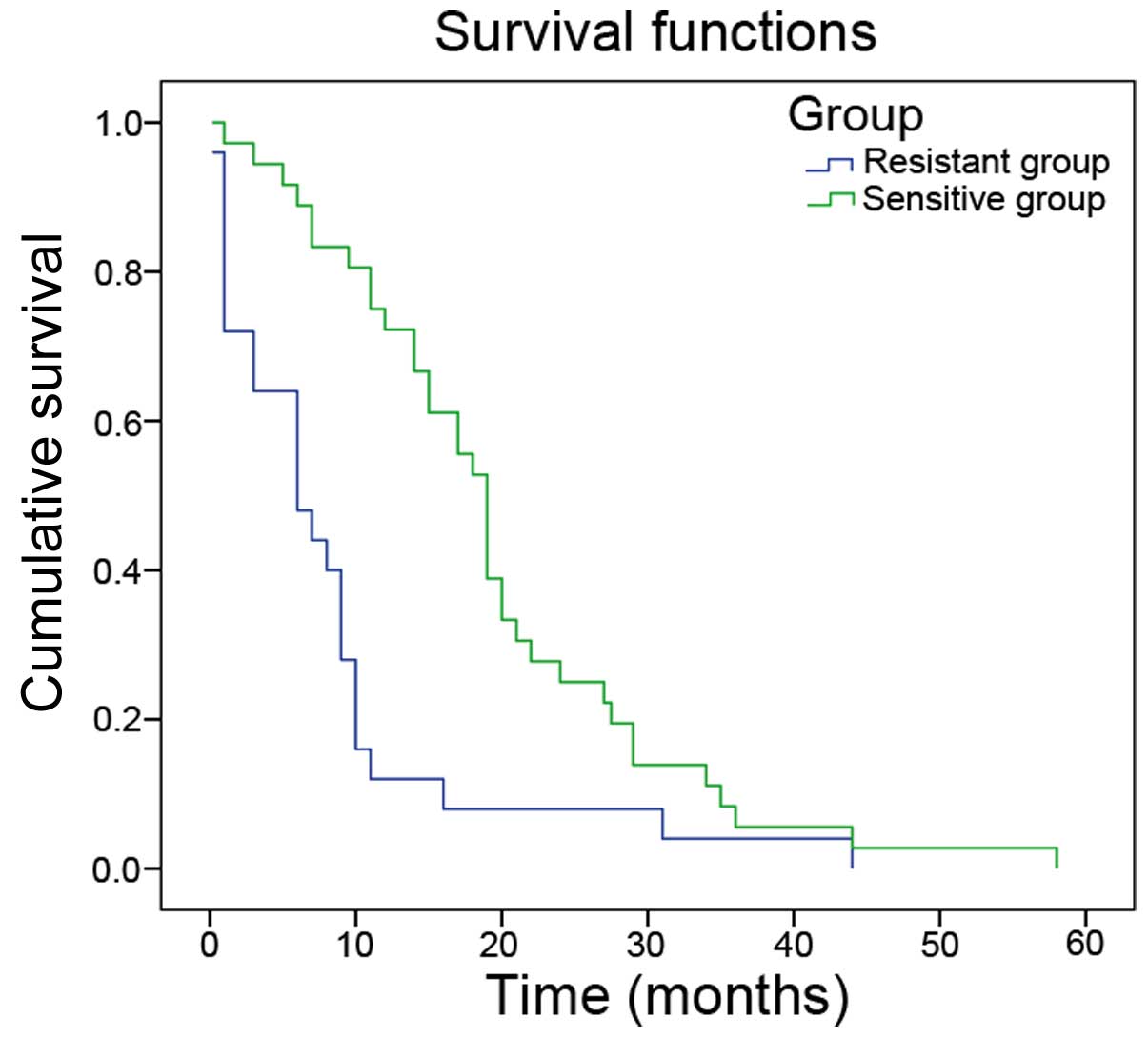
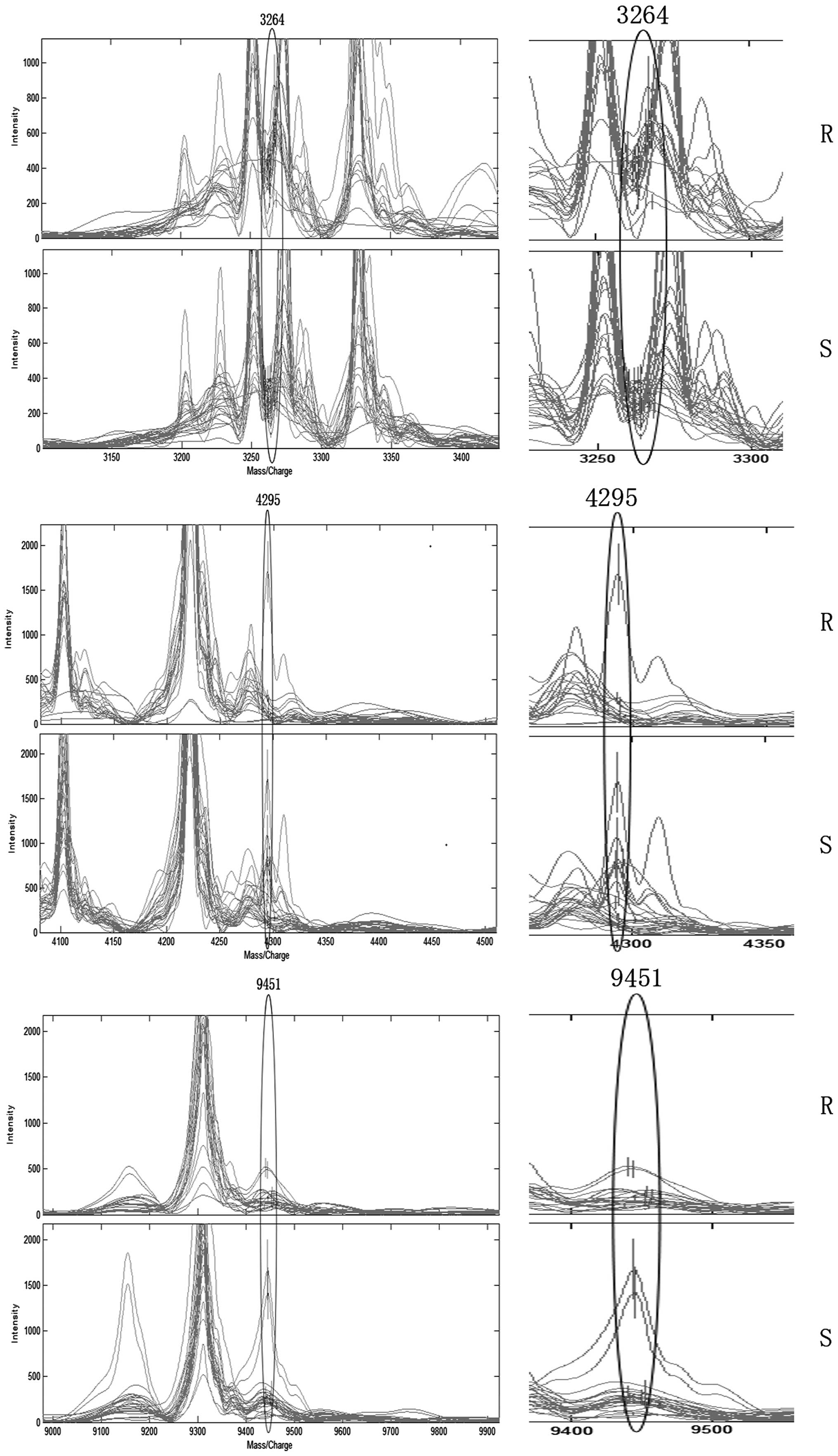
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mendelsohn J and Baselga J: Status of
epidermal growth factor receptor antagonists in the biology and
treatment of cancer. J Clin Oncol. 21:2787–2799. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukuoka M, Yano S, Giaccone G, Tamura T,
Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S,
Rischin D, et al: Multi-institutional randomized phase II trial of
gefitinib for previously treated patients with advanced
non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin
Oncol. 21:2237–2246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee DH, Han JY, Lee HG, Lee JJ, Lee EK,
Kim HY, Kim HK, Hong EK and Lee JS: Gefitinib as a first-line
therapy of advanced or metastatic adenocarcinoma of the lung in
never-smokers. Clin Cancer Res. 11:3032–3037. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Paez JG, Janne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jackman D, Pao W, Riely GJ, Engelman JA,
Kris MG, Jänne PA, Lynch T, Johnson BE and Miller VA: Clinical
definition of acquired resistance to epidermal growth factor
receptor tyrosine kinase inhibitors in non-small-cell lung cancer.
