Introduction
Thyroid cancer is one of the most common types of
endocrine malignancy, the incidence rate of which has rapidly
increased during previous years (1,2). It
has been estimated that the annual number of cases of thyroid
cancer diagnosed in the USA and the associated mortality rate are
12.9/100,000 and 0.5/100,000 individuals, respectively (3). Papillary thyroid cancer (PTC),
characterized by papillary architecture, psammoma bodies and
specific nuclear features, including nuclear orientation, nuclear
chromatin and nuclear grooving, is the major form of thyroid
cancer, accounting for 80–85% of all thyroid malignancies (4). Although PTC has a favorable
prognosis, certain cases exhibit aggressive clinical
characteristics, including invasion and metastasis, and the
management of PTC remains controversial (5,6).
Therefore, it is essential to investigate the molecular mechanisms
of PTC and identify potential biomarkers, which may be beneficial
for improving treatment.
The current understanding of the molecular
mechanisms of PTC has improved significantly, particularly with the
development of next generation sequencing and microarray
technology. It has been reported that mutations of BRAF (T1796A and
V599E) causing the activation of serine/threonine kinase may be an
alternative mechanism of oncogenic mitogen-activated protein kinase
pathway activation and are important in the development of PTC
(7–9). The chemokine receptor, CXCR7 not only
activates the phosphoinositide 3-kinase/AKT and nuclear factor-κB
signaling pathways, but it also downregulates the Notch signaling
pathway to regulate the growth and metastasis of PTC, according to
gene expression profiling analysis of PTC (10). Amyloid precursor protein is
overexpressed in PTC and may become a potential novel therapeutic
target for PTC (11). It has been
revealed that certain important upregulated microRNAs (miRs),
including miR-221, miR-222 and miR-146, and the regulation of KIT
are involved in the pathogenesis of PTC, based on previous
microarray analysis of GSE3467 (12). Zhu et al (13) showed that Trefoil factor 3,
cut-like homeobox 2 and forkhead box protein A2 are associated with
the development of PTC, and the differentially expressed genes
(DEGs) are primarily associated with positive regulation of gene
expression, gene transcription and metabolic processes.
In the previous study by Zhu et al (13) only the GSE3678 DNA microarray data
set was downloaded for DEG screening and subsequent enrichment
analysis. In the present study, two DNA microarray data sets,
GSE3467 and GSE3678, were combined for microarray analysis. The
DEGs found to be simultaneously differentially expressed in the
tumor samples of these two data sets were screened for enrichment
analysis and interaction network construction. The potential drugs
for the critical DEGs were also investigated. The present study may
aid the diagnosis and treatment of PTC.
Materials and methods
Gene expression profiles. The two gene expression
profiling data sets, GSE3467 and GSE3678, were downloaded from the
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (14). GSE3467 (12) consisted of 18 paired tumor and
normal thyroid tissue samples from nine patients with PTC. The
GSE3678 data set contained seven PTC samples and seven paired
normal thyroid tissue samples. The platform used for these two data
sets was the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133
Plus 2.0 array.
Data preprocessing and screening of DEGs. The probes
without annotation of the gene expression profiles were filtered,
and the probes were transformed into gene symbols. The average
value of a gene symbol corresponding to multiple probes was
calculated for the expression level analysis. The log2
transformation, background correction and data normalization were
performed using the GeneChip Robust Multi-array Analysis (GCRMA)
method within Bioconductor (http://www.bioconductor.org) (15). The DEGs were screened out by using
the Linear Models for Microarray Analysis (Limma) package in R
software (16) with the cut-off
criteria of adjusted P<0.01 and |log2 fold-change
(FC)|>2.
Functional and pathway enrichment analyses.
Functional and pathway enrichment analyses for the common DEGs were
performed using the Database for Annotation, Visualization and
Integrated Discovery online tools (17) based on the Gene Ontology (GO)
(18) and Kyoto Encyclopedia of
Genes and Genomes (19) databases.
The GO terms in three categories, including biological process,
cellular component and molecular function, were identified with
P<0.05. For pathway enrichment analysis, P≤0.05 was set as the
threshold.
Construction and analysis of the protein-protein
interaction (PPI) network. The physical PPIs and pathway
interactions were selected for PPI network construction using the
Gene Multiple Association Network Integration Algorithm (http://www.genemania.org/) (20) plug-in of Cytoscape software
(www.cytoscape.org; version 3.4.0). The
representative subnetwork containing nodes with high levels of
interconnection was further derived from the PPI network using
Molecular Complex Detection software (baderlab.org/Software/MCODE) (21). In addition, the potential drugs of
the hub DEGs in the subnetwork were searched using the DrugBank
database (www.drugbank.ca) (22) and the potential drug-like ligands,
which interacted with the genes, were screened from the diverse-lib
database in MTiOpenScreen
(bioserv.rpbs.univ-paris-diderot.fr/services/MTiOpenScreen)
(23) according to Lipinski's rule
of five (24).
