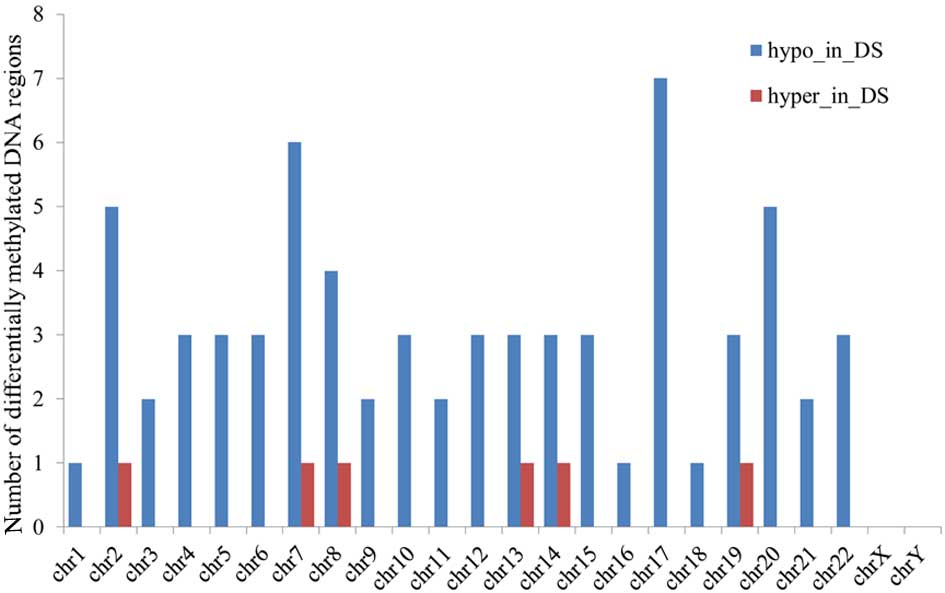
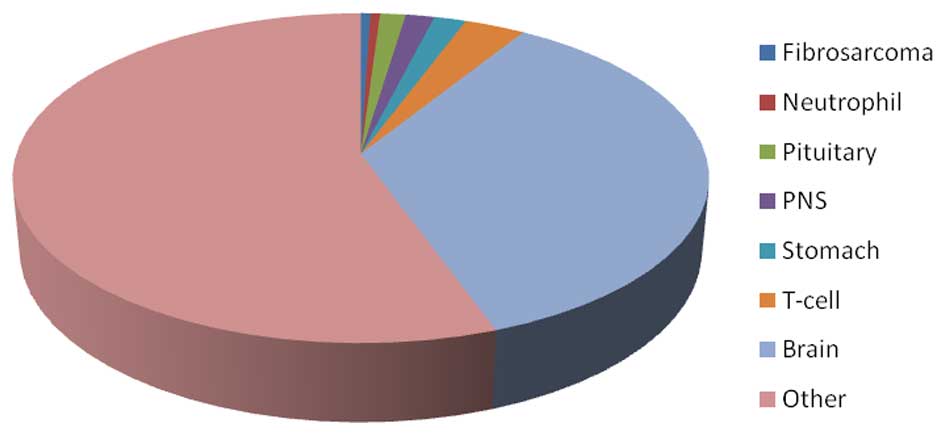
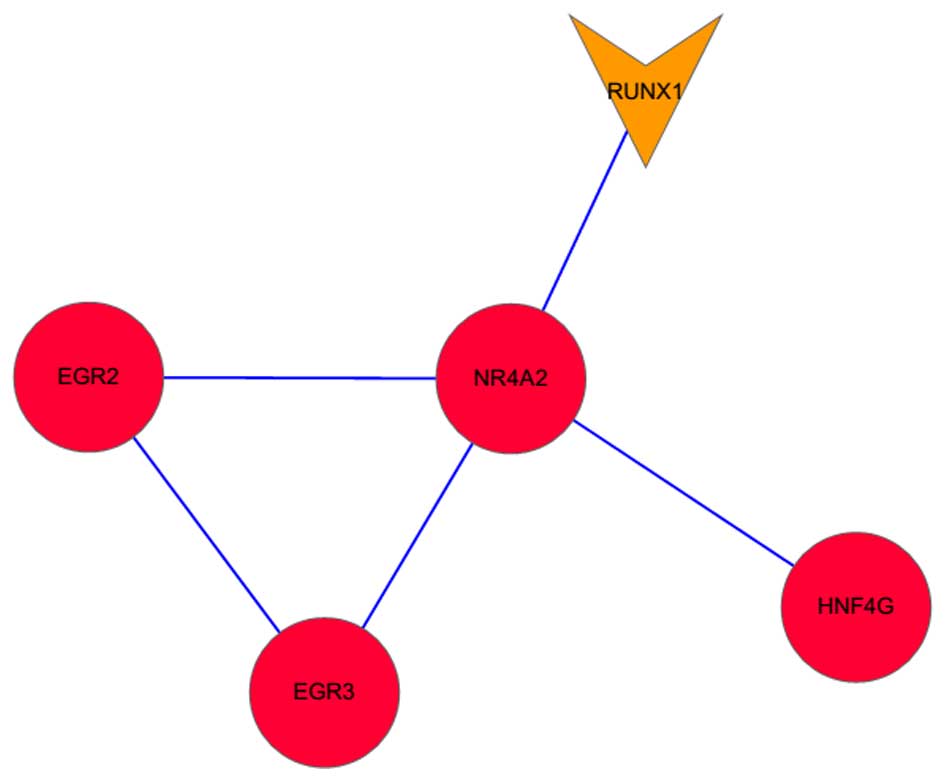
|
1
|
Palomaki GE, Kloza EM, Lambert-Messerlian
GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT,
Deciu C, Grody WW, et al: DNA sequencing of maternal plasma to
detect Down syndrome: An international clinical validation study.
Genet Med. 13:913–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Palomaki GE, Deciu C, Kloza EM,
Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom
D, Bombard AT, Grody WW, et al: DNA sequencing of maternal plasma
reliably identifies trisomy 18 and trisomy 13 as well as Down
syndrome: An international collaborative study. Genet Med.
14:296–305. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khocht A, Yaskell T, Janal M, Turner BF,
Rams TE, Haffajee AD and Socransky SS: Subgingival microbiota in
adult Down syndrome periodontitis. J Periodontal Res. 47:500–507.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosdi M, Kadir A, Sheikh RS, Hj Murat Z
and Kamaruzaman N: The comparison of human body electromagnetic
radiation between down syndrome and non down syndrome person for
brain, chakra and energy field stability score analysis. 2012 IEEE
Control and System Graduate Research Colloquium. Malaysia. pp.
370–375. 2012;
|
|
5
|
Ward O: John Langdon Down: The man and the
message. Downs Syndr Res Pract. 6:19–24. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cronk C, Crocker AC, Pueschel SM, Shea AM,
Zackai E, Pickens G and Reed RB: Growth charts for children with
Down syndrome: 1 month to 18 years of age. Pediatrics. 81:102–110.
