCGGGCTCGAGGGTGGGTGATCCAGGAAGTG-3,
R5′-CAGTACCGGAATGCCAAGCTTAGTAGCAGAAGCAGCCACAC-3′); 2.2 kb ADAMTS5
promoter (ADAMTS5-Luc,
F5′-GCGTGCTAGCCCGGGCTCGAGCCCCAGCGTAGCCAAAGTTA-3′, R
5′-CAGTACCGGAATGCCAAGCTTGCTTTATCCTGGGCAGGTGT-3′) with XhoI-
and HindIII restriction enzyme digestive sites. The cycling
conditions were as follows: An initial denaturation step at 94°C
for 5 min was followed by 35 cycles of denaturation at 94°C for 30
sec, annealing at 60°C for 30 sec and extension at 72°C for 40 sec,
and a final extension step at 72°C for 5 min. The pGL3-SV40 plasmid
(Promega Corporation) containing the firefly luciferase gene driven
by the SV40 promoter was used as a positive control. WP631 and
mithramycin A were purchased from Sigma-Aldrich; Merck Millipore
(Darmstadt, Germany). TNF-α and IL-1β were obtained from PeproTech,
Inc. (Rocky Hill, NJ, USA). Rabbit anti-MMP3 (catalog no. ab52915)
and rabbit anti-SDC4 (catalog no. ab24511) antibodies were
purchased from Abcam (Cambridge, UK); rabbit anti-Cox2 (catalog no.
12282) and rabbit anti-Sp1 (catalog no. 9389) antibodies were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA);
mouse anti-GAPDH (catalog no. AM4300) antibody was provided by
Thermo Fisher Scientific, Inc. (Waltham, MA, USA); and mouse
anti-β-tubulin (catalog no. N357) antibody and anti-mouse (catalog
no. RPN4201) and anti-rabbit (catalog no. RPN4301) horseradish
peroxidase (HRP)-conjugated IgG were purchased from GE Healthcare
Life Sciences (Chalfont, UK).
Isolation, culture and treatment of NP
cells
Consistent with the Institutional Review Board
guidelines of Sun Yat-sen University (Guangzhou, China), human
tissue samples were obtained from patients with thoracolumbar
fractures undergoing spine fusion; informed consent for sample
collection was obtained from each patient. A total of 8 male
Sprague-Dawley rats (age, 2–3 months; weight, 220.3±13.2 g) were
obtained from the Experimental Animal Center of Sun Yat-sen
University. Rats were housed in specific pathogen-free conditions,
at 19–28°C under a 12-h light/dark cycle and with ad libitum
access to food and water. The Animal Care and Use Committee of the
Sun Yat-sen University approved the experimental procedures.
Following anesthetisia with 10% chloral hydrate
(Wuhan Hechang Chemical Co., Ltd., Wuhan, China) (>3.5 ml/kg),
NP cells were isolated as described previously (21,22).
NP cells were cultured in Gibco Dulbecco's modified Eagle's medium
(DMEM; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100
U/ml penicillin and 100 U/ml streptomycin) at 37°C in a 5%
CO2 incubator. The medium was refreshed every 3 days.
Following growth to 80% confluence, the NP cells were treated with
50 ng/ml TNF-α or 10 ng/ml IL-1β for 4, 8 or 24 h, and cell RNA or
protein extraction was performed. WP631 (100 µM) or 1,000 nM
mithramycin A were added 1 h prior to addition of cytokines.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following the manufacturer's instructions, total RNA
was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and single-stranded cDNA was prepared by RT from
2,000 ng total RNA using Superscript III Reverse Transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Template cDNA and gene-specific primers
were added into the Fast SYBR Green master mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.). A 7900HT Fast Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used and
the cycling conditions were as follows: An initial denaturation
step at 95°C for 5 min was followed by 40 cycles of denaturation at
95°C for 5 sec, annealing at 58°C for 10 sec and extension at 72°C
for 15 sec, and a final extension step at 72°C for 10 min. mRNA
expression was quantified using the standard curve method (23). Hypoxanthine
phosphoribosyltransferase 1 and β-actin were used to normalize the
expression of rat and human samples, respectively. Each sample was
analyzed in duplicate. All primers were synthesized by Sangon
Biotech Co., Ltd. (Shanghai, China), and are listed in Table I.
 | Table IPrimer sequences for quantitative
polymerase chain reaction. |
Table I
Primer sequences for quantitative
polymerase chain reaction.