Results
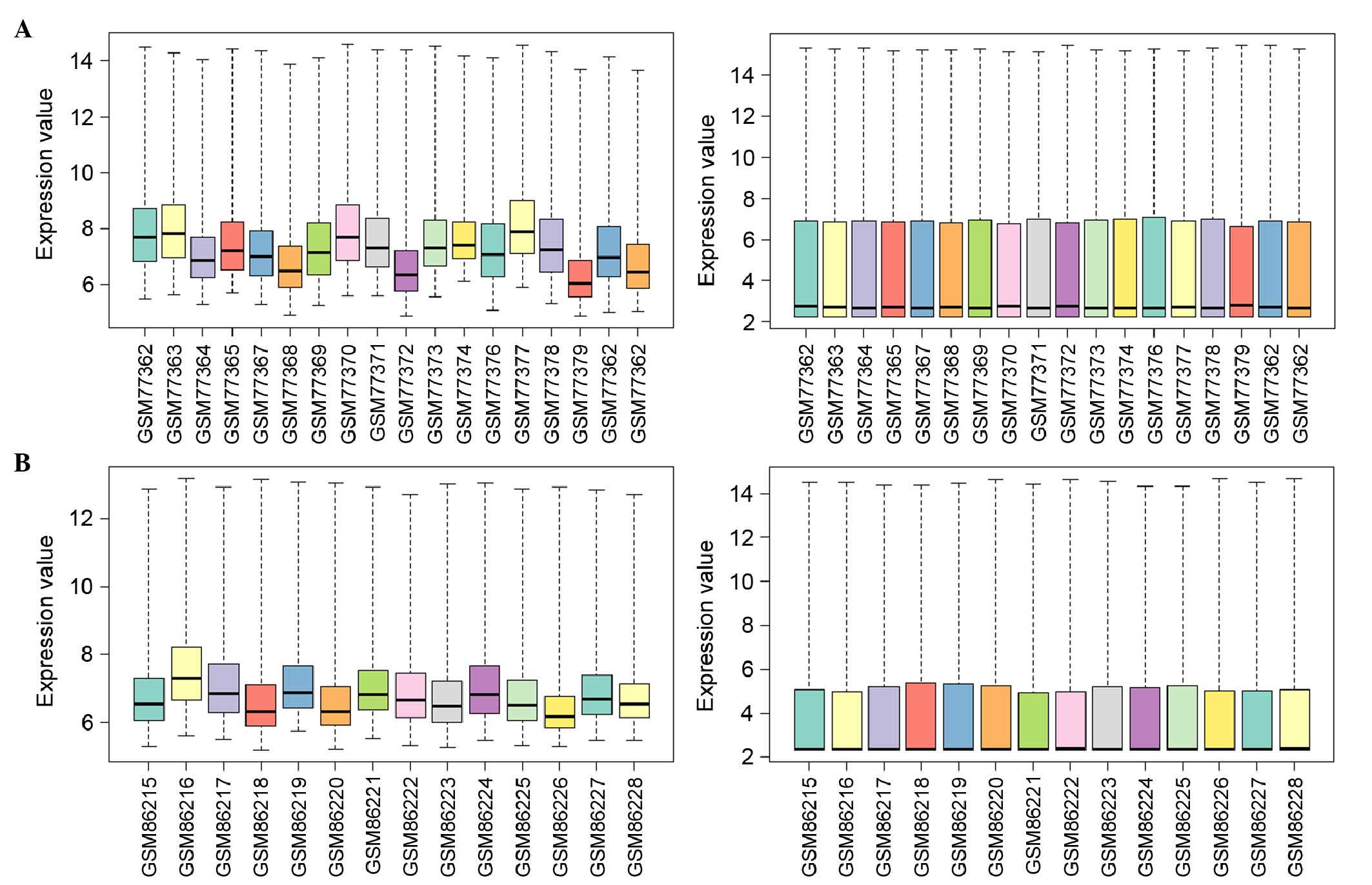
Identification of DEGs. Following data
preprocessing, the standardized expression profiling data showed
that the median values of gene expression were almost at the same
level, indicating that the degree of standardization was sufficient
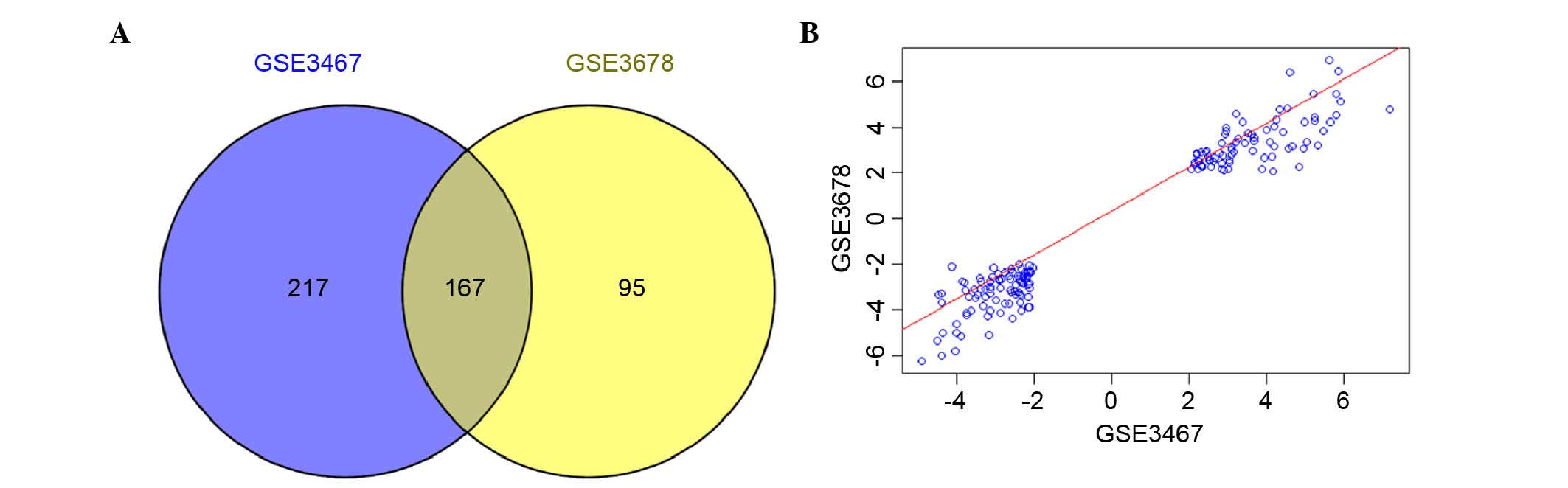
to be used for the subsequent analysis (Fig. 1). A total of 384 and 262 DEGs were
identified for GSE3467 and GSE3678, respectively. In addition, 167
genes were found to be simultaneously differentially expressed in
the tumor samples of these two data sets (Fig. 2A). Among these 167 common DEGs, 77
genes were upregulated and 90 genes were downregulated. The
correlation of the expression values for the 167 genes was 0.97
(P<2.2e-16; Fig. 2B).
