Introduction
Chronic obstructive pulmonary disease (COPD) is
characterized by persistent airflow limitation, which is usually
progressive and typically involves clinical or pathological
presentations (chronic bronchitis, emphysema and small airway
disease) (1). Accumulating
evidence over the past decade has demonstrated that the pathology
of COPD, in addition to bronchoconstriction, may be attributed to
inflammation of the airways (2).
Inflammation occurring in airway epithelium is a defense mechanism
to remove the injurious stimuli, which ensures the healing of
tissues and cells. However, prolonged inflammation often causes
progressive damage to the airway structure and function.
Clara cell protein (CC16), an anti-inflammatory
protein secreted by epithelial Clara cells of the airways, is
involved in the development of airway inflammatory diseases,
including COPD and asthma (3).
Reduced levels of CC16 in the bronchial epithelium of COPD patients
and reduction of CC16-positive epithelial cells in the small
airways of asthmatics contribute to aggravation of inflammatory
responses in chronic lung inflammation (4,5).
Induction of CC16 expression by gene transfection inhibits
interleukin (IL)-1β-induced IL-8 expression in BEAS-2B bronchial
epithelial cells by suppressing the transcriptional activity of
nuclear factor (NF)-κB (6). Our
previous study demonstrated that recombinant rat CC16 suppressed
lipopolysaccharide (LPS)-mediated inflammatory matrix
metalloproteinase (MMP)-9 production through inactivation of
nuclear factor κB (NF-κB) and p38 mitogen-activated protein kinase
signaling pathways in rat tracheal epithelial (RTE) cells (7). The anti-inflammatory properties of
CC16 may render it a useful strategy for treatment of inflammatory
respiratory disorders; This may be verified by the elucidation of
the underlying mechanism via protein profile analysis of
CC16-treated cells. Proteomics is an efficient method to identify
the associated proteins that are downregulated or overexpressed in
complex biological processes.
Mass spectrometry (MS)-based proteomics technologies
are powerful tools used for large-scale protein identification and
quantitation (8). Among these
techniques, label-free shotgun proteomics is highly effective for
the identification of peptides and, subsequently, to obtain a
global protein profile of a sample. Label-free shotgun proteomics
provides a unique opportunity to measure peptides present in a
sample and subsequently determine the abundance of the proteins
across various samples (9).
Notably, label-free shotgun proteomics is suitable for applications
in complex biological systems and generates faster, cleaner and
simpler results (10).
Consequently, numerous researchers are employing label-free shotgun
proteomics techniques for discovery studies (11,12).
In the present study, label-free quantitative shotgun proteomics
was combined with MS to compare the protein expression profiles in
RTE cells cultured in the absence or presence of LPS and
recombinant CC16 (rCC16), to examine the molecular mechanisms
underlying the anti-inflammatory action of rCC16.
Materials and methods
Cell culture and drug treatment
RTE cells were purchased from the Cell Culture
Center of the Chinese Academy of Medical Sciences (Beijing, China)
and cultured in Minimal Essential Medium with Earle's Balanced
Salts (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 20% fetal calf serum (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 µg/ml
streptomycin in a 5% CO2 humidified atmosphere at 37°C. rCC16 was
prepared as previously described (7) and stored at −70°C until use.
RTE cells were cultured in 12.5-cm diameter dishes
at 1×108 cells/dish, and divided into three groups. The
cells were washed with phosphate-buffered saline (PBS) and cultured
in serum-free media (control group), serum-free media supplemented
with 0.1 µg/ml LPS (Sigma-Aldrich; Merck Millipore, Darmstadt,
Germany) for 24 h (LPS treatment group) or pretreated with 2.0
µg/ml rCC16 in serum-free media for 2 h prior to 0.1 µg/ml LPS
treatment for a further 24 h (rCC16 treatment group) (7).
Sample collection and protein
extraction
Following drug treatment, RTE cells were trypsinized
with 0.25% trypsin-EDTA (Thermo Fisher Scientific, Inc.) and
cellular proteins were extracted according to standard protocols
(13). Briefly, all samples were
resuspended in 50 µl PBS, transferred to a 1.5-ml screw-capped tube
and centrifuged at 10,000 × g for 30 min at 4°C. Next, 100
µl lysis buffer (7 M urea and 2 M thiourea) was added into each of
the samples, which were then sonicated to extract total proteins.
Proteins were precipitated with trichloroacetic acid for 30 min on
ice and centrifuged at 40,000 × g for 30 min. Protein
concentrations were determined using the Qubit Protein assay kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol.
The protein extracts were diluted with 50 mM
NH4HCO3 to a final concentration 0.5 mg/ml.
Following the addition of 100 mmol/l dithiothreitol to a final
concentration of 10 mmol/l, the protein fractions were mixed at
56°C for 60 min, diluted 10X with 250 mmol/l 2-iodoacetamide and
incubated in the dark for 60 min. Finally, the samples were
digested with trypsin (substrate to enzyme mass to mass ratio,
50:1) at 37°C for 12 h. Digested supernatant fractions were stored
at −80°C prior to MS analysis.
Liquid chromatography (LC)-tandem
MS/MS analysis
The digested peptide mixtures were pressure-loaded
onto a fused silica capillary column packed with 3-µm dionex C18
material [reversed phase (RP); Phenomenex, Torrance, CA, USA]. The
RP sections of 100 Å were 15 cm long, and the column was washed
with buffer A (water; 0.1% formic acid) and buffer B (can; 0.1%
formic acid). Following desalting, a 5-mm, 300-µm C18 capture tip
was placed in line with an Agilent 1100 quaternary HPLC (Agilent
Technologies, Inc., Santa Clara, CA, USA) and analyzed using a
12-step separation.
The first step consisted of a 5-min gradient from 0
to 2% buffer B, followed by a 45-min gradient to 40% buffer B.
Next, the buffer B flowed by 3-min gradient from 40 to 80% and
10-min at 80% buffer B. Following a 2-min buffer B gradient from 80
to 2%, ~20 µg tryptic peptide mixture was loaded onto the column
and separated at a flow rate of 2 µl/min using a linear gradient.
As peptides were eluted from the microcapillary column, they were
electrosprayed directly into a micrOTOF-Q II™ mass spectrometer
(Bruker Scientific Technology Co., Ltd., Beijing, China) with the
application of a distal 180°C source temperature. The mass
spectrometer was operated in the MS/MS (auto) mode. Survey MS scans
were acquired in the time of flight (TOF)-Q II with the resolution
set to a value of 20,000. Each survey scan (50~2,500) was followed
by five data-dependent tandem MS/MS scans at 2 Hz normalized scan
speed.
Bioinformatics analysis
Tandem mass spectra were searched against the mascot
local host rat protein database version 2.1 (Matrix Science, Inc.,
Boston, MA, USA). The search results were then filtered using a
cutoff of 1% as the peptide false identification rate. Peptides
with a Z score <4 or Delta-Mass >5 ppm were rejected. The
minimum number of peptides to identify a protein was set to 1. The
default parameters for the quantification software, ProfileAnalysis
software version 2.0, were used throughout the analysis (Bruker
Scientific Technology Co., Ltd.).
Bioinformatics analysis was performed to categorize
proteins based on the biological process, cellular component and
molecular function using annotations in Protein Analysis Through
Evolutionary Relationships (PANTHER) database version 6.1
(www.pantherdb.org) (14), which is in compliance with gene
ontology (GO) standards. Signaling pathway analysis was performed
with the tools on the Kyoto Encyclopedia of Genes and Genome (KEGG)
database (www.genome.jp/kegg/pathway.html).