| Species | Gene | Sense (5′ to
3′) | Anti-sense (5′ to
3′) |
|---|
| Rat | MMP3 |
CAGGGAAAGTGACCCACATATT |
CGCCAAGTTTCAGAGGAAGA |
| MMP13 |
GAACCACGTGTGGAGTTATGA |
GCATCTACTTTGTCGCCAATTC |
| ADAMTS4 |
GGAGATCGTGTTTCCAGAGAAG |
CAAAGGCTGGTAATCGGTACA |
| ADAMTS5 |
GAATGTAGACCCTACAGCAACTC |
CACACTCCACACTTGTCATACT |
| Cox2 |
TCAACCAGCAGTTCCAGTATC |
GTGTACTCCTGGTCTTCAATGT |
| SDC4, |
CCCTTGGTGCCACTAGATAAC |
GACCTCATTCTCTTCCAGTTCC |
| Sp1 |
GAGGATGAGTAGACCAGCTTTG |
TGGAAGAGGGAGTAGGAACTTA |
| HPRT |
GCTGACCTGCTGGATTACAT |
CCCGTTGACTGGTCATTACA |
| Human | MMP3 |
GCAAGACAGCAAGGCATAGA |
CAGCAACAGTAGGATTGGAAGA |
| MMP13 |
GTTTGGTCCGATGTAACTCCTC |
GAAGTCGCCATGCTCCTTAAT |
| ADAMTS4 |
GGTGGTGGTGATAGGTATAAGTG |
TCAGGAAGAGGAAAGCAGAAC |
| ADAMTS5 |
CTGCCACCACACTCAAGAA |
TCCTCCCGAGTAAACAGGATAG |
| Cox2 |
CTATGTGCTAGCCCACAAAGA |
GCATCCACAGATCCCTCAAA |
| SDC4, |
GACTGGGATTGGATCACTTCTT |
ACTCCACACAACATCCGTTAG |
| Sp1 |
TCCAAGGCCTGGCTAATAATG |
GACAGGTAGCAAGGTGATGTT |
| β-actin |
GGACCTGACTGACTACCTCAT |
CGTAGCACAGCTTCTCCTTAAT |
Western blotting
Following treatment, the NP cells and medium were
collected and then treated with lysis buffer, containing 1X
protease inhibitor cocktail (Roche Diagnostics, Basel,
Switzerland), NaCl (5 mM), NaF (200 µM),
Na3VO4 (200 µM) and dithiothreitol
(0.1 mM). Following quantification by bicinchoninic acid assay,
total cell proteins (30 ng) were resolved by sodium dodecyl
sulfate-polyacrylamide gels on 10% gels, then transferred by
electroblotting to polyvinylidene difluoride membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Membranes were blocked in
5% non-fat dry milk for 1 h at room temperature in Tris-buffered
saline (50 mM Tris, pH 7.6, 150 mM NaCl) plus 0.1% Tween-20 and
incubated overnight at 4°C with the primary antibody (1:1,000
anti-MMP3; 1:1,000 anti-Cox2; 1:1,000 anti-SDC4; 1:1,000 anti-Sp1;
1:3,000 anti-GAPDH; or 1:3,000 anti-β-tubulin). Membranes were then
incubated with HRP-conjugated anti-rabbit or anti-mouse IgG
(1:2,000–1:5,000) for 1 h and the signals detected using enhanced
chemiluminescence (GE Healthcare Life Sciences). GAPDH and
β-tubulin were used as protein loading controls for rat and human
samples, respectively. Protein bands were quantified by
densitometric analysis using Kodak 1D software version 3.6 (Kodak,
Rochester, NY, USA).
Transfections and dual-luciferase
reporter assay
Rat NP cells were seeded in 48-well plates at a
density of 3×104 cells/ml with 2% Opti-Minimal Essential
Medium Reduced Serum (Gibco; Thermo Fisher Scientific, Inc.). The
following day, cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), 250 ng MMP3, ADAMTS4 or ADAMTS5 promoter
luciferase reporter plasmids and 250 ng of the internal control
plasmid, pRL-TK. Cells were stimulated with WP631 (100 µM)
or mithramycin A (1,000 nM) 48 h post-transfection. Cells were
harvested 24 h later, and firefly and Renilla luciferase activities
were measured using the Dual Luciferase Reporter Assay Kit (Promega
Corporation) according to the manufacturer's instructions.
Lentivirus production and
transduction
HEK 293T human embryonic kidney cells were seeded in
a humidified incubator at 37°C, 5% CO2 at a density of
3×106 cells per 10 cm plate in DMEM with 10%
heat-inactivated FBS. Following ~24 h incubation, cells were
transfected with lentiviral vectors and packaging plasmids (psPAX2
and pMD2.G) using Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Following 16 h incubation, the transfection medium
was replaced with DMEM with 10% heat-inactivated FBS. At 48 and 60
h post-transfection, the supernatant was harvested from HEK 293T
human embryonic kidney cells and was introduced into human NP cell
cultures (0.5×106 cells/plate) with 6 mg/ml Polybrene
(EMD Millipore, Billerica, MA, USA). A total of 24 h later, the
medium containing viral particles was removed and replaced with
DMEM containing 10% FBS and antibiotics. Cells and medium were
harvested from NP cells 5 days later for mRNA or protein
extraction.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed as described previously
(21,22). Briefly, human NP cells were
cultured in DMEM media supplemented with 10% FBS to 80% confluence.
The cells were then cross-linked, lysed and the chromatin was
sheared by sonication. Input DNA was generated by treating aliquots
with RNase, proteinase K, and heat, followed by ethanol
precipitation. DNA complexes were immunoprecipitated overnight
using antibodies for Sp1 and nonspecific rabbit IgG followed by
binding to protein G-agarose beads. Cross-links were reversed by
treatment with proteinase K and heat, and the DNA was purified
using DNA purification elution buffer (Active Motif, Carlsbad, CA,
USA). qPCR analysis was performed using a ChIP-IT quantitative PCR
analysis kit (cataolog no. 53040; Active Motif) using the following
primer pairs for putative Sp1 sites within the ADAMTS4 and ADAMTS5
promoters: (−577/−567) ADMATS4 motif, 5′-CTATTATCCATTCGGCTGCTAGA-3′
(sense) and 5′-ATGGGTGTATCATCGCTTCC-3′ (anti-sense); and −718/−708
ADAMTS5 motif, 5′-TTAAGAGGCAGCGTGCAG-3′ (sense) and
5′-GCGAGCGCTGTGAAGAT-3′ (anti-sense). The negative control primers
were purchased from Active Motif (catalog no. 71001).
Statistical analysis
All experiments were repeated independently at least
three times. Data are presented as the mean ± standard error.
Statistical analyses were performed in SPSS software version 13.0
(SPSS, Inc., Chicago, IL, USA). Differences between groups were
analyzed by one-way analysis of variance, followed by the post hoc
tests Student-Newman-Keuls and least significant difference.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Treatment with Sp1 inhibitors attenuates
catabolic gene expression induced by TNF-α
To observe the effect of Sp1 and GC-rich island
inhibitors on the expression of the catabolic enzyme genes MMP3,
ADAMTS4 and Cox2, 1 h prior to TNF-α stimulation, the NP cells were
treated with WP631 and mithramycin A, which serve as
transcriptional inhibitors by displacing Sp1 from promoters by
binding to GC-rich DNA motifs. The data revealed that MMP3, MMP13,
ADAMTS4, ADAMTS5, Cox2 and SDC4 mRNA expression was significantly
increased following stimulation with TNF-α compared with untreated
controls (P<0.05), whereas the mRNA expression was significantly
decreased following treatment with WP631 or mithramycin A compared
with TNF-α treatment only (P<0.05; Fig. 1A). Furthermore, western blot
results revealed that stimulation with TNF-α significantly
increased the intracellular protein expression of MMP3, Cox2 and
SDC4 and the protein expression of MMP3 in the medium compared with
unstimulated cells (P<0.05; Fig.