Functional and pathway enrichment analysis. The
common DEGs were primarily associated with the cellular components
of the plasma membrane (P=2.60E-05) and integral to the membrane
(P=0.002429). Certain common DEGs, including LC26A4, GABRB3, GABRB2
and CLCNKB, were significantly enriched in the anion transmembrane
transporter activity (P=4.45E-04) and chloride anion binding
(P=0.005756). In addition, SERPINA1, CITED1 and GHR were involved
with response to steroid hormone stimulus (P=0.001849). FN1, LAMB3
and COL13A1 were enriched in cell adhesion (P=0.011121; Table I).
 | Table I.Top ten enriched GO terms in the CC,
MF and BP categories for the common differentially expressed
genes. |
Table I.
Top ten enriched GO terms in the CC,
MF and BP categories for the common differentially expressed
genes.
| Category | ID | Term | Count | Genes | P-value |
|---|
| CC | GO:0044459 | Plasma membrane
part | 40 | GPR125, STARD13,
NRCAM, SDPR, TPO, PLA2R1, ENTPD1, DPP4, FN1, GHR, COL13A1, MET,
SLC34A2… | 2.60E-05 |
| CC | GO:000998 | Cell surface | 13 | SCUBE3, GPR125,
KIT, NCAM1, NRCAM, CD36, GPM6A, LAYN, CDON, TGFA, TPO, DPP4,
GHR | 9.53E-05 |
| CC | GO:0005886 | Plasma membrane | 54 | GABRB3, TPO, TGFA,
SLC4A4, GHR, PSD3, LIFR, SLC34A2, FN1, MUC1, COL13A1, MET,
SHANK2… | 3.56E-04 |
| CC | GO:0044420 | Extracellular matrix
part | 7 | SMOC2, LAMB3,
SCUBE3, COL13A1, TFF3, ENTPD1, FN1 | 7.74E-04 |
| CC | GO:0030054 | Cell junction | 14 | GABRB3, GABRB2,
COL13A1, PSD3, CLDN10, GABBR2, CDH3, ARHGAP24, SHANK2, CAMK2N1,
LAYN, CLDN1, DLG2, DPP4 | 0.001052 |
| CC | GO:0044421 | Extracellular region
part | 20 | SCUBE3, LGALS3,
COL13A1, CHI3L1, KIT, SMOC2, LAMB3, TNFRSF11B, TGFA, TFF3, ANGPTL1,
SERPINA1, CFI, GDF15, ENTPD1, PLA2R1, SFTPB, FN1, GHR,
BMP8A | 0.001350 |
| CC | GO:0031012 | Extracellular
matrix | 11 | SMOC2, LAMB3,
TNFRSF11B, LGALS3, SCUBE3, COL13A1, CHI3L1, TFF3, SERPINA1, ENTPD1,
FN1 | 0.001416 |
| CC | GO:0045211 | Postsynaptic
membrane | 7 | GABRB3, GABRB2,
PSD3, GABBR2, SHANK2, DLG2, CAMK2N1 | 0.001625 |
| CC | GO:0044456 | Synapse part | 9 | GABRB3, GABRB2,
PSD3, GABBR2, LRRK2, SHANK2, DLG2, ITPR1, CAMK2N1 | 0.002101 |
| CC | GO:0016021 | Integral to
membrane | 66 | GABBR2, TPO,
DPP6, SLC4A4, DPP4, GHR, SEL1L3, ENTPD1, MUC1, MET, TMEM163, LAYN,
BTBD11… | 0.002429 |
| MF | GO:0043168 | Anion binding | 7 | SLC26A4, GABRB3,
GABRB2, CLCNKB, CTSC, GHR, SLC34A2 | 2.15E-04 |
| MF | GO:0008509 | Anion transmembrane
transporter activity | 8 | SLC26A4, GABRB3,
GABRB2, SLC26A7, ABCC3, CLCNKB, SLC4A4, SLC34A2 | 4.45E-04 |
| MF | GO:0015103 | Inorganic anion
transmembrane transporter activity | 4 | SLC26A4,
