Introduction
Cancer stem cells (CSCs) are a small population of
cells present in tumors, which exhibit stem cell-like properties,
including self-renewal and multi-lineage differentiation potential
(1,2). CSCs have been reported to serve an
important role in tumor recurrence, metastasis and chemotherapeutic
resistance in breast cancer (3,4).
Targeting CSCs is considered a promising therapeutic strategy for
the treatment of breast cancer (5), and identification of the signaling
pathways that regulate breast CSCs may facilitate the development
of therapeutic agents that target breast CSCs.
The hedgehog (Hh) signaling pathway is known to
regulate cell proliferation and self-renewal in normal stem cells
during embryonic development, as well as in malignant stem cells
(6–8). The Hh signaling pathway is activated
by binding of Hh ligands, including sonic hedgehog, desert hedgehog
and Indian hedgehog, to the Patched (PTCH) receptor. PTCH receptor
activation subsequently results in Smoothened activation, which
eventually leads to regulation of the expression of Gli
transcription factors that are responsible for cancer cell
proliferation, apoptosis and invasion (9). Previous studies have demonstrated
that Hh signaling regulates CSCs in several types of human cancer,
including breast cancer (10),
glioblastoma (11), glioma
(12) and myeloid leukemia
(13). In breast CSCs, the Hh
signaling pathway has an important role in maintaining the cluster
of differentiation (CD)44+/CD24−
subpopulation and the side population of breast cancer cells
(14). Activation of the Hh
signaling pathway by Hh ligands and Gli1 or Gli2 overexpression
promotes self-renewal of breast CSCs via modulation of Bmi-1
expression (10). However, it
remains to be elucidated as to whether Hh signaling activation
regulates breast CSCs and contributes to clinical outcomes in
patients with breast cancer.
The cell adhesion molecules CD44 and CD24 are
expressed on breast cancer cells, and are associated with cell
adhesion, tumor initiation, development and metastasis (15). The
CD44+/CD24− phenotype is often used as a
marker to isolate breast CSCs from solid tumors (16). Breast cancer cells with the
CD44+/CD24− phenotype exhibit stem cell-like
properties (16), and are
associated with enhanced invasion and metastasis (17,18).
Furthermore, it has been reported that the
CD44+/CD24− phenotype contributes to relapse
and poor prognosis in patients with breast cancer (19,20).
It has previously been demonstrated that components of the Hh
signaling pathway are highly upregulated in breast cancer cells
with the CD44+/CD24− phenotype, and that the
Hh signaling pathway is essential for maintaining this population
of breast cancer cells (14).
However, the role of the Hh signaling pathway in breast cancer
patients with the CD44+/CD24− phenotype
remains to be determined.
The present study used immunohistochemistry to
investigate the expression of PTCH, Gli1 and CD44/CD24 in 266
patients with breast cancer. The aim of the present study was to
investigate the association between the expression of PTCH and
Gli1, which are the main components of the Hh signaling pathway, in
breast cancer patients with the CD44+/CD24−
phenotype, and to analyze the correlation of their expression with
clinicopathological features and prognosis of breast cancer
patients with the CD44+/CD24− phenotype.
Materials and methods
Patients and tissue samples
The Medical Ethics Committee of China Medical
University (Shenyang, China) approved this retrospective study. Due
to the retrospective nature of the present study, the Medical
Ethics Committee waived the requirement for written informed
consent by the patients. Human breast tissues were obtained from
266 female patients with sporadic breast cancer, who underwent
surgery at the First Hospital of China Medical University between
2006 and 2010. The diagnosis of breast cancer was confirmed by
pathological staining. A total of 232 patients had invasive ductal
carcinoma, and 34 patients had invasive lobular carcinoma. The
histological grade of the cancer was determined according to the
World Health Organization grading system (21). Clinicopathological data, including
patient age, menopausal status, tumor size and lymph node
metastasis were retrospectively retrieved from medical records.