1B). However, treatment with WP631 or mithramycin A
significantly suppressed this increased expression (P<0.05;
Fig. 1B).
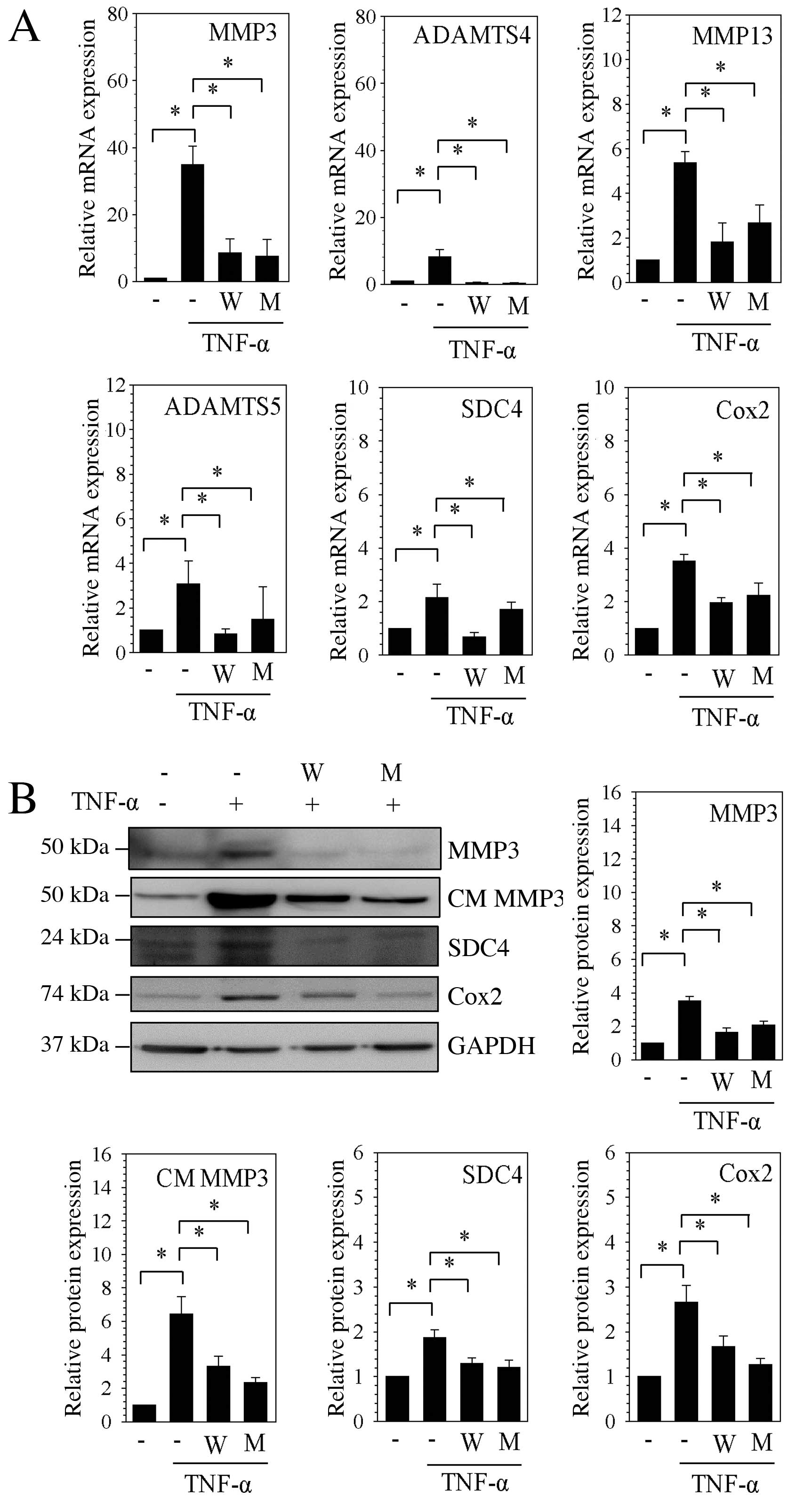 | Figure 1Expression of catabolic factors
induced by TNF-α in nucleus pulposus cells was suppressed by
specificity protein-1 transcription factor inhibitors. (A) mRNA
expression levels of MMP3, ADAMTS4, MMP13, ADAMTS5, SDC4 and Cox2
were assessed by reverse transcription-quantitative polymerase
chain reaction following stimulation with TNF-α, and treatment with
WP631 or mithramycin A. (B) Western blot analysis of protein
expression levels of MMP3, CM MMP3, Cox2 and SDC4, following
stimulation with TNF-α and treatment with WP631 or mithramycin A.
Densitometric quantification was relative to GAPDH. Data are
presented as the mean ± standard error of three independent
experiments performed in triplicate, relative to the unstimulated
control; *P<0.05, comparison indicated by brackets.
MMP, matrix metalloproteinases; W, WP631; M, mithramycin A; TNF-α,
tumor necrosis factor-α; ADAMTSs, disintegrins and
metalloproteinases with thrombospondin motifs; SDC4, syndecan 4;
Cox2, cyclooxygenase 2; CM, conditioned medium; GAPDH,
glyceraldehyde 3-phosphate dehydrogenase. |
Sp1 transcription factor controls MMP3,
ADAMTS4 and ADAMTS5 promoter activities in NP cells
To further confirm the involvement of Sp1 in the
regulation of catabolic factors, the luciferase activity of pGL3
firefly luciferase reporter constructs containing the promoter
region of MMP3, ADAMTS4 and ADAMTS5 was assessed following
stimulation with TNF-α or IL-1β, and treatment with WP631 or
mithramycin A. Stimulation with TNF-α and IL-1β resulted in
significantly increased MMP3 promoter activity compared with
unstimulated rat NP cells (P<0.05), while WP631 and mithramycin
A significantly suppressed this MMP3 promoter induction (P<0.05;
Fig. 2A). TNF-α and IL-1β
stimulation had no significant effect on ADAMTS4 and ADAMTS5
promoter activities (Fig. 2B and
C), despite TNF-α stimulation having been demonstrated to
increase ADAMTS4 and ADAMTS5 mRNA and protein expression levels
(Fig. 1). However, treatment with
WP631 and mithramycin A did result in significantly reduced ADAMTS4
and ADAMTS5 promoter activity compared with untreated cells that
had been stimulated with TNF-α or IL-1β (P<0.05; Fig. 2B and C, respectively).