SLC26A7, SLC4A4, SLC34A2 | 0.003514 |
| MF | GO:0031404 | Chloride ion
binding | 5 | SLC26A4, GABRB3,
GABRB2, CLCNKB, CTSC | 0.005756 |
| MF | GO:0022838 | Substrate specific
channel activity | 10 | GABRB3, GPM6A,
GABRB2, SLC26A7, AQP4, RYR2, CLCNKB, KCNJ2, KCNIP4, ITPR1 | 0.012085 |
| MF | GO:0015267 | Channel
activity | 10 | GABRB3, GPM6A,
GABRB2, SLC26A7, AQP4 RYR2, CLCNKB, KCNJ2, KCNIP4, ITPR1 | 0.014860 |
| MF | GO:0022803 | Passive
transmembrane transporter activity | 10 | GABRB3, GPM6A,
GABRB2, SLC26A7, AQP4, RYR2, CLCNKB, KCNJ2, KCNIP4, ITPR1 | 0.015074 |
| MF | GO:0030246 | Carbohydrate
binding | 9 | NOD1, LGALS3,
GALNT7, LAYN, COL13A1, MRC2, CHI3L1, PLA2R1, FN1 | 0.017677 |
| MF | GO:0016917 | GABA receptor
activity | 3 | GABRB3, GABRB2,
GABBR2 | 0.020798 |
| MF | GO:0004714 | Transmembrane
receptor protein tyrosine kinase activity | 4 | MET, KIT, IRS1,
EPHB1 | 0.024558 |
| BP | GO:0007167 | Enzyme linked
receptor protein signaling pathway | 11 | TIAM1, MET,
LIFR, ANGPTL1, PPM1 L, KIT, GDF15, IRS1, EPHB1, CITED1,
GHR | 0.001120 |
| BP | GO:0009725 | Response to hormone
stimulus | 10 | KRT19,
TNFRSF11B, AVPR1A, FABP4, TFF3, SERPINA1, IRS1, CITED1, GHR,
SLC34A2 | 0.006298 |
| BP | GO:0043627 | Response to
estrogen stimulus | 6 | KRT19,
TNFRSF11B, SERPINA1, CITED1, GHR, SLC34A2 | 0.002681 |
| BP | GO:0048545 | Response to steroid
hormone stimulus | 8 | KRT19,
TNFRSF11B, AVPR1A, FABP4, SERPINA1, CITED1, GHR, SLC34A2 | 0.001849 |
| BP | GO:0001570 | Vasculogenesis | 4 | HEY2, ZFPM2,
CITED1, CITED2 | 0.006079 |
| BP | GO:0001666 | Response to
hypoxia | 6 | PDE5A, RYR2,
SERPINA1, ITPR1, DPP4, CITED2 | 0.007532 |
| BP | GO:0042060 | Wound healing | 7 | CD36, SERPINA1,
CDH3, ENTPD1, PAPSS2, PROS1, FN1 | 0.007962 |
| BP | GO:0060350 | Endochondral bone
morphogenesis | 3 | COL13A1, RUNX2,
GHR | 0.009053 |
| BP | GO:0070482 | Response to oxygen
levels | 6 | PDE5A, RYR2,
SERPINA1, ITPR1, DPP4, CITED2 | 0.009277 |
| BP | GO:0007155 | Cell adhesion | 14 | PLXNC1, MPZL2,
COL13A1, CLDN10, CDH3, CDH6, NCAM1, NRCAM, LAMB3, CD36, CDON,
CLDN1, ENTPD1, FN1 | 0.011121 |
In addition, two significant pathways were enriched
for the common DEGs (Table II).
The ALDH1A3, AOX1, HGD, ADH1B and TPO genes were significantly
involved in the tyrosine metabolism pathway (P=0.001). Another five
common DEGs, including NRCAM, NCAM1, CLDN1, CLDN10 and CDH3, were
enriched in the cell adhesion molecules pathway (P=0.05).
 | Table II.Enriched pathways for the common
differentially expressed genes. |
Table II.
Enriched pathways for the common
differentially expressed genes.