The identified proteins in the various groups were
analyzed for their molecular function, biological process or
pathway terms in PANTHER using the binomial test (15). Protein-protein interactions were
obtained from the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database version 9.0 (string-db.org/), which contains known and predicted
physical and functional protein-protein interactions (16). STRING in protein mode was used, and
only interactions based on experimental protein-protein
interactions and curated databases with confidence levels >0.5-
were retained.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To verify the differential patterns of protein
expression obtained by LC-MS/MS analysis from cells treated with
rCC16 and LPS or LPS only, or untreated control cells, total RNA
was extracted from each of the groups of cells using
TRIzol® reagent (CWBIO, Beijing, China). cDNA was
synthesized using the SuperRT cDNA Synthesis Kit (cat. no. CW0741M;
CWBIO, Beijing, China). Briefly, 20 µl reverse transcription
mixture containing 4 µl deoxynucleotide triphosphate (dNTP) mix
(2.5 mM each dNTP), 2 µl primer mix, 4 µl SuperRT buffer (5X), 1 µl
SuperRT (200 U/µl), 1 µg RNA template and RNase-free water was
prepared. The reaction conditions were as follows: Incubation at
42°C for 50 min followed by 85°C for 5 min using the PTC-100
Peltier Thermal Cycler (MJ Research, Inc., Waltham, MA, USA). qPCR
was performed using an Applied Biosystems® Real-Time PCR
Instrument (Thermo Fisher Scientific, Inc.) and a SYBR®
Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's protocol. qPCR was conducted in a
20 µl reaction mixture containing 1 µl cDNA under the following
conditions: Denaturation at 95°C for 1 min, followed by 40 cycles
of denaturation at 95°C for 10 sec and annealing at 60°C for 40 sec
in triplicates. The 2-ΔΔCq method (17) was used to calculate the relative
levels of target gene expression, and GAPDH expression was used as
an internal control. The sequences of the primers used for qPCR are
listed in Table I.
 | Table I.Quantitative profiles of the
differentially expressed proteins involved in label-free
quantitative proteomics analysis and primer sequences. |
Table I.
Quantitative profiles of the
differentially expressed proteins involved in label-free
quantitative proteomics analysis and primer sequences.
|
|
|
|
|
| Sequences
(5′-3′) |
|---|
|
|
|
|
|
|
|
|---|
| NCBI number | Protein | Unit peptides | Protein ratio
LPS:RTE | Protein ratio
rCC16+LPS:LPS | Forward | Reverse |
|---|
| gi|6756041 | Tyrosine
3-monooxygenase | 8 | 1.961 | 0.854 |
CGAGCGATACGACGAAAT |
GCTGATTATTCTCCAGGATGC |
| gi|206440 | Statin-related
protein | 5 | 0.540 | 1.384 |
GAAGAATGTGTCTGTCAAGGAC |
GGGTGGTTCAGGATGATAAC |
| gi|40849846 | UDP
glycosyltransferase 1 family polypeptide A8 | 4 | 0.813 | 3.431 |
ATGGTCTACATTGGTGGGA |
CGCTTTCTTCTCTGGAATCT |
| gi|13928704 | Myosin heavy chain
10, non-muscle | 4 | 5.167 | 0.539 |
CAGATTCCTCTCCAACGG |
GCAGCACTGAAGACACGA |
| gi|34852966 | Elongation factor
1-α-1 | 3 | 0.533 | 1.617 |
TTGTTGCTGCTGGTGTTG |
TGGTGACTCAGTGAAATCCA |
| gi|34879484 | Actin-related
protein 3 homolog | 4 | 1.517 | 0.648 |
GCATCCTGGACCTCAAGA |
CAGCGATTGGAATGTGTTT |
| gi|34862480 | Caspase recruitment
domain protein 12 | 4 | 0.826 | 1.782 |
CCACGGAGGATGAGCAGTA |
GGAGGTTCTTCAGATTACCCA |
| gi|34870011 | DNA-dependent
protein kinase catalytic subunit | 3 | 0.709 | 1.304 |
AGTGTTAGGTGAGGTTCATCCT |
CAGTCCCTTTAGACAGCCA |
| gi|34852713 | ATPase,
H+-transporting, V1 subunit A, isoform 1 | 1 | 0.793 | 1.368 |
GAGAAGCCTCCATTTACACTG |
GGTATGCTGGGTATCCACTAA |
| gi|34855075 | Putative
adenosylhomocysteinase 3 | 2 | 0.613 | 1.417 |
CACTAAGGAATGGCTGGAGTT |
CATCAACCTCAGAATCGCA |
| gi|57708 | Acidic ribosomal
phosphoprotein P0 | 4 | 1.326 | 0.741 |
GACTACACTTTCCCACTGGC |
TCCTCTGACTCTTCCTTTGC |
|
| GAPDH |
|
|
|
GTGCCAGCCTCGTCTCATAG |
CTTTGTCACAAGAGAAGGCAG |
Statistical analysis
Data are presented as mean ± standard deviation for
three independent experiments. Comparisons between groups were
performed with the Student's t-test. Statistical analyses
were performed in GraphPad Prism software version 5 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Proteomic analysis
Protein samples obtained from the LPS-treated, rCC16
+ LPS-treated and control RTE cells were subjected to quantitative
proteomic analysis using label-free shotgun proteomics involving RP
fractionation and high-resolution Fourier transform MS. The
resulting mass spectra were searched against the rat RefSeq protein
database of 25,050 protein sequence inputs using the
ProfileAnalysis search algorithm. In total, 613 non-redundant
proteins were identified based on the identification of one or more
unique peptides, and 122 of these proteins were revealed to be
differentially expressed in the three treatment groups. Among the
differentially expressed proteins, 87 were upregulated and 23
downregulated in LPS-treated RTE cells compared with control cells;
and 75 were upregulated and 17 downregulated in cells treated with
LPS + rCC16 compared with those treated with LPS alone.