None of the patients underwent radiation therapy or chemotherapy
prior to surgery. Following surgery, 195 patients were followed up
for 48–77 months. The chemotherapy regimens of these patients
included CEF (cyclophosphamide + epimbicin + fluorouracil, n=151),
CAF (cyclophosphamide + Adriamycin + fluorouracil, n=18) and CET
(cyclophosphamide + epimbicin + taxol, n=26).
Immunohistochemistry
Immunohistochemical staining was performed as
previously described (22).
Briefly, sections (4 µm) were obtained from formalin-fixed and
paraffin-embedded tissue blocks. Sections were deparaffinized with
xylene, rehydrated in a graded alcohol series, and heated in
citrate buffer solution (pH 6) for 10 min to retrieve antigens. To
suppress endogenous peroxidase activity, the sections were treated
with 3% H2O2 at 37°C for 20 min. To block nonspecific protein
binding sites, sections were incubated in 10% normal goat serum at
37°C for 30 min. For immunohistochemical staining of PTCH and Gli1,
the sections were incubated with primary antibodies against PTCH
(rabbit anti-human polyclonal antibodies; 1:100 dilution; cat. no.
ab39266; Abcam, Cambridge, UK) or Gli1 (rabbit anti-human
polyclonal antibodies; 1:200 dilution; cat. no. ab92611; Abcam)
overnight at 4°C. For double immunohistochemical staining of CD44
and CD24, sections were incubated with primary antibodies against
CD44 (clone 156-3C11; mouse anti-human monoclonal antibodies; 1:800
dilution; cat. no. MA5-13890; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and CD24 (Clone SN3b; mouse anti-human monoclonal
antibodies; 1:400 dilution; cat. no. MA5-11828; Thermo Fisher
Scientific, Inc.) overnight at 4°C. Sections in which primary
antibodies were replaced with PBS were used as a negative control.
Sections were subsequently incubated with biotinylated secondary
antibodies (1:1,000 dilution) for 30 min at 37°C, followed by
incubation with streptavidin-horseradish peroxidase for an
additional 20 min (LSAB kit; Dako, Glostrup, Denmark). For PTCH and
Gli1, sections were stained with 3,3-diaminobenzidine (DAB;
Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and
counterstained with hematoxylin. For CD44 and CD24 staining, CD24
was detected with Permanent Red (from the Double SP kit; Maixin
Biotech. Co., Ltd., Fuzhou, China) and CD44 with DAB. Subsequently,
the sections were dehydrated and mounted. Images from each section
were captured using a Digital Sight digital camera under a Nikon
Eclipse 80i microscope (Nikon Corporation, Tokyo, Japan).
Evaluation of
immunohistochemistry
Immunoreactivity was evaluated by two independent
investigators blinded to the patients' clinicopathological
characteristics, according to the percentage of stained cells and
the intensity of immunoreactivity (23,24).
Immunoreactive intensity was scored as follows: 0, no staining; 1,
weak staining; 2, moderate staining; and 3, strong staining. The
percentage of stained cells was scored as follows: 0, <5%
stained cells; 1, 5–25% stained cells; 2, 26–50% stained cells; 3,
51–75% stained cells; and 4, >75% stained cells. The final
immunoreactive score was calculated by multiplying the intensity
score with the score for the percentage of stained cells, and was
used to generate the receiver operating characteristic (ROC) curve
analysis. The ROC was used to determine the cutoff value for
discriminating tumors with positive expression of PTCH, Gli1, CD44
and CD24, from those with negative expression, as previously
described by Kim et al (25).