| Pathway_ID | Name | Count | Genes | P-value |
|---|
| hsa00350 | Tyrosine
metabolism | 5 | ALDH1A3, AOX1,
HGD, ADH1B, TPO | 0.001 |
| hsa04514 | Cell adhesion
molecules | 5 | NRCAM, NCAM1,
CLDN1, CLDN10, CDH3 | 0.050 |
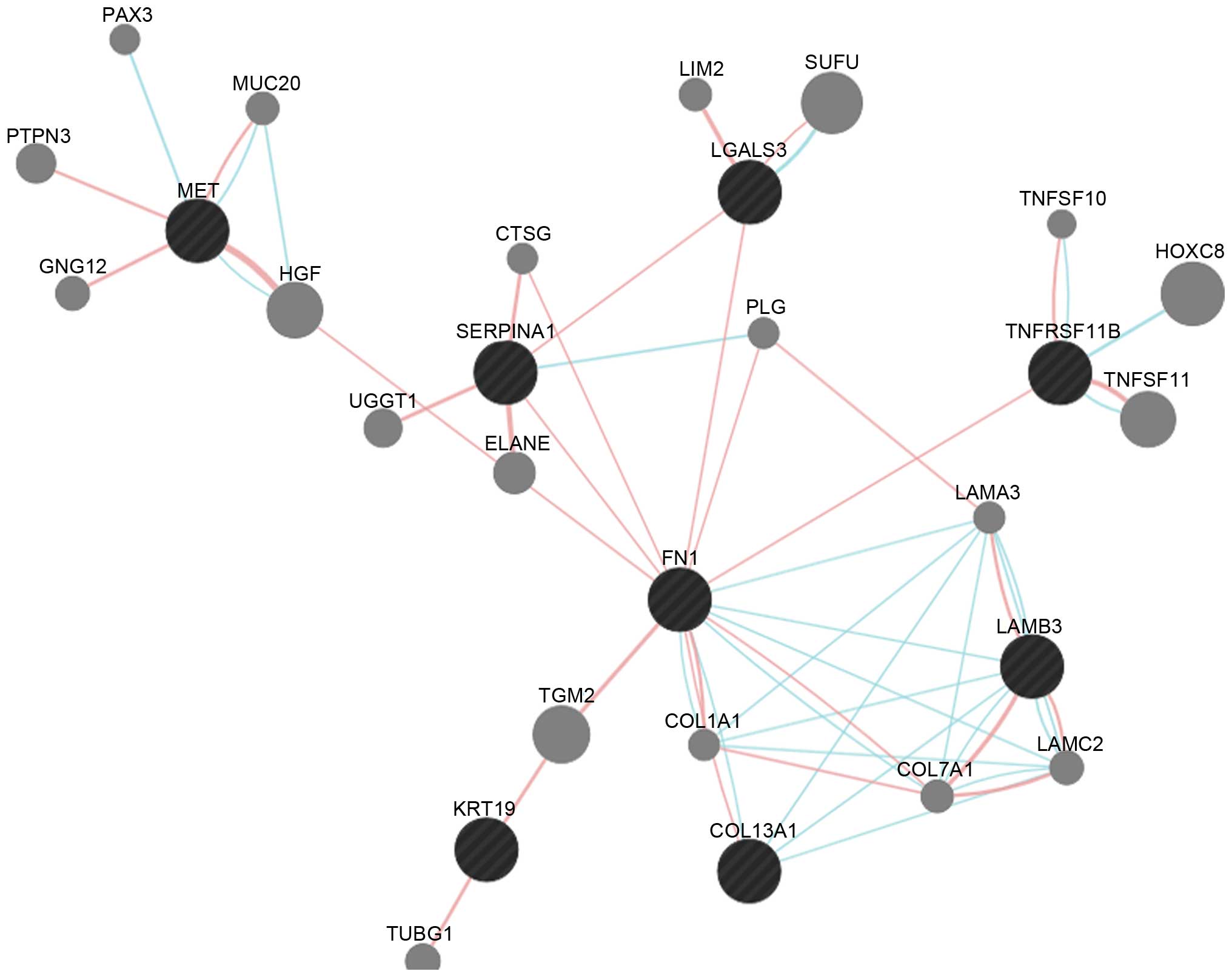
Interaction network analysis. There were eight
common DEGs, of which MET, SERPINA1, LGALS3, FN1, LAMB3, KRT19 and
COL13A1 were upregulated, and TNFRSF11B was downregulated) in the
constructed subnetwork (Fig. 3).
The FN1 DEG can interact with SERPINA1, LGALS3, TNFRSF11B and
LAMB3. There were also pathway interactions between any two of the
FN1, COL13A1 and LAMB3 DEGs.
Potential drug identification. The potential drugs
for FN1 and SERPINA1 were examined further, as these two DEGs
showed higher degrees in the interaction subnetwork and may be
critical in the regulatory process. The results revealed that two
drugs, lanoteplase and ocriplasmin, were found for FN1, and four
potential drugs, β-mercaptoethanol, recombinant α 1-antitrypsin,
PPL-100 and API, were found for SERPINA1 (Table III). Lanoteplase is currently
under clinical investigation and ocriplasmin, whose pharmacological
action involves the cleavage of fibronectin protein, has been
approved by the Food and Drug Administration (25). For SERPINA1, β-mercaptoethanol is
during experimental stage and the other three drugs are under
clinical investigation.
 | Table III.Potential drugs for FN1 and
SERPINA1 |
Table III.