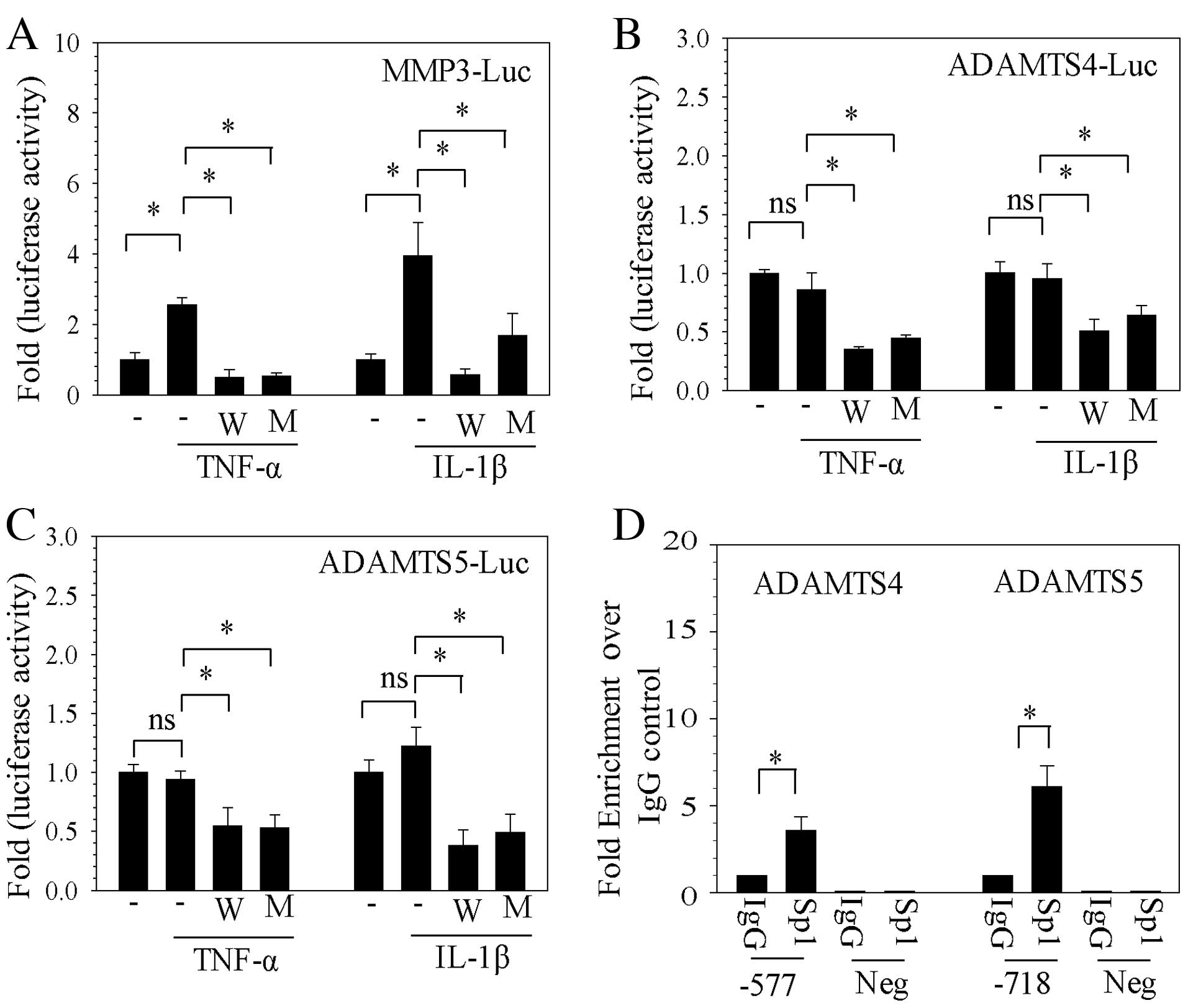 | Figure 2Sp1 controls catabolic factor promoter
activity in NP cells. Firefly luciferase was used as a reporter of
transcriptional regulation of the promoter regions of (A) MMP3, (B)
ADAMTS4 and (C) ADAMTS5, following stimulation with TNF-α or IL-1β
and treatment with WP631 or mithramycin A. (D) Sp1 binding to Sp1
motifs at −577/−567 bp of ADAMTS4 and −718/−708 bp ADAMTS5
promoters in human NP cells was analyzed by genomic chromatin
immunoprecipitation analysis with Sp1 and IgG antibody pulldowns.
Negative primers revealed minimal amplification from experimental
and IgG control samples. Data are presented as the mean ± standard
error of three independent experiments performed in triplicate.
*P<0.05, comparison indicated by brackets. Sp1,
specificity protein-1 transcription factor; NP, nucleus pulposus;
MMP, matrix metalloproteinases; Luc, luciferase; W, WP631; M,
mithramycin A; TNF-α, tumor necrosis factor-α; IL-1β,
interleukin-1β; ADAMTSs, disintegrins and metalloproteinases with
thrombospondin motifs; IgG, immunoglobulin G; Neg, negative control
primers. |
To determine whether Sp1 binds to the catabolic gene
promoters, ChIP analysis was performed to assess Sp1 binding to the
cognate Sp1 motifs in the promoter regions of ADAMTS4 and ADAMTS5.
As presented in Fig. 2D, using the
Sp1 antibody for ChIP resulted in ~3.6-fold enrichment in binding
of Sp1 to a site located at −577/−567 bp in the human ADAMTS4
promoter compared with the IgG control (P= 0.039; Fig. 2D), while ~6.1-fold enrichment of
Sp1 binding compared with IgG control was detected at the motif
located at −718/−708 bp in the human ADAMTS5 promoter (P=0.027;
Fig. 2D). Negative control primers
demonstrated almost undetectable amplification (Fig. 2D).