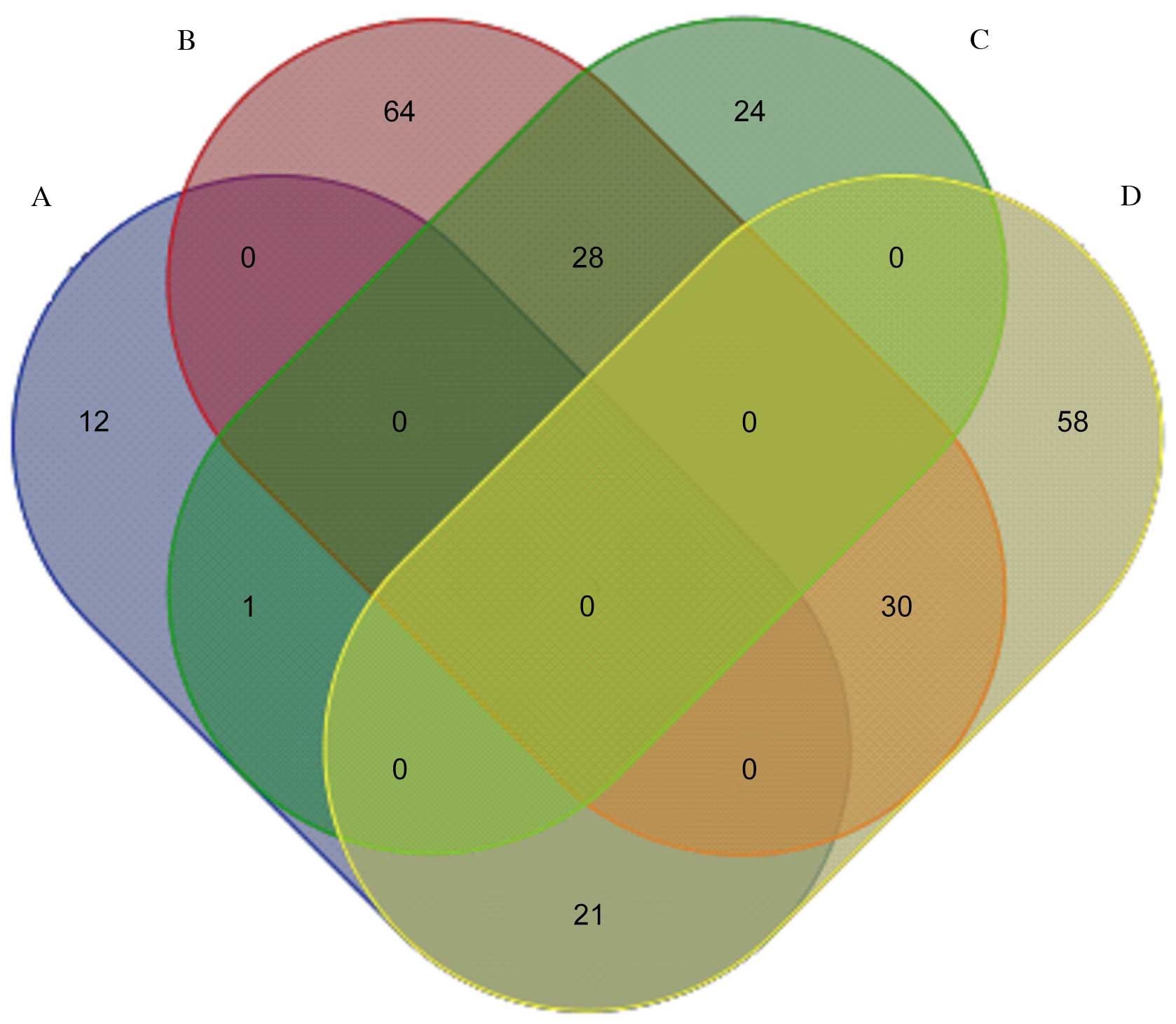
Fig. 1 presents a
pairwise comparison of the identified proteins obtained from
LPS-treated, rCC16 + LPS-treated and control RTE cells. A total of
28 proteins highly expressed in the LPS-treated group were
downregulated in the rCC16 + LPS-treated group, and 21 proteins
with low expression in the LPS-treated group were upregulated in
the rCC16 + LPS-treated group. Of these 49 proteins, seven were
downregulated in LPS-treated RTE cells, but upregulated in rCC16 +
LPS-treated RTE cells, including uridine diphosphate (UDP)
glycosyltransferase 1 family polypeptide A8 (UGT1A8),
statin-related protein, elongation factor 1-α 1 (EF-1-α-1),
adenosine triphosphatase (ATPase, H+-transporting, V1
subunit A, isoform 1; Atp6v1a1), DNA-dependent protein kinase
catalytic subunit (DNA-PKcs), putative adenosylhomocysteinase
(SAHH) 3 and caspase recruitment domain (CARD) protein 12. In
addition, five proteins were observed to be upregulated in
LPS-treated RTE cells and downregulated in rCC16 + LPS-treated RTE
cells: Matrix metalloproteinase 9 (MMP-9), actin-related protein 3
homolog (Arp3), acidic ribosomal phosphoprotein P0, myosin heavy
chain (MHC) type 10 (non-muscle) and tyrosine 3-monooxygenase
(Fig. 2).
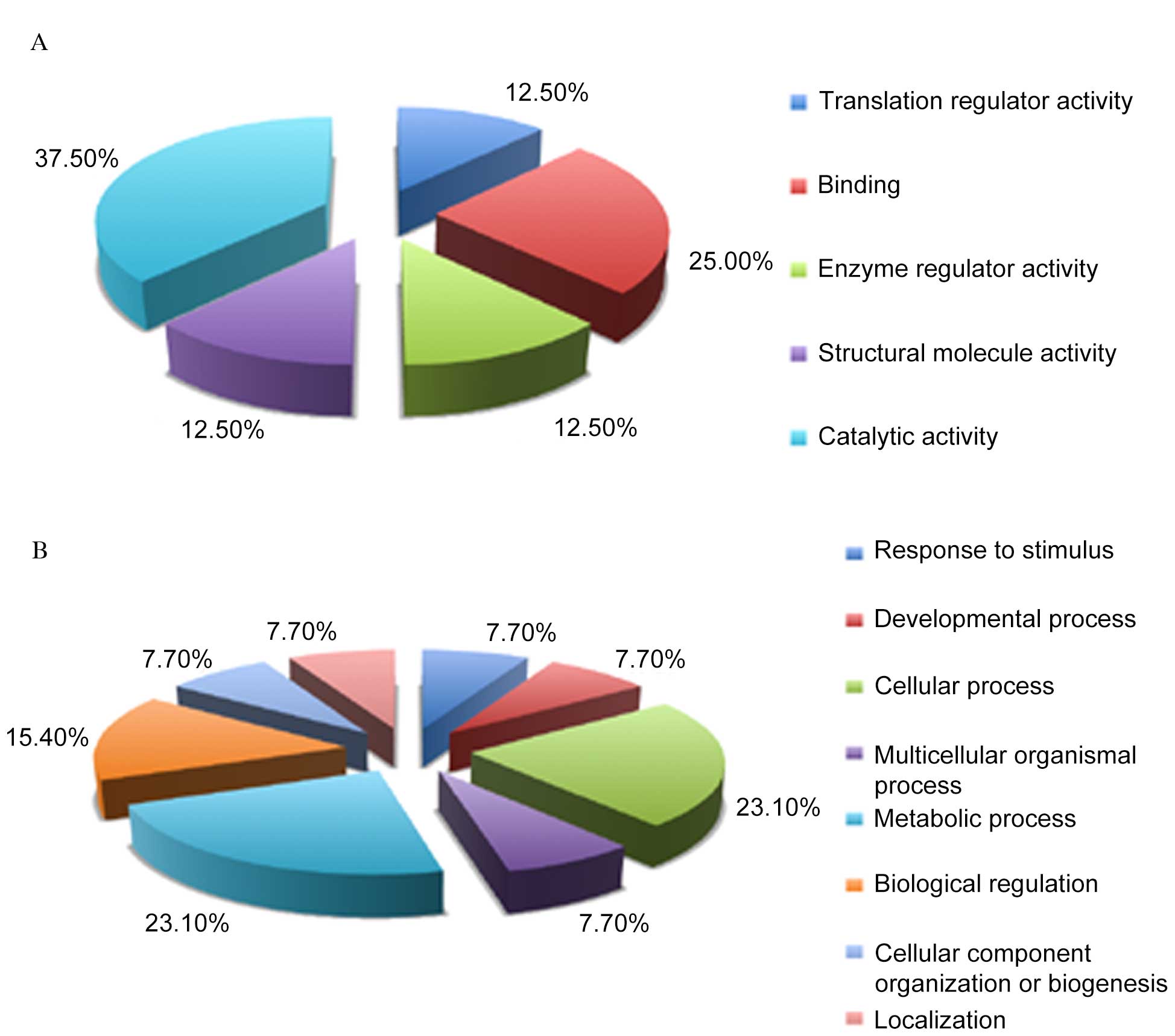
Categorization of the 12
differentially expressed proteins based on GO annotation
Differentially expressed proteins were categorized
as those that were downregulated in LPS-treated RTE cells, but
upregulated in rCC16+LPS-treated RTE cells, or those which were
upregulated in LPS-treated RTE cells but downregulated in
rCC16+LPS-treated RTE cells (Fig.