1988.PubMed/NCBI
|
|
7
|
Myrelid A, Gustafsson J, Ollars B and
Annerén G: Growth charts for Down's syndrome from birth to 18 years
of age. Arch Dis Child. 87:97–103. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lott IT: Neurological phenotypes for Down
syndrome across the life span. Prog Brain Res. 197:1012012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roizen NJ and Patterson D: Down's
syndrome. Lancet. 361:1281–1289. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Freeman SB, Bean LH, Allen EG, Tinker SW,
Locke AE, Druschel C, Hobbs CA, Romitti PA, Royle MH, Torfs CP, et
al: Ethnicity, sex, and the incidence of congenital heart defects:
A report from the national down syndrome project. Genet Med.
10:173–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hernandez D and Fisher EM: Down syndrome
genetics: Unravelling a multifactorial disorder. Hum Mol Genet.
5:1411–1416. 1996.PubMed/NCBI
|
|
12
|
Patterson D and Costa AC: Down syndrome
and genetics-a case of linked histories. Nat Rev Genet. 6:137–147.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Arron JR, Winslow MM, Polleri A, Chang CP,
Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, et al: NFAT
dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome
21. Nature. 441:595–600. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Altafaj X, Dierssen M, Baamonde C, Martí
E, Visa J, Guimerà J, Oset M, González JR, Flórez J, Fillat C and
Estivill X: Neurodevelopmental delay, motor abnormalities and
cognitive deficits in transgenic mice overexpressing Dyrk1A
(minibrain), a murine model of Down's syndrome. Hum Mol Genet.