Potential drugs for FN1 and
SERPINA1
| Target | Drug | State | Pharmacological
action |
|---|
| FN1 | Lanoteplase |
Investigational | Unknown |
|
| Ocriplasmin | Approved | Cleavage |
|
SERPINA1 |
β-mercaptoethanol | Experimental | Unknown |
|
| Recombinant α
1-antitrypsin |
Investigational | Unknown |
|
| PPL-100 |
Investigational | Unknown |
|
| API |
Investigational | Unknown |
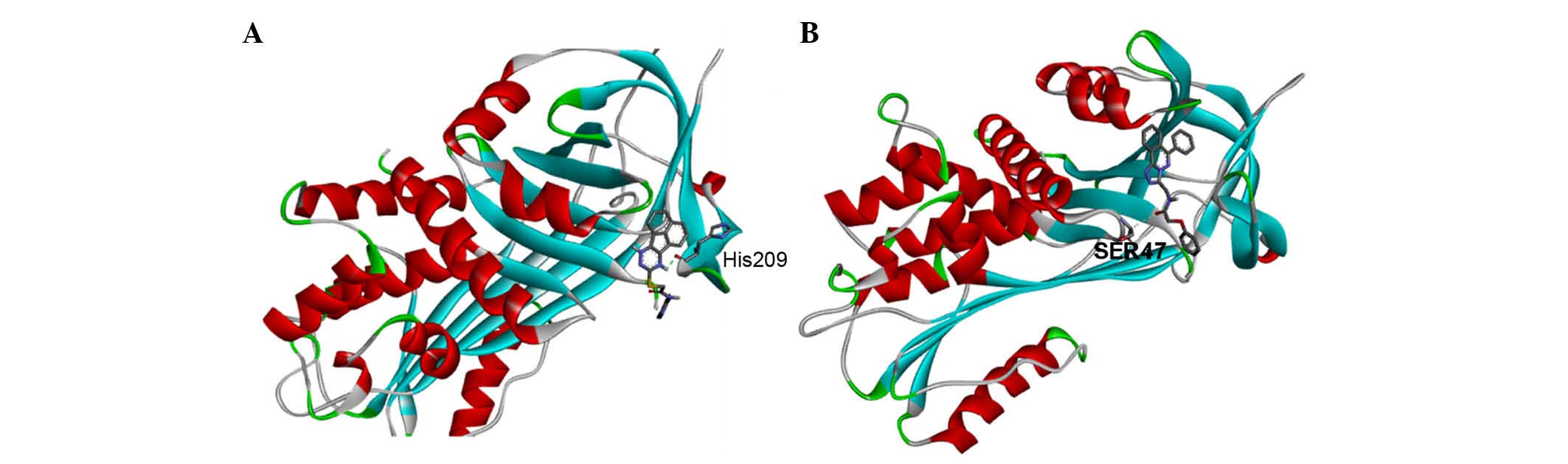
In addition, five potential drug-like compounds with
binding energies below −8.4 were screened out from the diverse-lib
database in MTiOpenScreen (Table
IV). The 24350490 compound, with a binding energy of −8.7
kcal/mol, has six hydrogen bond acceptors and one hydrogen bond
donor. Another compound, 17506143, with a binding energy of −8.6
kcal/mol has seven hydrogen bond acceptors and one hydrogen bond
donor. The structural interactions between SERPINA1 and these two
drug-like compounds are shown in Fig.
4. The binding pocket of SERPINA1 contained His209 and
SER47.
 | Table IV.Five drug-like compounds for
SERPINA1. |
Table IV.
Five drug-like compounds for
SERPINA1.
| Compound | Energy
(kcal/mol) | nRot (count) | Lead-like
(yes/no) | HBA (count) | HBD (count) | LogP | MW (g/mol) | TPSA
(Å2) |
|---|
| 24350490 | −8.7 | 4 | Y | 6 | 1 | 3.49 | 405.86 | 105.96 |
| 17506143 | −8.6 | 6 | Y | 7 | 1 | 3.50 | 409.44 | 81.41 |
| 26649374 | −8.5 | 1 | Y | 7 | 2 | 2.43 | 366.41 | 87.32 |
| 49671559 | −8.4 | 4 | Y | 7 | 0 | 3.14 | 445.92 | 92.10 |
| 26639702 | −8.4 | 4 | Y | 6 | 0 | 3.51 | 389.47 | 94.93 |
Discussion
In the present study, 167 genes were identified to
be simultaneously differentially expressed in the tumor samples of
GSE3467 and GSE3678. The subsequent pathway enrichment analysis
revealed that ALDH1A3, AOX1, HGD, ADH1B and TPO were significantly
involved in tyrosine metabolism. Several studies have revealed that
the protooncogene, RET, which encodes a tyrosine kinase cell
surface receptor, is involved in the oncogenesis of PTC by
activating its tyrosine kinase, either by rearrangement or mutation
(26,27). It has been confirmed that the mRNA
expression level of TPO is markedly suppressed leading to low
thyroid peroxidase activity in PTC (28). Thyroid peroxidase oxidizes iodide
to form active iodine for addition onto tyrosine residues on
thyroglobulin for the production of thyroid hormones (29). In the present study, TPO was also
found to be significantly downregulated in the tumor samples.
Therefore, ALDH1A3, AOX1, HGD, ADH1B and TPO may be important in
PTC via their involvement in the tyrosine metabolism pathway. By
contrast, the NRCAM, NCAM1, CLDN1, CLDN10 and CDH3 were
significantly enriched in the pathway of cell adhesion molecules.
Molecules associated with cell adhesion, for example ICAM-1, are
known to be involved in papillary growth and proliferative capacity
(30). The pathway enrichment
analysis indicated that these common DEGs are involved in the
pathogenesis and progression of PTC by regulating the pathways of
tyrosine metabolism and cell adhesion molecules.
The representative subnetwork derived from the
interaction network in the present study contained seven
upregulated common DEGs (MET, SERPINA1, LGALS3, FN1, LAMB3 and
COL13A1) and one downregulated DEG (TNFRSF11B). The MET, SERPINA1,
LGALS3 and FN1 DEGs have been identified to be the potent
biomarkers for PTC in previous studies (31,32).