2). To further understand these differentially expressed
proteins, a GO analysis was performed with the PANTHER
classification system to determine the molecular functions and
associated biological processes of these 12 proteins. Based on
their molecular functions, these differentially expressed proteins
may be classified into five groups: Translation regulator activity
(12.5%), binding (25%), enzyme regular activity (12.5%), structural
molecule activity (12.5%) and catalytic activity (37.5%; Fig. 3A). Accordingly, these proteins were
classified into eight groups of biological processes: Response to
stimulus (7.7%), developmental process (7.7%), cellular process
(23.1%), multicellular organismal process (7.7%), metabolic process
(23.1%), biological regulation (15.4%), cellular component
organization or biogenesis (7.7%) and localization (7.7%; Fig. 3B). These data suggest the possible
molecular components and pathways involved in the transcription and
energy metabolism activities of LPS and rCC16-responsive RTE
cells.
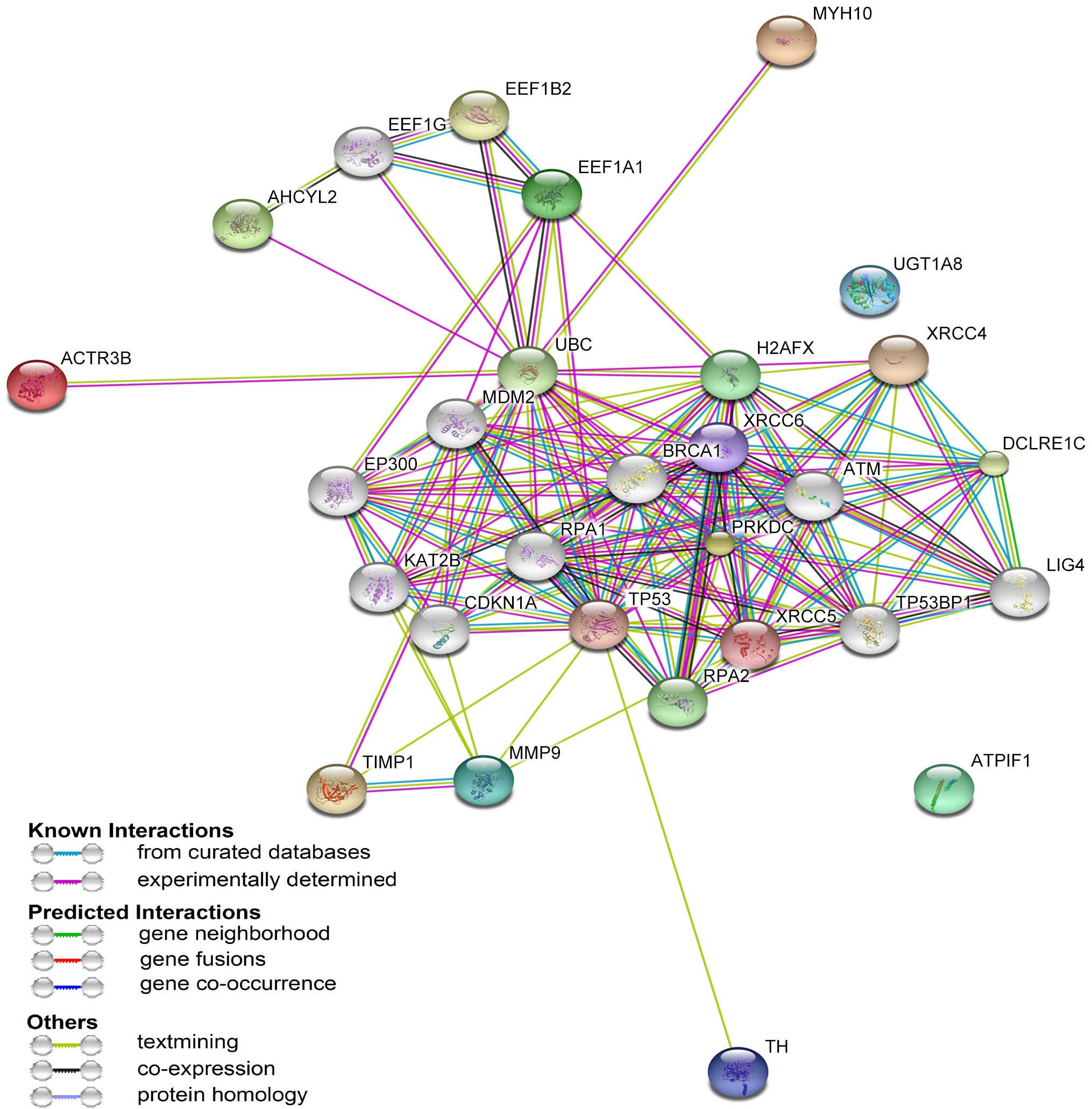
STRING protein-protein analysis of
differentially expressed proteins
STRING is a database program for protein-protein
interaction analysis to generate a network of interactions from a
variety of sources, including various interaction databases, text
mining, genetic interactions and shared pathway interactions. To
further characterize the functions associated with the marked
differences in the expression of the 12 proteins, these proteins
were analyzed using STRING software version 9.0- Known and
Predicted Protein-Protein Interactions. As presented in Fig. 4, the 12 differentially expressed
proteins were connected with numerous proteins associated with
inflammation, including tumor protein p53 (TP53) and ataxia
telangiectasia-mutated kinase (ATM) (18). These data indicate that an
interacting network of proteins is associated with the molecular
mechanisms underlying the anti-inflammatory activity of CC16.