J Clin Oncol. 28:357–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hashida S, Yamamoto H, Shien K, Ohtsuka T,
Suzawa K, Maki Y, Furukawa M, Soh J, Asano H, Tsukuda K, et al:
Hsp90 inhibitor NVP-AUY922 enhances the radiation sensitivity of
lung cancer cell lines with acquired resistance to EGFR-tyrosine
kinase inhibitors. Oncol Rep. 33:1499–1504. 2015.PubMed/NCBI
|
|
8
|
Lin Y, Wang X and Jin H: EGFR-TKI
resistance in NSCLC patients: Mechanisms and strategies. Am J
Cancer Res. 4:411–435. 2014.PubMed/NCBI
|
|
9
|
Chen MC, Chen CH, Wang JC, Tsai AC, Liou
JP, Pan SL and Teng CM: The HDAC inhibitor, MPT0E028, enhances
erlotinib-induced cell death in EGFR-TKI-resistant NSCLC cells.
Cell Death Dis. 4:e8102013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Milan E, Lazzari C, Anand S, Floriani I,
Torri V, Sorlini C, Gregorc V and Bachi A: SAA1 is over-expressed
in plasma of non small cell lung cancer patients with poor outcome
after treatment with epidermal growth factor receptor
tyrosine-kinase inhibitors. J Proteomics. 76:91–101. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang JJ, Chen HJ, Yan HH, Zhang XC, Zhou
Q, Su J, Wang Z, Xu CR, Huang YS, Wang BC, et al: Clinical modes of
EGFR tyrosine kinase inhibitor failure and subsequent management in
advanced non-small cell lung cancer. Lung Cancer. 79:33–39. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arcila ME, Oxnard GR, Nafa K, Riely GJ,
Solomon SB, Zakowski MF, Kris MG, Pao W, Miller VA and Ladanyi M:
Rebiopsy of lung cancer patients with acquired resistance to EGFR
inhibitors and enhanced detection of the T790M mutation using a
locked nucleic acid-based assay. Clin Cancer Res. 17:1169–1180.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukui T, Ohe Y, Tsuta K, Furuta K,
Sakamoto H, Takano T, Nokihara H, Yamamoto N, Sekine I, Kunitoh H,
et al: Prospective study of the accuracy of EGFR mutational
analysis by high-resolution melting analysis in small samples
obtained from patients with non-small cell lung cancer. Clin Cancer
Res. 14:4751–4757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weber B, Meldgaard P, Hager H, Wu L, Wei
W, Tsai J, Khalil A, Nexo E and Sorensen BS: Detection of EGFR
mutations in plasma and biopsies from non-small cell lung cancer
patients by allele-specific PCR assays. BMC Cancer. 14:2942014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Q, Shi J, Cheng M, Li G, Cao D and
Jiang G: Preparation of graphene-encapsulated magnetic microspheres
for protein/peptide enrichment and MALDI-TOF MS analysis. Chem
Commun (Camb). 48:1874–1876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mantini D, Petrucci F, Pieragostino D, Del
Boccio P, Sacchetta P, Candiano G, Ghiggeri GM, Lugaresi A,
Federici G, Di Ilio C and Urbani A: A computational platform for
MALDI-TOF mass spectrometry data: Application to serum and plasma
samples. J Proteomics. 73:562–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yanagisawa K, Shyr Y, Xu BJ, Massion PP,
Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S,
et al: Proteomic patterns of tumour subsets in non-small-cell lung
cancer. Lancet. 362:433–439. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song QB, Hu WG, Wang P, Yao Y and Zeng HZ:
Identification of serum biomarkers for lung cancer using magnetic
bead-based SELDI-TOF-MS. Acta Pharmacol Sin. 32:1537–1542. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu LH, Shan BE, Tian ZQ, Sang MX, Ai J,
Zhang ZF, Meng J, Zhu H and Wang SJ: Potential biomarkers for
esophageal carcinoma detected by matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry. Clin Chem
Lab Med. 48:855–861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
de Noo ME, Deelder A, van der Werff M,
Ozalp A, Mertens B and Tollenaar R: MALDI-TOF serum protein
profiling for the detection of breast cancer. Onkologie.
29:501–506. 2006.PubMed/NCBI
|
|
21
|
Mountain CF: Revisions in the
international system for staging lung cancer. Chest. 111:1710–1717.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the eastern cooperative oncology group. Am J Clin
Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Artimo P, Jonnalagedda M, Arnold K,
Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A,
Gasteiger E, et al: ExPASy: SIB bioinformatics resource portal.