The expression levels of FN1 and MET have been also reported to be
upregulated in papillary thyroid carcinoma (33). The functional enrichment analysis
revealed that FN1 was significantly enriched in the plasma
membrane, extracellular matrix and cell adhesion. Vasko et
al (34) showed that
epithelial-to-mesenchymal transition is common in PTC invasion. It
has been demonstrated that the genes differentially expressed in
aggressive PTC are primarily associated with cell adhesion
(35). In addition, FN1 can
interact with SERPINA1, LGALS3, TNFRSF11B and LAMB3, according to
the subnetwork. These results suggested that FN1 might be involved
in PCT by regulating the epithelial-to-mesenchymal transition and
interacting with other DEGs. Lanoteplase and ocriplasmin were
identified to be the potential drugs for FN1. It has been reported
that fibronectin is upregulated in PTC (36). Therefore, ocriplasmin may be of
assistance in the prevention or treatment of PTC due to the
cleavage of fibronectin.
Another common DEG with a higher degree, SERPINA1,
was significantly associated with the functions of response to
steroid hormone stimulus and response to estrogen stimulus in the
present study. Vierlinger et al (37) showed that SERPINA1 can be
considered as a single marker for PTC. In papillary thyroid tumors
and normal thyroid tissue, estrogen and progesterone receptors have
been identified in thyroid tissue responsive to steroid hormones
(38). Kawabata et al
(39) reported estrogen receptor α
(ERα) is expressed at high levels in papillary thyroid cancer,
suggesting that ERα may be involved in the development, physiology
and pathology of papillary carcinoma. Therefore, upregulated
expression levels of SERPINA1 may contribute to the response to
steroid hormones in PTC. In the present study, four potential drugs
were selected for SERPINA1. As His209 and SER47 are critical for
the binding of compounds by forming hydrogen bonds (40), the amino acid around the pocket,
which binds with compounds, may be critical for the function of
SERPINA1. Therefore, β-mercaptoethanol and recombinant α
1-antitrypsin may be the potential drugs for PTC. However, there
were a number of limitations to the present study. The gene
expression profiles of PTC at different stages require further
analysis. In addition, the gene and protein expression levels of
important DEGs may be better confirmed using experimental
studies.
In conclusion, the common DEGs identified from
GSE3467 and GSE3678 were found to have the potential for use as
candidate biomarkers for PTC. The upregulated expression levels of
FN1 and SERPINA1 may be involved in PTC by regulating the
epithelial-to-mesenchymal transition and response to steroid
hormones, respectively. Furthermore, ocriplasmin, β-mercaptoethanol
and recombinant α 1-antitrypsin may be potential drugs for the
treatment of PTC. In further investigations, confirmation of the
expression levels of important genes is required, as is the
examination of PTC at different stages.
References
|
1
|
Nguyen QT, Lee EJ, Huang MG, Park YI,
Khullar A and Plodkowski RA: Diagnosis and treatment of patients
with thyroid cancer. Am Health Drug Benefits. 8:30–40.
2015.PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institute, . SEER stat
fact sheets: Thyroid cancer. http://seer.cancer.gov/statfacts/html/thyro.htmlAccessed:
May 20, 2015.
|
|
4
|
Schneider DF and Chen H: New developments
in the diagnosis and treatment of thyroid cancer. CA Cancer J Clin.
63:374–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu XM, Wan Y, Sippel RS and Chen H: Should
all papillary thyroid microcarcinomas be aggressively treated? An
analysis of 18,445 cases. Ann Surg. 254:653–660. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mazeh H and Chen H: Advances in surgical
therapy for thyroid cancer. Nat Rev Endocrinol. 7:581–588. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cohen Y, Xing M, Mambo E, Guo Z, Wu G,
Trink B, Beller U, Westra WH, Ladenson PW and Sidransky D: BRAF
mutation in papillary thyroid carcinoma. J Natl Cancer Inst.
95:625–627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soares P, Trovisco V, Rocha AS, Lima J,
Castro P, Preto A, Máximo V, Botelho T, Seruca R and
Sobrinho-Simões M: BRAF mutations and RET/PTC rearrangements are
alternative events in the etiopathogenesis of PTC. Oncogene.
22:4578–4580. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukushima T, Suzuki S, Mashiko M, Ohtake
T, Endo Y, Takebayashi Y, Sekikawa K, Hagiwara K and Takenoshita S:
BRAF mutations in papillary carcinomas of the thyroid. Oncogene.
22:6455–6457. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Teng X and Liu Z, Zhang L and Liu
Z: Gene expression profile analyze the molecular mechanism of CXCR7
regulating papillary thyroid carcinoma growth and metastasis. J Exp
Clin Cancer Res. 34:162015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Fan Y, Deng Z, Wu B and Zheng Q:
Amyloid precursor protein as a potential marker of malignancy and
prognosis in papillary thyroid carcinoma. Oncol Lett. 3:1227–1230.
2012.PubMed/NCBI
|
|
12
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu X, Yao J and Tian W: Microarray
technology to investigate genes associated with papillary thyroid
carcinoma. Mol Med Rep. 11:3729–3733. 2015.PubMed/NCBI
|
|
14
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res. 35:(Database
issue). D760–D765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Gentleman R, Varey VJ, Huber W, Irizarry
RA and Dudoit S: 1st. Springer-Verlag; New York, NY: pp. 397–420.