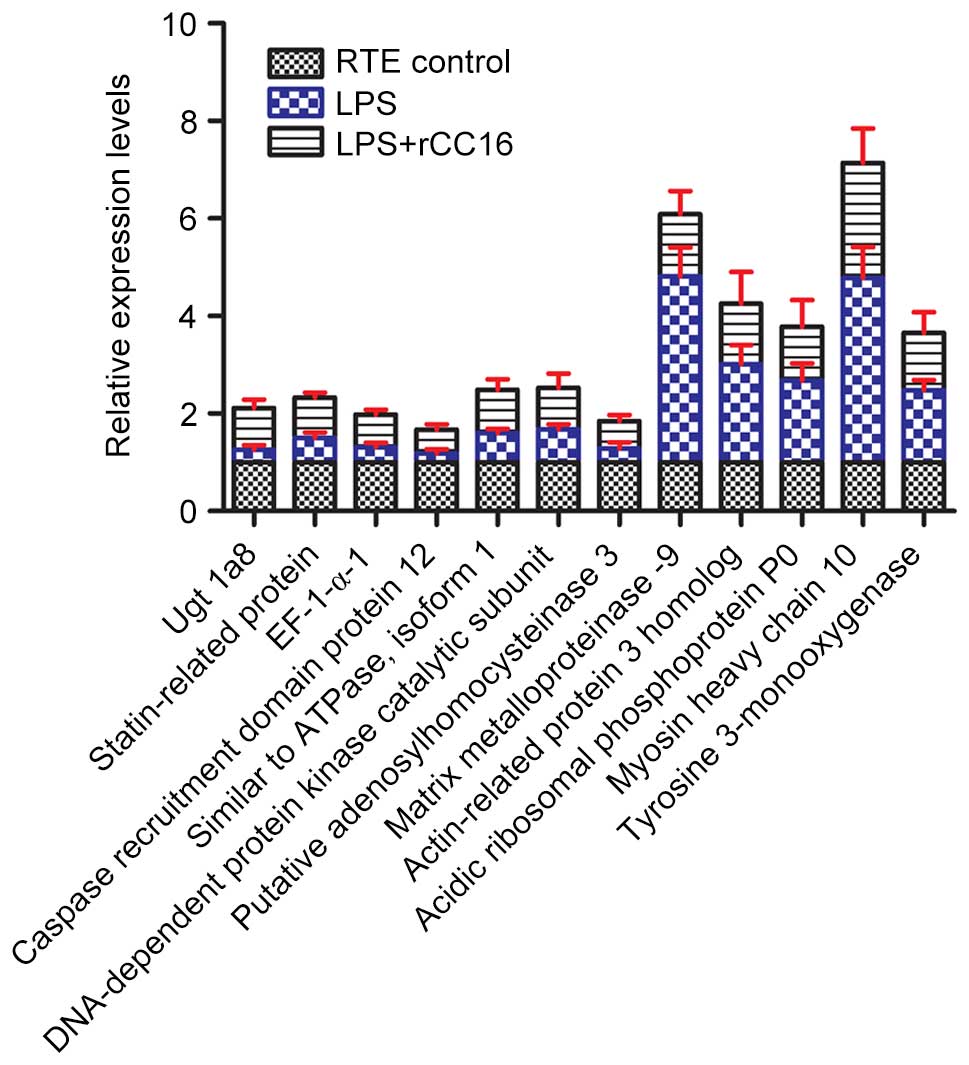
RT-qPCR analysis of differentially
expressed proteins
To further determine the differential expression
patterns of these proteins in RTE cells cultured in the absence or
presence of LPS and rCC16, RT-qPCR was performed for
transcriptional analyses of the differentially expressed proteins
at the mRNA level, (with the exception of MMP-9, the expression of
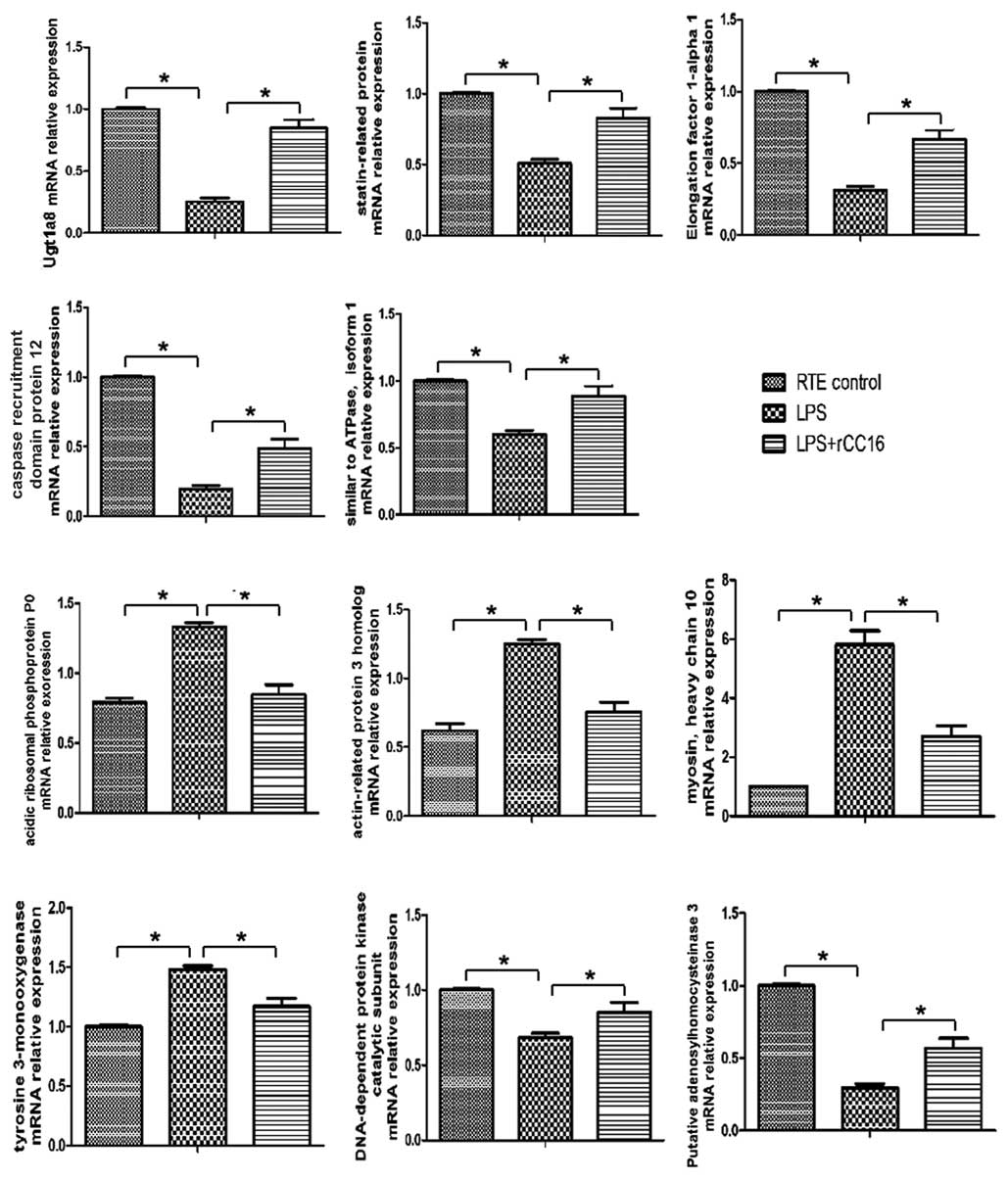
which has been previously described by our laboratory) (7). As presented in Fig. 5, all 11 genes exhibited similar
patterns of expression to their corresponding proteins, including
tyrosine 3-monooxygenase, statin-related protein, UGT1A8, MHC type
10, EF-1-α-1, Arp3, CARD protein 12, acidic ribosomal
phosphoprotein P0, DNA-PKcs, Atp6v1a1 and putative SAHH 3. Certain
variations may exist between the levels of qPCR-determined gene
expression and their proteins, including, for example, EF-1-α-1,
CARD protein 12 and DNA-PKcs. These variations between protein and
mRNA expression levels may be due to alterations in the regulation
of protein translation and degradation (19). In addition, specific processes that
occur prior to translation may influence the mRNA translation
efficiency. For example, the poly (A) tail length can affect
transcript stability and therefore the abundance of a specific mRNA
sequence (20).
Discussion
CC16 is an anti-inflammatory protein involved in the
development of airway inflammatory diseases, including COPD. The
present study has identified a differential pattern of protein
expressions in RTE cells cultured in the presence or absence of LPS
and rCC16 using label-free shotgun proteomics, and verified this at
the mRNA level by RT-qPCR. The differentially expressed proteins of
various molecular functions and associated biological processes are
connected with numerous proteins involved in inflammation in a
unique protein-protein interacting network. The present study
provides a basis for the further understanding of the role of CC16
and its anti-inflammation activity in the development of airway
diseases, including COPD.
Inflammation is one of mechanisms underlying the
development of COPD. Therefore, reducing the inflammatory response
is regarded as the primary interference measure for COPD patients.
It is well-known that CC16 is a secretory protein with
anti-inflammatory characteristics (21). Reduced levels of CC16 in the
bronchial epithelium or sputum supernatants of COPD patients may be
a contributing factor in airway inflammations (4,22).
Consequently, administration of CC16 may be an effective treatment
for COPD. Our previous study demonstrated that rCC16 inhibited the
expression of COPD-contributing MMP-9 in LPS-treated RTE cells via
the attenuation of NF-κB activation (7). The present study has identified a
network of proteins involved in the mechanisms underlying the
anti-inflammatory activity of rCC16.