Statistical analysis
Analyses were performed using SPSS 11.5 (SPSS Inc.,
Chicago, IL, USA). Pearson χ2 or Fisher's exact
probability tests were used to evaluate the association between
PTCH, Gli1 and CD44+/CD24− expression, and
the clinicopathological characteristics of the patients with breast
cancer. Spearman rank correlation analysis was used to assess the
association between PTCH and Gli1 expression and
CD44+/CD24−expression. Survival probabilities
were estimated using the Kaplan-Meier method and were assessed by a
log-rank test. Disease-free survival (DFS) was calculated as the
time between the first day of diagnosis and the occurrence of local
recurrence or distant metastasis. Overall survival (OS) was
calculated as the time between the first day of diagnosis and
disease-related mortality. Univariate and multivariate Cox
proportional hazards regression models were used for assessing the
association between potential confounding variables and prognosis
(OS or DFS). Mann-Whitney U test was used to compared the
expression of PTCH and Gli1 in breast cancer with the
CD44+/CD2− phenotype with
non-CD44+/CD24− phenotype. P≤0.05 was
considered to indicate a statistically significant difference.
Results
Clinicopathological
characteristics
Table I summarizes
the clinicopathological characteristics of the 266 patients with
breast cancer. The average age of the patients was 50.8 years
(range, 29–74 years). The majority of these patients had a tumor
that was diagnosed as invasive ductal carcinoma (87.2%), was <2
cm in size (61.4%) and was graded as histological Grade II (62.6%).
Lymph node metastasis occurred in 99 (37.2%) of the 266 patients.
Follow-up information was available for 195 patients with breast
cancer. Relapses occurred in 144 cases and breast cancer-associated
mortality occurred in 25 cases. The 5-year survival rate was 84.8%.
The mean OS and DFS were 72.5 and 55.3 months, respectively.
 | Table I.Clinicopathological characteristics
of 266 patients with breast cancer. |
Table I.
Clinicopathological characteristics
of 266 patients with breast cancer.
| Clinicopathological
feature | Number | % |
|---|
| Age |
|
| ≤50
years | 138 | 51.9 |
| >50
years | 128 | 48.1 |
| Menopausal
status |
|
|
Premenopausal | 148 | 55.6 |
|
Postmenopausal | 118 | 44.4 |
| Histologic
type |
|
|
Invasive ductal carcinoma | 232 | 87.2 |
|
Invasive lobular
carcinoma | 34 | 12.8 |
| Tumor
sizea |
|
| ≤2.0
cm | 145 | 61.4 |
|
>2.0, ≤5.0 cm | 91 | 38.6 |
| Lymph node
metastasis |
|
|
Negative | 167 | 62.8 |
|
Positive | 99 | 37.2 |
| Histological
gradeb |
|
| I | 42 | 21.2 |
| II | 124 | 62.6 |
|
III | 32 | 16.2 |
Expression of PTCH, Gli1, CD44 and
CD24 in breast cancer tissues
The expression of PTCH, Gli1, CD44 and CD24 was
detected in 266 breast cancer tissues using immunohistochemistry. A
ROC curve analysis was performed to determine an optimal cutoff
score for the expression of PTCH, Gli1, CD44 and CD24 in breast
cancer samples, based on the sensitivity and specificity for each
clinicopathological parameter. The parameter with the biggest area
under the curve was selected. According to the criteria, OS, DFS,
OS and lymph node metastasis were selected to determine the cutoff
values for PTCH, Gli1, CD44 and CD24, respectively. Cutoff scores
of 2.5, 2.5, 3.5 and 3.5 were determined for PTCH, Gli1, CD44 and
CD24 expression, respectively (Fig.
1). Since the final immunoreactive scores were integers,
negative and positive immunoreactivity were defined by a final
score of <3 and ≥3 for PTCH and Gli1, and <4 and ≥4 for CD44
and CD24.