2005, View Article : Google Scholar
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The Gene Ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:(Database issue). D258–D261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic acids Res. 38:(Web Server issue). W214–W220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wishart DS, Knox C, Guo AC, Shrivastava S,
Hassanali M, Stothard P, Chang Z and Woolsey J: DrugBank: A
comprehensive resource for in silico drug discovery and
exploration. Nucleic Acids Res. 34:(Database issue). D668–D672.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Labbé CM, Rey J, Lagorce D, Vavruša M,
Becot J, Sperandio O, Villoutreix BO, Tufféry P and Miteva MA:
MTiOpenScreen: A web server for structure-based virtual screening.
Nucleic Acids Res. 43:W448–W454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collen D and Lijnen HR: Tissue-type
plasminogen activator: A historical perspective and personal
account. J Thromb Haemostasis. 2:541–546. 2004. View Article : Google Scholar
|
|
26
|
Komminoth P: The RET proto-oncogene in
medullary and papillary thyroid carcinoma. Molecular features,
pathophysiology and clinical implications. Virchows Arch. 431:1–9.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eberhardt NL, Grebe SK, McIver B and Reddi
HV: The role of the PAX8/PPARgamma fusion oncogene in the
pathogenesis of follicular thyroid cancer. Mol Cell Endocrinol.
321:50–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tanaka T, Umeki K, Yamamoto I, Sugiyama S,
Noguchi S and Ohtaki S: Immunohistochemical loss of thyroid
peroxidase in papillary thyroid carcinoma: Strong suppression of
peroxidase gene expression. J Pathol. 179:89–94. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruf J and Carayon P: Structural and
functional aspects of thyroid peroxidase. Arch Biochem Biophys.
445:269–277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Buitrago D, Keutgen X, Crowley M, Filicori
F, Aldailami H, Hoda R, Liu YF, Hoda RS, Scognamiglio T, Jin M, et
al: Intercellular adhesion molecule-1 (ICAM-1) is upregulated in
aggressive papillary thyroid carcinoma. Ann Surg Oncol. 19:973–980.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jarząb B, Wiench M, Fujarewicz K, Simek K,
Jarzab M, Oczko-Wojciechowska M, Wloch J, Czarniecka A, Chmielik E,
Lange D, et al: Gene expression profile of papillary thyroid
cancer: Sources of variability and diagnostic implications. Cancer
Res. 65:1587–1597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griffith OL, Melck A, Jones SJ and Wiseman
SM: Meta-analysis and meta-review of thyroid cancer gene expression
profiling studies identifies important diagnostic biomarkers. J
Clin Oncol. 24:5043–5051. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
da Silveira Mitteldorf CA, de
Sousa-Canavez JM, Leite KR, Massumoto C and Camara-Lopes LH: FN1,
GALE, MET, and QPCT overexpression in papillary thyroid carcinoma:
Molecular analysis using frozen tissue and routine fine-needle
aspiration biopsy samples. Diagn Cytopathol. 39:556–561. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vasko V, Espinosa AV, Scouten W, He H,
Auer H, Liyanarachchi S, Larin A, Savchenko V, Francis GL, de la
Chapelle A, et al: Gene expression and functional evidence of
epithelial-to-mesenchymal transition in papillary thyroid carcinoma
invasion. Proc Natl Acad Sci USA. 104:2803–2808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Yuan Z, Fan Y, Deng X and Zheng Q:
Integrated analyses of microRNA and mRNA expression profiles in
aggressive papillary thyroid carcinoma. Mol Med Rep. 8:1353–1358.
2013.PubMed/NCBI
|
|
36
|
Wasenius VM, Hemmer S, Kettunen E,
Knuutila S, Franssila K and Joensuu H: Hepatocyte growth factor
receptor, matrix metalloproteinase-11, tissue inhibitor of
metalloproteinase-1, and fibronectin are up-regulated in papillary
thyroid carcinoma: A cDNA and tissue microarray study. Clin Cancer
Res. 9:68–75. 2003.PubMed/NCBI
|
|
37
|
Vierlinger K, Mansfeld MH, Koperek O,
Nöhammer C, Kaserer K and Leisch F: Identification of SERPINA1 as
single marker for papillary thyroid carcinoma through microarray
meta analysis and quantification of its discriminatory power in
independent validation. BMC Med Genomics. 4:302011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lewy-Trenda I: Estrogen and progesterone
receptors in neoplastic and non-neoplastic thyroid lesions. Pol J
Pathol. 53:67–72. 2002.PubMed/NCBI
|
|
39
|
Kawabata W, Suzuki T, Moriya T, Fujimori
K, Naganuma H, Inoue S, Kinouchi Y, Kameyama K, Takami H,
Shimosegawa T and Sasano H: Estrogen receptors (alpha and beta) and
17beta-hydroxysteroid dehydrogenase type 1 and 2 in thyroid
disorders: Possible in situ estrogen synthesis and actions. Mod
Pathol. 16:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Derewenda ZS and Derewenda U: The
structure and function of platelet-activating factor
acetylhydrolases. Cell Mol Life Sci. 54:446–455. 1998. View Article : Google Scholar : PubMed/NCBI
|