For example, exposure of RTE cells to LPS increased
MHC type 10 expression that was abolished by further rCC16
treatment. Systemic muscle inflammation is known to be a major
factor in COPD, particularly following exacerbations (23). The changes of MHC perturb the
function and structure of muscle (24) and types IIx, IIa and I of MHC
isoforms are expressed in human skeletal muscle (25). The downregulation of the type I
isoform, and upregulation of the type IIa isoform, has been
demonstrated in patients with COPD (26). In addition, reduction of MHC IIa
fibers has been reported in LPS-treated fetal membranes
(chorioamnionitis) (27). While
the reason for rCC16-reduced MHC type 10 expression remains
unclear, carbonylation of MHC is well known to be degraded by the
ubiquitin-proteasome pathway (24). Therefore, rCC16 may be involved in
the process of carbonylation of MHC type 10. In addition, the
cytoskeletal protein Arp3 is important in airway smooth muscle
contraction as a subunit of the Arp2/3 complex that initiates the
branching of actin filaments (28). A previous study revealed that the
carbachol-stimulated cytoskeletal recruitment of Arp3 was
upregulated at short muscle lengths and downregulated at long
muscle lengths (29). In the
present study, Arp3 was increased in LPS-treated RTE cells, and
decreased by further rCC16 treatment. Similarly, rCC16 reduced the
levels of LPS-induced expression of tyrosine 3-monooxygenase, which
encodes the protein, 14-3-3 η. The latter belongs to the 14-3-3
family of proteins that mediate signal transduction and are
required for mitotic progression. Upregulation of 14-3-3 η has been
evident in sporadic Creutzfeldt-Jakob disease, joint inflammation
and head-and-neck squamous cell carcinoma (30–32).
In accordance with this, the results of the present study
demonstrated that rCC16 enhanced the expression of DNA-PKcs, which
is crucial for DNA double-strand break repair and normal cell cycle
progression (33). High-dose CC16
has been revealed to delay the growth of immortalized bronchial
epithelial cells and inhibit platelet-derived growth factor-induced
airway smooth muscle cell proliferation (34,35).
CC16 knockdown in vivo results in augmented inflammation due
to polymorphonuclear leukocyte recruitment to the air space in mice
exposed to LPS (36). Therefore,
rCC16 may target the signaling molecules involved in cytoskeletal
recruitment and mitotic progressions of bronchial epithelial cells
and inflammatory cells.
In the present study, a number of proteins were
identified that were downregulated by LPS and upregulated by rCC16
in RTE cells, including statin-related protein, CARD protein 12,
eukaryotic translation EF-1-α-1, SAHH and acidic ribosomal
phosphoprotein P0. Statin-related protein is a 57 kDa nuclear
protein present in non-dividing cells (37) and involved in the process of
myopathy (38). Although the
functions of statin-related protein remain to be fully elucidated,
an anti-inflammatory activity via tumor necrosis factor-α reduction
has been reported for the statin-related rosuvastatin protein
(39). CARD proteins are scaffold
proteins that function as key regulators of NF-κB signaling by
providing a link between membrane receptors and NF-κB
transcriptional subunits (40).
CARD11 and CARD14-mediated alterations in NF-κB signaling are
thought to be an early event in the development of cutaneous
squamous cell carcinoma and psoriasis, respectively (40,41).
CARD9 in macrophages mediates necrotic smooth muscle cell-induced
inflammation by activating NF-κB (42). Other CARD proteins, including
CARD16, CARD17 and CARD18, negatively regulate inflammation by
inhibiting the activation of caspase-1, which may induce the
expression of IL-1β without altering NF-κB signaling (43,44).
EF-1-α-1 is a ubiquitously expressed GTPase that couples the
hydrolysis of guanosine triphosphate (GTP) to guanosine diphosphate
(GDP), with the delivery of aminoacyl transfer RNAs to the ribosome
during protein translation (45,46).
In addition, it is involved in cell proliferation, cytoskeletal
organization and protein degradation (47–49).
High levels of EF-1-α-1 have been associated with survival
(50) and downregulation of
EF-1-α-1 expression resulted in cell death (51). SAHH 3 is an
NAD+-dependent tetrameric enzyme that catalyzes the
breakdown of S-adenosylhomocysteine to adenosine and homocysteine,
and is therefore important in cell growth and regulation of gene
expression (52). Loss of SAHH
function may result in global inhibition of cellular
methyltransferase enzymes due to S-adenosylhomocysteine
accumulation (52). Acidic
ribosomal phosphoprotein P0 is a structural component of the
ribosome of all organisms. It interacts with trans-acting factor
Mrt4 during ribosome assembly and is important for protein
synthesis (53). These data
together suggest that rCC16 may function as an anti-inflammatory
effector by altering the expression of tyrosine 3-monooxygenase,
statin-related protein, CARD, eEF1A1, SAHH and acidic ribosomal
phosphoprotein P0. The effect of CC16 on these proteins requires
further study.
In addition, the present study demonstrated that
rCC16 promoted expression of Atp6v1a1 and UGT1A8. Atp6v1a1 belongs
to the respiratory electron transport chain (ETC) of oxidative
phosphorylation in energy metabolism. Downregulated expression of
Atp6v1a1 inhibits the efficiency of ETC and ATP synthesis (54). This is consistent with the
knowledge that the disorder of skeletal muscle energy metabolism is
important in COPD, and that systemic inflammation alters the state
of energy metabolism in peripheral muscles, causing them to
dysfunction (55). While UGTs are
well known for their drug-metabolizing and detoxification
potentials in drug toxicity (56),
UGT1A8 has in addition been associated with metabolism of
mycophenolic acid (57),
raloxifene (58) and piceatannol,
which is a dietary polyphenol present in grapes and wine and may
possess anticancer and anti-inflammatory activities (59). Therefore, the anti-inflammatory
action of rCC16 may be associated with the activities of skeletal
muscle energy metabolism and UGT1A8.
The differentially expressed proteins of diverse
functions and biological processes were linked to numerous other
inflammation-associated proteins, including TP53 and ATM by
protein-protein interaction analysis using STRING software.
Expression of inflammatory cytokine genes may be directly induced
by TP53- and ATM-dependent mechanisms upon radiation exposure in
human monocytes and macrophages (18). In addition, activation of
pro-inflammatory pathways during COPD involves enhanced expression
of TP53 and TP21 in lung fibroblasts (60). However, ATM deficiency in mice
delays the acute inflammatory response following myocardial
infarction (61). While TP53 and
ATM pathways are always integrated with NF-κB signaling in
inflammation (62–64), their role in the anti-inflammatory
effect of CC16 remains to be determined.
In conclusion, the results of the present study
demonstrate that label-free differential proteomics is able to
reveal the protein profiles of the anti-inflammatory action of CC16
on RTE cells. The differential proteins and their interacting
network of proteins identified in the present study may provide a
basis for elucidating the molecular mechanisms underlying the
anti-inflammatory effects of CC16. Further studies into the role of
these proteins in COPD pathogenesis and its cellular and tissue
models may lead to the development of novel diagnostic and
therapeutic strategies for this disease.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81200032), the
Research Project Supported by Shanxi Scholarship Council of China
(grant no. 2015-101), the Fund Program for the Scientific
Activities of Selected Returned Overseas Professionals in Shanxi
Province (grant no. 2016-097) and the Research Fund for Doctoral
Program of Shanxi Medical University (grant no. 03201539).