Nucleic Acids Res. 40:(Web Server issue). W597–W603. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fung ET, Wright GL Jr and Dalmasso EA:
Proteomic strategies for biomarker identification: Progress and
challenges. Curr Opin Mol Ther. 2:643–650. 2000.PubMed/NCBI
|
|
25
|
Skytt A, Thysell E, Stattin P, Stenman UH,
Antti H and Wikström P: SELDI-TOF MS versus prostate specific
antigen analysis of prospective plasma samples in a nested
case-control study of prostate cancer. Int J Cancer. 121:615–620.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Castan-Laurell I, Dray C, Attané C, Duparc
T, Knauf C and Valet P: Apelin, diabetes, and obesity. Endocrine.
40:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee DK, Cheng R, Nguyen T, Fan T,
Kariyawasam AP, Liu Y, Osmond DH, George SR and O'Dowd BF:
Characterization of apelin, the ligand for the APJ receptor. J
Neurochem. 74:34–41. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krist J, Wieder K, Klöting N, Oberbach A,
Kralisch S, Wiesner T, Schön MR, Gärtner D, Dietrich A, Shang E, et
al: Effects of weight loss and exercise on apelin serum
concentrations and adipose tissue expression in human obesity. Obes
Facts. 6:57–69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sorli SC, Le Gonidec S, Knibiehler B and
Audigier Y: Apelin is a potent activator of tumour neoangiogenesis.
Oncogene. 26:7692–7699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berta J, Kenessey I, Dobos J, Tovari J,
Klepetko W, Jan Ankersmit H, Hegedus B, Renyi-Vamos F, Varga J,
Lorincz Z, et al: Apelin expression in human non-small cell lung
cancer: Role in angiogenesis and prognosis. J Thorac Oncol.
5:1120–1129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Antushevich H, Pawlina B, Kapica M,
Krawczynska A, Herman AP, Kuwahara A, Kato I and Zabielski R:
Influence of fundectomy and intraperitoneal or intragastric
administration of apelin on apoptosis, mitosis, and DNA repair
enzyme OGG1,2 expression in adult rats gastrointestinal tract and
pancreas. J Physiol Pharmacol. 64:423–428. 2013.PubMed/NCBI
|
|
32
|
Bouchon A, Hernandez-Munain C, Cella M and
Colonna M: A DAP12-mediated pathway regulates expression of CC
chemokine receptor 7 and maturation of human dendritic cells. J Exp
Med. 194:1111–1122. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takaki R, Watson SR and Lanier LL: DAP12:
An adapter protein with dual functionality. Immunol Rev.
214:118–129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma J, Jiang T, Tan L and Yu JT: TYROBP in
Alzheimer's disease. Mol Neurobiol. 51:820–826. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sfakianakis S, Bei ES, Zervakis M, Vassou
D and Kafetzopoulos D: On the identification of circulating tumor
cells in breast cancer. IEEE J Biomed Health Inform. 18:773–782.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Ma D, Li X, Deng C, Shi Q, You X,
Leng X, Li M, Tang F, Zhang F and Li Y: Gene expression profiles of
peripheral blood mononuclear cells in primary biliary cirrhosis.
Clin Exp Med. 14:409–416. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Spinella F, Caprara V, Di Castro V, Rosanò
L, Cianfrocca R, Natali PG and Bagnato A: Endothelin-1 induces the
transactivation of vascular endothelial growth factor receptor-3
and modulates cell migration and vasculogenic mimicry in melanoma
cells. J Mol Med (Berl). 91:395–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rosanò L, Spinella F and Bagnato A:
Endothelin 1 in cancer: Biological implications and therapeutic
opportunities. Nat Rev Cancer. 13:637–651. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zuco V, Cassinelli G, Cossa G, Gatti L,
Favini E, Tortoreto M, Cominetti D, Scanziani E, Castiglioni V,
Cincinelli R, et al: Targeting the invasive phenotype of
cisplatin-resistant non-small cell lung cancer cells by a novel
histone deacetylase inhibitor. Biochem Pharmacol. 94:79–90. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arun C, DeCatris M, Hemingway DM, London
NJ and O'Byrne KJ: Endothelin-1 is a novel prognostic factor in
non-small cell lung cancer. Int J Biol Markers. 19:262–267.
2004.PubMed/NCBI
|
|
41
|
Kappers MH, Smedts FM, Horn T, van Esch
JH, Sleijfer S, Leijten F, Wesseling S, Strevens H, Jan Danser AH
and van den Meiracker AH: The vascular endothelial growth factor
receptor inhibitor sunitinib causes a preeclampsia-like syndrome
with activation of the endothelin system. Hypertension. 58:295–302.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bagnato A, Loizidou M, Pflug BR, Curwen J
and Growcott J: Role of the endothelin axis and its antagonists in
the treatment of cancer. Br J Pharmacol. 163:220–233. 2011.
View Article : Google Scholar : PubMed/NCBI
|