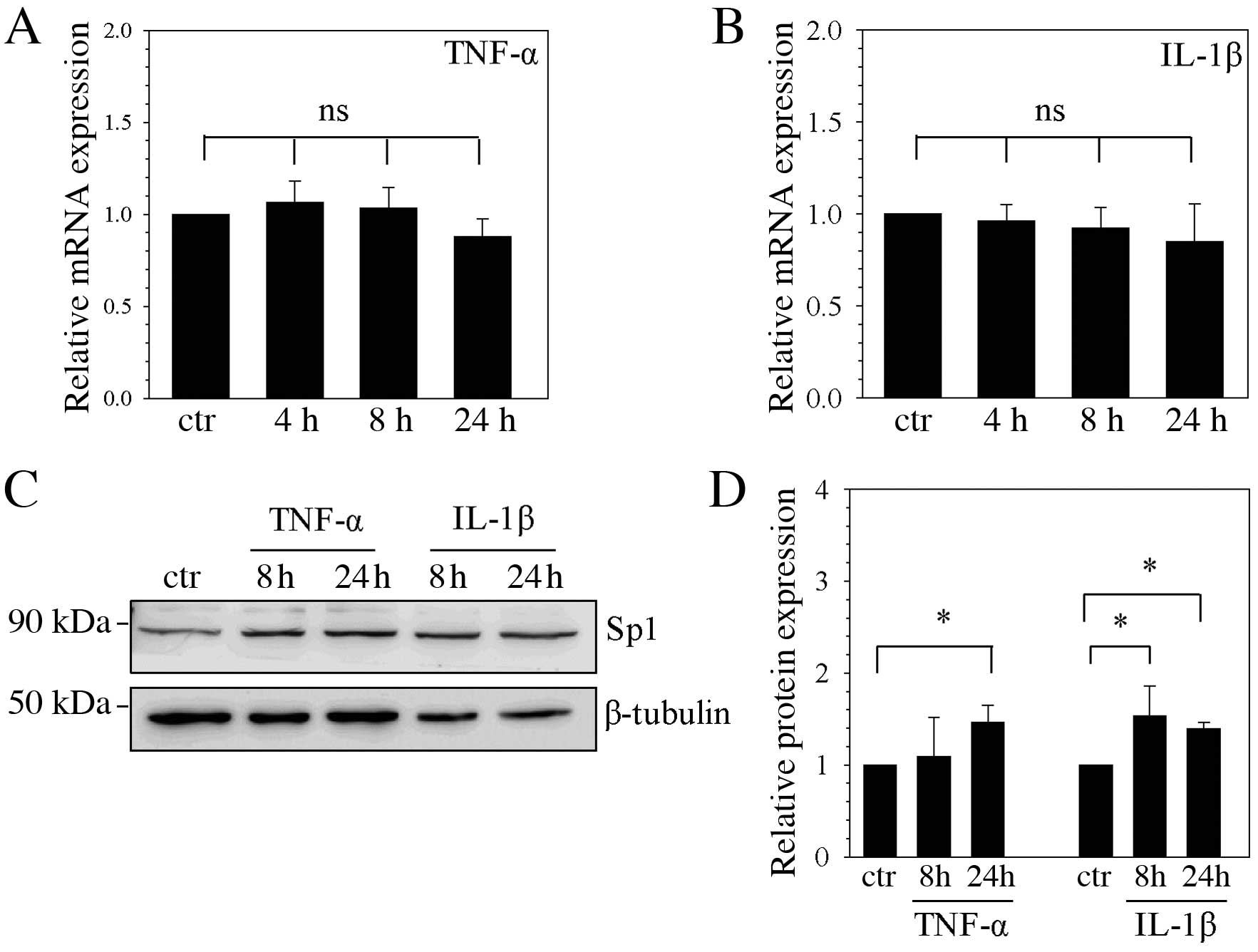
Regulation of Sp1 mRNA and protein
following TNF-α and IL-1β stimulation
To determine the effect of cytokine stimulation on
the expression of Sp1 mRNA and protein, NP cells were stimulated
with TNF-α and IL-1β. Sp1 mRNA expression levels remained unchanged
at 4, 8 and 24 h following stimulation with TNF-α (Fig. 3A) and IL-1β (Fig. 3B), compared with unstimulated
control cells. However, Sp1 protein expression levels were
significantly higher following 24 h TNF-α stimulation compared with
the unstimulated control (P=0.040; Fig. 3C and D), and were significantly
higher following 8 and 24 h of IL-1β stimulation compared with the
unstimulated control (P=0.039 and P=0.048, respectively; Fig. 3C and D).
Stable silencing of Sp1 decreases
catabolic gene expression induced by TNF-α
To further examine whether the Sp1 transcription
factor controls catabolic gene expression in human NP cells, Sp1
loss-of-function studies were performed using lentiviral delivery
of gene-targeting shRNA. As observed in rat NP cells, no
significant difference in Sp1 mRNA expression was observed in
response to stimulation with TNF-α in human NP cells transduced
with control shRNA compared with unstimulated cells transduced with
control shRNA (Fig. 4A). mRNA
expression levels in human NP cells stimulated with TNF-α and
transduced with Sp1 shRNA were ~78% reduced, compared with
TNF-α-stimulated cells transduced with control shRNA (P=0.001;
Fig. 4A). In TNF-α-stimulated NP
cells in which Sp1 was suppressed (cells transduced with Sp1
shRNA), MMP3, MMP13, ADAMTS4, ADAMTS5 and SDC4 mRNA expression was
significantly reduced compared with TNF-α-stimulated cells
transduced with control shRNA (P<0.05; Fig. 4A). Furthermore, western blot
analysis was used to investigate protein expression levels of Sp1,
the cell protein, Cox2, and condition medium MMP3 in Sp1-silenced
cells (cells transduced with Sp1 shRNA). Sp1 protein expression was
significantly reduced in cells transduced with Sp1 shRNA and
stimulated with TNF-α compared with TNF-α-stimulated cells
transduced with control shRNA (P=0.001; Fig. 4B). Consistent with the changes in
mRNA expression levels of catabolic genes, including MMP3 and
ADAMTS4, Cox2 expression and the level of MMP3 protein in the
medium in cells transduced with Sp1 shRNA and stimulated with TNF-α
was ~58 and ~44% lower than in TNF-α-stimulated cells transduced
with control shRNA (P=0.023 and P=0.032, respectively; Fig. 4B).