References
|
1
|
Vestbo J, Hurd SS, Agustí AG, Jones PW,
Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ,
Nishimura M, et al: Global strategy for the diagnosis, management,
and prevention of chronic obstructive pulmonary disease: GOLD
executive summary. Am J Respir Crit Care Med. 187:347–365. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gosens R, Zaagsma J, Meurs H and Halayko
AJ: Muscarinic receptor signaling in the pathophysiology of asthma
and COPD. Respir Res. 7:732006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen LC, Zhang Z, Myers AC and Huang SK:
Cutting edge: Altered pulmonary eosinophilic inflammation in mice
deficient for Clara cell secretory 10-kDa protein. J Immunol.
167:3025–3028. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pilette C, Godding V, Kiss R, Delos M,
Verbeken E, Decaestecker C, De Paepe K, Vaerman JP, Decramer M and
Sibille Y: Reduced epithelial expression of secretory component in
small airways correlates with airflow obstruction in chronic
obstructive pulmonary disease. Am J Respir Crit Care Med.
163:185–194. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shijubo N, Itoh Y, Yamaguchi T, Imada A,
Hirasawa M, Yamada T, Kawai T and Abe S: Clara cell
protein-positive epithelial cells are reduced in small airways of
asthmatics. Am J Respir Crit Care Med. 160:930–933. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Long XB, Hu S, Wang N, Zhen HT, Cui YH and
Liu Z: Clara cell 10-kDa protein gene transfection inhibits NF-κB
activity in airway epithelial cells. PLoS One. 7:e359602012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pang M, Wang H, Bai JZ, Cao D, Jiang Y,
Zhang C, Liu Z, Zhang X, Hu X, Xu J and Du Y: Recombinant rat CC16
protein inhibits LPS-induced MMP-9 expression via NF-κB pathway in
rat tracheal epithelial cells. Exp Biol Med (Maywood).
240:1266–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cravatt BF, Simon GM and Yates JR III: The
biological impact of mass-spectrometry-based proteomics. Nature.
450:991–1000. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Domon B and Aebersold R: Options and
considerations when selecting a quantitative proteomics strategy.
Nat Biotechnol. 28:710–721. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bauer KM, Lambert PA and Hummon AB:
Comparative label-free LC-MS/MS analysis of colorectal
adenocarcinoma and metastatic cells treated with 5-fluorouracil.
Proteomics. 12:1928–1937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clough T, Thaminy S, Ragg S, Aebersold R
and Vitek O: Statistical protein quantification and significance
analysis in label-free LC-MS experiments with complex designs. BMC
Bioinformatics. 13:(Suppl 16). S62012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niehl A, Zhang ZJ, Kuiper M, Peck SC and
Heinlein M: Label-free quantitative proteomic analysis of systemic
responses to local wounding and virus infection in Arabidopsis
thaliana. J Proteome Res. 12:2491–2503. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guerrera IC, Quetier I, Fetouchi R, Moreau
F, Vauloup-Fellous C, Lekbaby B, Rousselot C, Chhuon C, Edelman A,
Lefevre M, et al: Regulation of interleukin-6 in head and neck
squamous cell carcinoma is related to papillomavirus infection. J
Proteome Res. 13:1002–1011. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thomas PD, Campbell MJ, Kejariwal A, Mi H,
Karlak B, Daverman R, Diemer K, Muruganujan A and Narechania A:
PANTHER: A library of protein families and subfamilies indexed by
function. Genome Res. 13:2129–2141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho RJ and Campbell MJ: Transcription,
genomes, function. Trends Genet. 16:409–415. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:(Database
Issue). D412–D416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Candeias SM and Testard I: The many
interactions between the innate immune system and the response to
radiation. Cancer Lett. 368:173–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Sousa Abreu R, Penalva LO, Marcotte EM
and Vogel C: Global signatures of protein and mRNA expression
levels. Mol Biosyst. 5:1512–1526. 2009.PubMed/NCBI
|
|
20
|
Lackner DH, Beilharz TH, Marguerat S, Mata
J, Watt S, Schubert F, Preiss T and Bähler J: A network of multiple
regulatory layers shapes gene expression in fission yeast. Mol
Cell. 26:145–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Irander K, Palm JP, Borres MP and Ghafouri
B: Clara cell protein in nasal lavage fluid and nasal nitric
oxide-biomarkers with anti-inflammatory properties in allergic
rhinitis. Clin Mol Allergy. 10:42012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lensmar C, Nord M, Gudmundsson GH, Roquet
A, Andersson O, Jörnvall H, Eklund A, Grunewald J and Agerberth B:
Decreased pulmonary levels of the anti-inflammatory Clara cell 16
kDa protein after induction of airway inflammation in asthmatics.
Cell Mol Life Sci. 57:976–981. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Spruit MA, Gosselink R, Troosters T,
Kasran A, Gayan-Ramirez G, Bogaerts P, Bouillon R and Decramer M:
Muscle force during an acute exacerbation in hospitalised patients
with COPD and its relationship with CXCL8 and IGF-I. Thorax.
58:752–756. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamada T, Mishima T, Sakamoto M, Sugiyama
M, Matsunaga S and Wada M: Oxidation of myosin heavy chain and
reduction in force production in hyperthyroid rat soleus. J Appl
Physiol (1985). 100:1520–1526. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Serrano AL, Pérez M, Lucía A, Chicharro
JL, Quiroz-Rothe E and Rivero JL: Immunolabelling, histochemistry
and in situ hybridisation in human skeletal muscle fibres to detect
myosin heavy chain expression at the protein and mRNA level. J
Anat. 199:329–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Maltais F, Sullivan MJ, LeBlanc P,
Schachat FH, Simard C, Blank JM and Jobin J: Altered expression of
myosin heavy chain in the vastus lateralis muscle in patients with
COPD. Eur Respir J. 13:850–854. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song Y, Karisnan K, Noble PB, Berry CA,
Lavin T, Moss TJ, Bakker AJ, Pinniger GJ and Pillow JJ: In utero
LPS exposure impairs preterm diaphragm contractility. Am J Respir
Cell Mol Biol. 49:866–874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Goley ED and Welch MD: The ARP2/3 complex:
An actin nucleator comes of age. Nat Rev Mol Cell Biol. 7:713–726.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim HR, Liu K, Roberts TJ and Hai CM:
Length-dependent modulation of cytoskeletal remodeling and
mechanical energetics in airway smooth muscle. Am J Respir Cell Mol
Biol. 44:888–897. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schubert KO, Focking M and Cotter DR:
Proteomic pathway analysis of the hippocampus in schizophrenia and
bipolar affective disorder implicates 14-3-3 signaling, aryl
hydrocarbon receptor signaling and glucose metabolism: Potential
roles in GABAergic interneuron pathology. Schizophr Res. 167:64–72.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yun J, Jeong BH, Kim HJ, Park YJ, Lee YJ,
Choi EK, Carp RI and Kim YS: A polymorphism in the YWHAH gene
encoding 14-3-3 eta that is not associated with sporadic
Creutzfeldt-Jakob disease (CJD). Mol Biol Rep. 39:3619–3625. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kilani RT, Maksymowych WP, Aitken A, Boire
G, St-Pierre Y, Li Y and Ghahary A: Detection of high levels of 2
specific isoforms of 14-3-3 proteins in synovial fluid from
patients with joint inflammation. J Rheumatol. 34:1650–1657.