10:1915–1923. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Voronov SV, Frere SG, Giovedi S, Pollina
EA, Borel C, Zhang H, Schmidt C, Akeson EC, Wenk MR, Cimasoni L, et
al: Synaptojanin 1-linked phosphoinositide dyshomeostasis and
cognitive deficits in mouse models of Down's syndrome. Proc Natl
Acad Sci USA. 105:9415–9420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin S, Lee YK, Lim YC, Zheng Z, Lin XM, Ng
DP, Holbrook JD, Law HY, Kwek KY, Yeo GS and Ding C: Global DNA
hypermethylation in down syndrome placenta. PLoS Genet.
9:e10035152013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krueger F and Andrews SR: Bismark: A
flexible aligner and methylation caller for Bisulfite-Seq
applications. Bioinformatics. 27:1571–1572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hebestreit K, Dugas M and Klein HU:
Detection of significantly differentially methylated regions in
targeted bisulfite sequencing data. Bioinformatics. 29:1647–1653.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu H and Lachenbruch PA: Paired t test.
Wiley Encyclopedia of Clinical Trials. 1–3. 2008.
|
|
22
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:(Web Server issue). W169–W175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:(Database
issue). D412–D416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Y, Mullaney KA, Peterhoff CM, Che S,
Schmidt SD, Boyer-Boiteau A, Ginsberg SD, Cataldo AM, Mathews PM
and Nixon RA: Alzheimer's-related endosome dysfunction in Down
syndrome is Abeta-independent but requires APP and is reversed by
BACE-1 inhibition. Proc Natl Acad Sci USA. 107:1630–1635. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Gameren-Oosterom H, Fekkes M, van
Wouwe JP, Detmar SB, Oudesluys-Murphy AM and Verkerk PH: Problem
behavior of individuals with Down syndrome in a nationwide cohort
assessed in late adolescence. J Pediatr. 163:1396–1401. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bourquin JP, Subramanian A, Langebrake C,
Reinhardt D, Bernard O, Ballerini P, Baruchel A, Cavé H, Dastugue
N, Hasle H, et al: Identification of distinct molecular phenotypes
in acute megakaryoblastic leukemia by gene expression profiling.
Proc Natl Acad Sci USA. 103:3339–3344. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Edwards H, Xie C, LaFiura KM, Dombkowski
AA, Buck SA, Boerner JL, Taub JW, Matherly LH and Ge Y: RUNX1
regulates phosphoinositide 3-kinase/AKT pathway: Role in
chemotherapy sensitivity in acute megakaryocytic leukemia. Blood.
114:2744–2752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Patel A, Rees SD, Kelly MA, Bain SC,
Barnett AH, Thalitaya D and Prasher VP: Association of variants
within APOE, SORL1, RUNX1, BACE1 and ALDH18A1 with dementia in
Alzheimer's disease in subjects with Down syndrome. Neurosci Lett.
487:144–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sekiya T, Kashiwagi I, Inoue N, Morita R,
Hori S, Waldmann H, Rudensky AY, Ichinose H, Metzger D, Chambon P
and Yoshimura A: The nuclear orphan receptor Nr4a2 induces Foxp3
and regulates differentiation of CD4+ T cells. Nat Commun.
2:2692011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okada M, Hibino S, Someya K and Yoshmura
A: Regulation of regulatory T cells: Epigenetics and plasticity.
Adv Immunol. 124:249–273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jankovic J, Chen S and Le WD: The role of
Nurr1 in the development of dopaminergic neurons and Parkinson's
disease. Prog Neurobiol. 77:128–138. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Le W, Pan T, Huang M, Xu P, Xie W, Zhu W,
Zhang X, Deng H and Jankovic J: Decreased NURR1 gene expression in
patients with Parkinson's disease. J Neurol Sci. 273:29–33. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan Y, Tan X, Wu X, Shao B, Wu X, Cao J,
Xu J, Jin W, Li L, Xu W, et al: Involvement of early growth
response-2 (Egr-2) in lipopolysaccharide-induced neuroinflammation.
J Mol Histol. 44:249–257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Umetani N, Mori T, Koyanagi K, Shinozaki
M, Kim J, Giuliano AE and Hoon DS: Aberrant hypermethylation of ID4
gene promoter region increases risk of lymph node metastasis in T1
breast cancer. Oncogene. 24:4721–4727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peddada S, Yasui DH and LaSalle JM:
Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are
neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum
Mol Genet. 15:2003–2014. 2006. View Article : Google Scholar : PubMed/NCBI
|