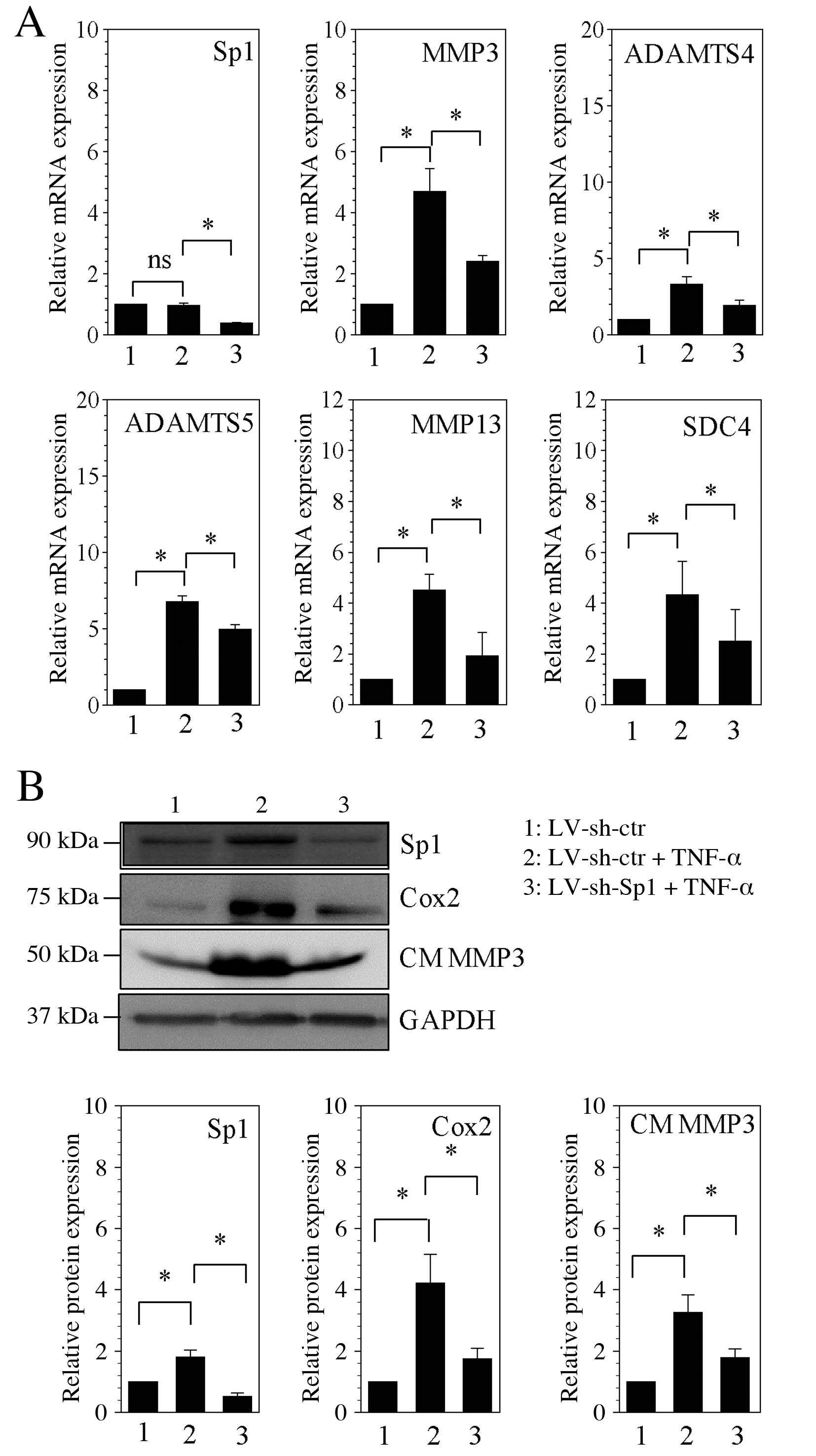 | Figure 4Stable silencing of Sp1 decreases
catabolic factors induction by TNF-α in human NP cells. (A) mRNA
expression levels of Sp1, MMP3, ADAMTS4, ADAMTS5, MMP13 and SDC4
were assessed by reverse transcription-quantitative polymerase
chain reaction following lentiviral transfection and stimulation
with TNF-α. (B) Western blot analysis of protein expression levels
of Sp1, Cox2 and CM MMP3 following lentiviral transfection and
stimulation with TNF-α, with quantification relative to GAPDH. Data
are presented as the mean ± standard error of three independent
experiments performed in triplicate, relative to LV-sh-ctr.
*P<0.05, comparison indicated by brackets. Sp1,
specificity protein-1 transcription factor; ns, no significance;
MMP, matrix metalloproteinase; ADAMTSs, disintegrins and
metalloproteinases with thrombospondin motifs; SDC4, syndecan 4;
LV, len-tivirus; sh, short hairpin RNA; ctr, control; TNF-α, tumor
necrosis factor-α; Cox2, cyclooxygenase 2; CM, conditioned medium;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
Discussion
The experiments described in the present study
demonstrated that in NP cells, Sp1 transcription factor inhibitors
and knockdown suppressed the TNF-α-induced expression of the
catabolic factors MMP3, ADAMTS-4, Cox2 and SDC4. Furthermore, Sp1
was essential for the maintenance of catabolic factor
transcriptional expression due to direct interaction with cognate
binding sites in their promoters. In addition, Sp1 transcription
factor protein expression is induced by stimulation with
inflammatory cytokines in NP cells.
Intervertebral disc degeneration is generally
believed to be a consequence of increased catabolism of the ECM.