2007.PubMed/NCBI
|
|
33
|
Davis AJ, Chen BP and Chen DJ: DNA-PK: A
dynamic enzyme in a versatile DSB repair pathway. DNA Repair
(Amst). 17:21–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei Y, Xu YD, Yin LM, Wang Y, Ran J, Liu
Q, Ma ZF, Liu YY and Yang YQ: Recombinant rat CC10 protein inhibits
PDGF-induced airway smooth muscle cells proliferation and
migration. Biomed Res Int. 2013:6909372013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Linnoila RI, Szabo E, DeMayo F, Witschi H,
Sabourin C and Malkinson A: The role of CC10 in pulmonary
carcinogenesis: From a marker to tumor suppression. Ann N Y Acad
Sci. 923:249–267. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Snyder JC, Reynolds SD, Hollingsworth JW,
Li Z, Kaminski N and Stripp BR: Clara cells attenuate the
inflammatory response through regulation of macrophage behavior. Am
J Respir Cell Mol Biol. 42:161–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ann DK, Wechsler A, Lin HH and Wang E:
Isoproterenol downregulation of statin-related gene expression in
the rat parotid gland. J Cell Sci. 100:641–647. 1991.PubMed/NCBI
|
|
38
|
Apostolopoulou M, Corsini A and Roden M:
The role of mitochondria in statin-induced myopathy. Eur J Clin
Invest. 45:745–754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McGuire TR, Kalil AC, Dobesh PP, Klepser
DG and Olsen KM: Anti-inflammatory effects of rosuvastatin in
healthy subjects: A prospective longitudinal study. Curr Pharm Des.
20:1156–1160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Watt SA, Purdie KJ, den Breems NY, Dimon
M, Arron ST, McHugh AT, Xue DJ, Dayal JH, Proby CM, Harwood CA, et
al: Novel CARD11 mutations in human cutaneous squamous cell
carcinoma lead to aberrant NF-κB regulation. Am J Pathol.
185:2354–2363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Harden JL, Lewis SM, Pierson KC,
Suárez-Fariñas M, Lentini T, Ortenzio FS, Zaba LC, Goldbach-Mansky
R, Bowcock AM and Lowes MA: CARD14 expression in dermal endothelial
cells in psoriasis. PLoS One. 9:e1112552014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu Y, Wang Y, Shi H, Jia L, Cheng J, Cui
W, Li H, Li P and Du J: CARD9 mediates necrotic smooth muscle
cell-induced inflammation in macrophages contributing to neointima
formation of vein grafts. Cardiovasc Res. 108:148–158. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Druilhe A, Srinivasula SM, Razmara M,
Ahmad M and Alnemri ES: Regulation of IL-1beta generation by
Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment
domain proteins. Cell Death Differ. 8:649–657. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lamkanfi M, Denecker G, Kalai M, D'hondt
K, Meeus A, Declercq W, Saelens X and Vandenabeele P: INCA, a novel
human caspase recruitment domain protein that inhibits
interleukin-1beta generation. J Biol Chem. 279:51729–51738. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hershey JW: Translational control in
mammalian cells. Annu Rev Biochem. 60:717–755. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Thornton S, Anand N, Purcell D and Lee J:
Not just for housekeeping: Protein initiation and elongation
factors in cell growth and tumorigenesis. J Mol Med (Berl).
81:536–548. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lamberti A, Caraglia M, Longo O, Marra M,
Abbruzzese A and Arcari P: The translation elongation factor 1A in
tumorigenesis, signal transduction and apoptosis: Review article.
Amino Acids. 26:443–448. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gross SR and Kinzy TG: Translation
elongation factor 1A is essential for regulation of the actin
cytoskeleton and cell morphology. Nat Struct Mol Biol. 12:772–778.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chuang SM, Chen L, Lambertson D, Anand M,
Kinzy TG and Madura K: Proteasome-mediated degradation of
cotranslationally damaged proteins involves translation elongation
factor 1A. Mol Cell Biol. 25:403–413. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lamberti A, Longo O, Marra M, Tagliaferri
P, Bismuto E, Fiengo A, Viscomi C, Budillon A, Rapp UR, Wang E, et
al: C-Raf antagonizes apoptosis induced by IFN-alpha in human lung
cancer cells by phosphorylation and increase of the intracellular
content of elongation factor 1A. Cell Death Differ. 14:952–962.
2007.PubMed/NCBI
|
|
51
|
Kobayashi Y and Yonehara S: Novel cell
death by downregulation of eEF1A1 expression in tetraploids. Cell
Death Differ. 16:139–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang Y, Kavran JM, Chen Z, Karukurichi KR,
Leahy DJ and Cole PA: Regulation of S-adenosylhomocysteine
hydrolase by lysine acetylation. J Biol Chem. 289:31361–31372.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rodríguez-Mateos M, García-Gómez JJ,
Francisco-Velilla R, Remacha M, de la Cruz J and Ballesta JP: Role
and dynamics of the ribosomal protein P0 and its related
trans-acting factor Mrt4 during ribosome assembly in Saccharomyces
cerevisiae. Nucleic Acids Res. 37:7519–7532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ji B, La Y, Gao L, Zhu H, Tian N, Zhang M,
Yang Y, Zhao X, Tang R, Ma G, et al: A comparative proteomics
analysis of rat mitochondria from the cerebral cortex and
hippocampus in response to antipsychotic medications. J Proteome
Res. 8:3633–3641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Davidsen PK, Herbert JM, Antczak P, Clarke
K, Ferrer E, Peinado VI, Gonzalez C, Roca J, Egginton S, Barberá JA
and Falciani F: A systems biology approach reveals a link between
systemic cytokines and skeletal muscle energy metabolism in a
rodent smoking model and human COPD. Genome Med. 6:592014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Meech R, Miners JO, Lewis BC and Mackenzie
PI: The glycosidation of xenobiotics and endogenous compounds:
Versatility and redundancy in the UDP glycosyltransferase
superfamily. Pharmacol Ther. 134:200–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vu D, Tellez-Corrales E, Yang J, Qazi Y,
Shah T, Naraghi R, Hutchinson IV and Min DI: Genetic polymorphisms
of UGT1A8, UGT1A9 and HNF-1α and gastrointestinal symptoms in renal
transplant recipients taking mycophenolic acid. Transpl Immunol.
29:155–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun D, Jones NR, Manni A and Lazarus P:
Characterization of raloxifene glucuronidation: Potential role of
UGT1A8 genotype on raloxifene metabolism in vivo. Cancer Prev Res
(Phila). 6:719–730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Miksits M, Maier-Salamon A, Vo TP, Sulyok
M, Schuhmacher R, Szekeres T and Jäger W: Glucuronidation of
piceatannol by human liver microsomes: Major role of UGT1A1, UGT1A8
and UGT1A10. J Pharm Pharmacol. 62:47–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
D'Anna C, Cigna D, Costanzo G, Ferraro M,
Siena L, Vitulo P, Gjomarkaj M and Pace E: Cigarette smoke alters
cell cycle and induces inflammation in lung fibroblasts. Life Sci.
126:10–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Daniel LL, Daniels CR, Harirforoosh S,
Foster CR, Singh M and Singh K: Deficiency of ataxia telangiectasia
mutated kinase delays inflammatory response in the heart following
myocardial infarction. J Am Heart Assoc. 3:e0012862014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Schuliga M: NF-kappaB signaling in chronic
inflammatory airway disease. Biomolecules. 5:1266–1283. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Pal S, Bhattacharjee A, Ali A, Mandal NC,
Mandal SC and Pal M: Chronic inflammation and cancer: Potential
chemoprevention through nuclear factor kappa B and p53 mutual
antagonism. J Inflamm (Lond). 11:232014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Osorio FG, Bárcena C, Soria-Valles C,
Ramsay AJ, de Carlos F, Cobo J, Fueyo A, Freije JM and López-Otín
C: Nuclear lamina defects cause ATM-dependent NF-κB activation and
link accelerated aging to a systemic inflammatory response. Genes
Dev. 26:2311–2324. 2012. View Article : Google Scholar : PubMed/NCBI
|