Over the last decade, the research on intervertebral disc
degeneration has emphasized the mechanism of ECM degradation. MMPs,
ADAMTSs and Cox2 have been proposed to be the major catabolic
enzymes in the intervertebral disc (8,24).
SDC4, a cell-surface heparan sulfate proteoglycan, participates in
inflammation and mediates the expression of MMP3, ADAMTS4 and
ADAMTS5 in NP cells (4,5,8).
TNF-α induces the expression of catabolic factors including MMP3,
MMP13, ADAMTS4, ADAMTS5, Cox2 and SDC4 in NP cells and articular
chondrocytes (3,5,6,8,9,21,25,26).
Sp1 is also involved in the expression of ECM-related genes in
various cells (15,16). The present study revealed that the
expression of Sp1 mRNA was unaffected by TNF-α and IL-1β
stimulation in human and rat NP cells, as previously reported in
bovine NP cells (27). However,
Sp1 protein expression was induced by TNF-α and IL-1β stimulation,
as reported in articular chondrocytes (28). In articular chondrocytes, Sp1
mediates the inductive activity of TNF-α (29–31).
To the best of our knowledge, the present study presents the first
evidence that the Sp1 inhibitors, WP631 and mithramycin A, suppress
the TNF-α-induced expression of catabolic factors in NP cells.
Furthermore, loss of function experiments also revealed that Sp1
knockdown decreased cytokine-induced expression of MMP3, MMP13,
ADAMTS4, ADAMTS5, Cox2 and SDC4 in NP cells.
There are numerous Sp1 binding sites in MMPs,
ADAMTSs, Cox2 and SDC4 promoters (17,18,32,33),
but the present study limited its investigations to MMP3, ADAMTS4
and ADAMTS5 promoter constructs containing Sp1 sites. Stimulation
of NP cells with TNF-α and IL-1β resulted in increased expression
from MMP3 promoter constructs, as previously reported (10), however, Sp1 inhibitors suppressed
the induction by TNF-α or IL-1β. Although TNF-α and IL-1β
stimulation had no effect on transcription from the ADAMTS4 and
ADAMTS5 promoters, this may have been due to the promoter
constructs being too short or lacking effective binding sites for
TNF-α downstream mediators. However, Sp1 inhibitors reduced the
baseline activity of ADAMTS4 and ADAMTS5 promoter constructs during
stimulation with TNF-α and IL-1β. To further support the functional
studies of ADAMTS4 and ADAMTS5 promoter constructs, genomic ChIP
analysis demonstrated that the enriched binding of Sp1 to the
ADAMTS4 and ADAMTS5 promoter fragment that contained the −577/−567
and 718/−708 bp binding sites, respectively, controls its basal
transcription. The present study therefore revealed that TNF-α
induces increased ADAMTS4 and ADAMTS5 mRNA expression, but did not
affect transcription from the ADAMTS4 and ADAMTS5 promoter reporter
constructs.
The pharmacological and genetic evidence presented
in the present study demonstrated that Sp1 is involved in the
TNF-α-induced expression of catabolic factors, including MMP3 and
ADAMTS4. Therapeutic targeting of the Sp1 transcription factor to
block the activity these catabolic factors may therefore present a
novel strategy to prevent degradation of intervertebral discs by
catabolic factors during the pathogenesis of intervertebral disc
degeneration.
Acknowledgments
This work was supported by grants from the National
Natural Science Foundation of China (grant nos. 81101385 and
81572197), the Natural Science Foundation of Guangdong Province,
China (grant no. 2016A030313189) and the Science and Technology
Planning Project of Guangdong Province (grant nos. 2012B031800359,
2012B031800360 and 2014A020212058).
References
|
1
|
Stemple DL: Structure and function of the
notochord: An essential organ for chordate development.
Development. 132:2503–2512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng H, Danfelter M, Strömqvist B and
Heinegård D: Extracellular matrix in disc degeneration. J Bone
Joint Surg Am. 88(Suppl 2): S25–S29. 2006. View Article : Google Scholar
|
|
3
|
Roughley PJ: Biology of intervertebral
disc aging and degeneration: Involvement of the extracellular
matrix. Spine (Phila Pa 1976). 29:2691–2699. 2004. View Article : Google Scholar
|
|
4
|
Boos N, Weissbach S, Rohrbach H, Weiler C,
Spratt KF and Nerlich AG: Classification of age-related changes in
lumbar intervertebral discs: 2002 Volvo Award in basic science.
Spine (Phila Pa 1976). 27:2631–2644. 2002. View Article : Google Scholar
|
|
5
|
Podichetty VK: The aging spine: The role
of inflammatory mediators in intervertebral disc degeneration. Cell
Mol Biol (Noisy-le-grand). 53:4–18. 2007.
|
|
6
|
Tian Y, Yuan W, Fujita N, Wang J, Wang H,
Shapiro IM and Risbud MV: Inflammatory cytokines associated with
degenerative disc disease control aggrecanase-1 (ADAMTS-4)
expression in nucleus pulposus cells through MAPK and NF-κB. Am J
Pathol. 182:2310–2321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jimbo K, Park JS, Yokosuka K, Sato K and
Nagata K: Positive feedback loop of interleukin-1beta upregulating
production of inflammatory mediators in human intervertebral disc
cells in vitro. J Neurosurg Spine. 2:589–595. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Wang H, Yang H, Li J, Cai Q,
Shapiro IM and Risbud MV: Tumor necrosis factor-α- and
interleukin-1β-dependent matrix metalloproteinase-3 expression in
nucleus pulposus cells requires cooperative signaling via syndecan
4 and mitogen-activated protein kinase-NF-κB axis: Implications in
inflammatory disc disease. Am J Pathol. 184:2560–2572. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Studer RK, Aboka AM, Gilbertson LG,
Georgescu H, Sowa G, Vo N and Kang JD: p38 MAPK inhibition in
nucleus pulposus cells: A potential target for treating
intervertebral disc degeneration. Spine (Phila Pa 1976).
32:2827–2833. 2007. View Article : Google Scholar
|
|
12
|
Lin TH, Tang CH, Wu K, Fong YC, Yang RS
and Fu WM: 15-deox y-Δ(12,14)-prostaglandin-J2 and ciglitazone
inhibit TNF-α-induced matrix metalloproteinase 13 production via
the antagonism of NF-κB activation in human synovial fibroblasts. J
Cell Physiol. 226:3242–3250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park HY, Kim TH, Kim CG, Kim GY, Kim CM,
Kim ND, Kim BW, Hwang HJ and Choi YH: Purpurogallin exerts
anti-inflammatory effects in lipopolysaccharide-stimulated BV2
microglial cells through the inactivation of the NF-κB and MAPK
signaling pathways. Int J Mol Med. 32:1171–1178. 2013.PubMed/NCBI
|
|
14
|
Kadonaga JT, Courey AJ, Ladika J and Tjian
R: Distinct regions of Sp1 modulate DNA binding and transcriptional
activation. Science. 242:1566–1570. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang L, Zhou Y, Xiong M, Fang L, Wen P,
Cao H, Yang J, Dai C and He W: Sp1 mediates microRNA-29c-regulated
type I collagen production in renal tubular epithelial cells. Exp
Cell Res. 319:2254–2265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Domenzain-Reyna C, Hernández D,
Miquel-Serra L, Docampo MJ, Badenas C, Fabra A and Bassols A:
Structure and regulation of the versican promoter: The versican
promoter is regulated by AP-1 and TCF transcription factors in
invasive human melanoma cells. J Biol Chem. 284:12306–12317. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sylvester J, Ahmad R and Zafarullah M:
Role of Sp1 transcription factor in Interleukin-1-induced ADAMTS-4
(aggrecanase-1) gene expression in human articular chondrocytes.
Rheumatol Int. 33:517–522. 2013. View Article : Google Scholar
|
|
18
|
Xu K and Shu HK: Transcription factor
interactions mediate EGF-dependent COX-2 expression. Mol Cancer
Res. 11:875–886. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang WJ, Song MJ, Park EY, Lee JJ, Park
JH, Park K, Park JH and Kim HP: Transcription factors Sp1 and Sp3
regulate expression of human ABCG2 gene and chemoresistance
phenotype. Mol Cells. 36:368–375. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang SC, Lin XL, Li J, Zhang TT, Wang HY,
Shi JW, Yang S, Zhao WT, Xie RY, Wei F, et al: MicroRNA-122
triggers mesenchymal-epithelial transition and suppresses
hepatocellular carcinoma cell motility and invasion by targeting
RhoA. PLoS One. 9:e1013302014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye W, Zhou J, Markova DZ, Tian Y, Li J,
Anderson DG, Shapiro IM and Risbud MV: Xylosyltransferase-1
expression is refractory to inhibition by inflammatory cytokines
tumor necrosis factor α and IL-1β in nucleus pulposus cells: Novel
regulation by AP-1, Sp1, and Sp3. Am J Pathol. 185:485–495. 2015.
View Article : Google Scholar :
|
|
22
|
Soutoglou E and Talianidis I: Coordination
of PIC assembly and chromatin remodeling during
differentiation-induced gene activation. Science. 295:1901–1904.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnson GL, Bibby DF, Wong S, Agrawal SG
and Bustin SA: A MIQE-compliant real time PCR assay for Aspergillus
detection. PLoS One. 7:e400222012. View Article : Google Scholar
|
|
24
|
Wang WJ, Yu XH, Wang C, Yang W, He WS,
Zhang SJ, Yan YG and Zhang J: MMPs and ADAMTSs in intervertebral
disc degeneration. Clin Chim Acta. 448:238–446. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu K, Chen W, Wang X, Peng Y, Liang A,
Huang D, Li C and Ye W: Autophagy attenuates the catabolic effect
during inflammatory conditions in nucleus pulposus cells, as
sustained by NF-κB and JNK inhibition. Int J Mol Med. 36:661–668.
2015.PubMed/NCBI
|
|
26
|
Echtermeyer F, Bertrand J, Dreier R,
Meinecke I, Neugebauer K, Fuerst M, Lee YJ, Song YW, Herzog C,
Theilmeier G and Pap T: Syndecan-4 regulates ADAMTS-5 activation
and cartilage breakdown in osteoarthritis. Nat Med. 15:1072–1076.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Séguin CA, Pilliar RM, Roughley PJ and
Kandel RA: Tumor necrosis factor-alpha modulates matrix production
and catabolism in nucleus pulposus tissue. Spine (Phila Pa 1976).
30:1940–1948. 2005. View Article : Google Scholar
|
|
28
|
Séguin CA, Pilliar RM, Madri JA and Kandel
RA: TNF-alpha induces MMP2 gelatinase activity and MT1-MMP
expression in an in vitro model of nucleus pulposus tissue
degeneration. Spine (Phila Pa 1976). 33:356–365. 2008. View Article : Google Scholar
|
|
29
|
Liacini A, Sylvester J, Li WQ and
Zafarullah M: Mithramycin downregulates proinflammatory
cytokine-induced matrix metalloproteinase gene expression in
articular chondrocytes. Arthritis Res Ther. 7:R777–R783. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hashimoto K, Otero M, Imagawa K, de Andrés
MC, Coico JM, Roach HI, Oreffo RO, Marcu KB and Goldring MB:
Regulated transcription of human matrix metalloproteinase 13
(MMP13) and interleukin-1β (IL-1B) genes in chondrocytes depends on
methylation of specific proximal promoter CpG sites. J Biol Chem.
288:10061–10066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu N, Opalenik S, Liu J, Jansen ED, Giro
MG and Davidson JM: Real time visualization of MMP-13 promoter
activity in transgenic mice. Matrix Biol. 21:149–161. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mizui Y, Yamazaki K, Kuboi Y, Sagane K and
Tanaka I: Characterization of 5′-flanking region of human
aggrecanase-1 (ADAMTS4) gene. Mol Biol Rep. 27:167–173. 2000.
View Article : Google Scholar
|
|
33
|
Chuang CW, Pan MR, Hou MF and Hung WC:
Cyclooxygenase-2 up-regulates CCR7 expression via AKT-mediated
phosphorylation and activation of Sp1 in breast cancer cells. J
Cell Physiol. 228:341–348. 2013. View Article : Google Scholar
|