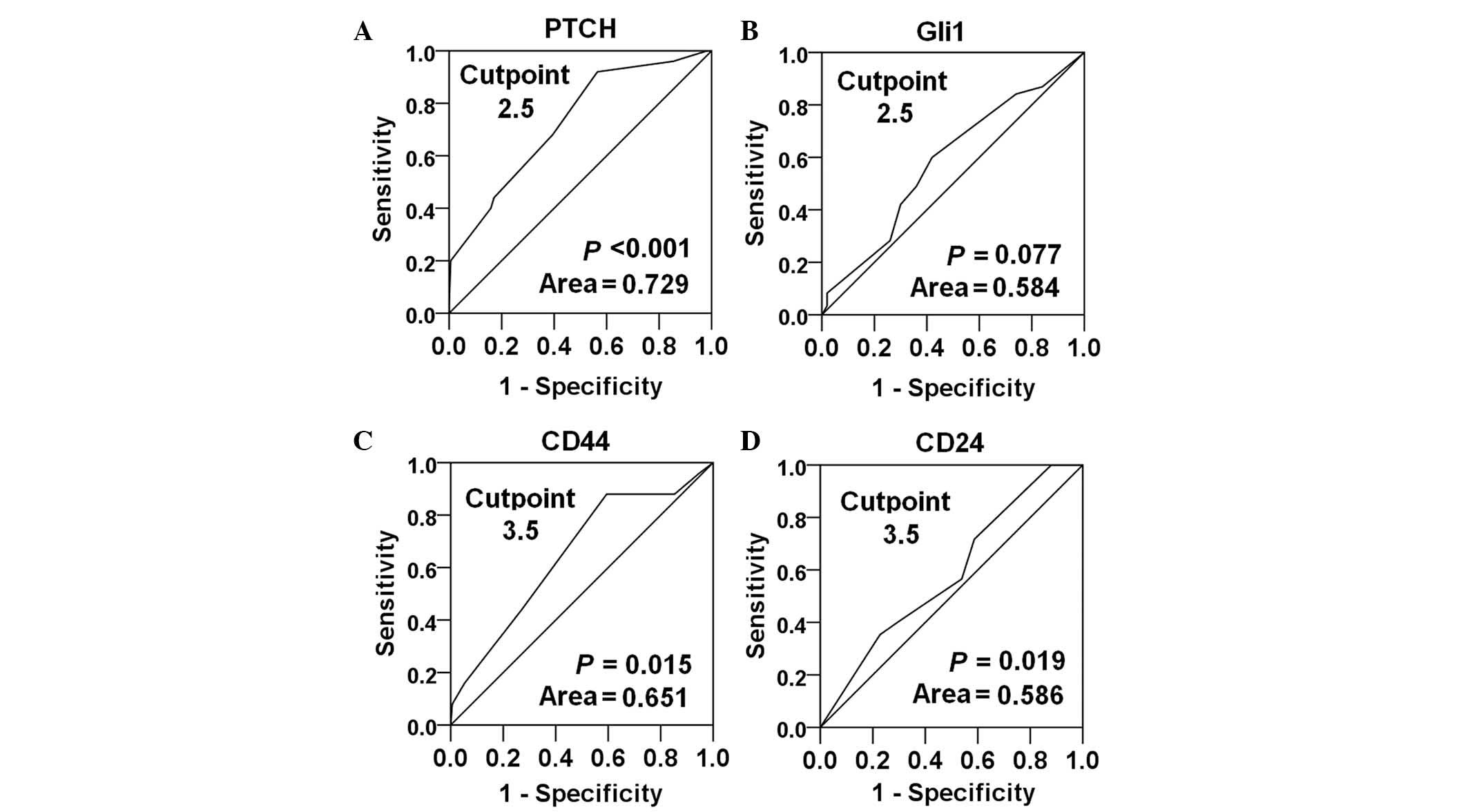 | Figure 1.Receiver operating characteristic
curves were used to determine the cutoff score for the expression
of (A) PTCH, (B) Gli1, (C) CD44 and (D) CD24 in patients with
breast cancer. The sensitivity and specificity for OS, DFS, OS and
lymph node metastasis were plotted for PTCH, Gli1, CD44 and CD24
expression, respectively. The areas under the curve and P-values
are indicated. PTCH, Patched; CD, cluster of differentiation; OS,
overall survival; DFS, disease-free survival. |
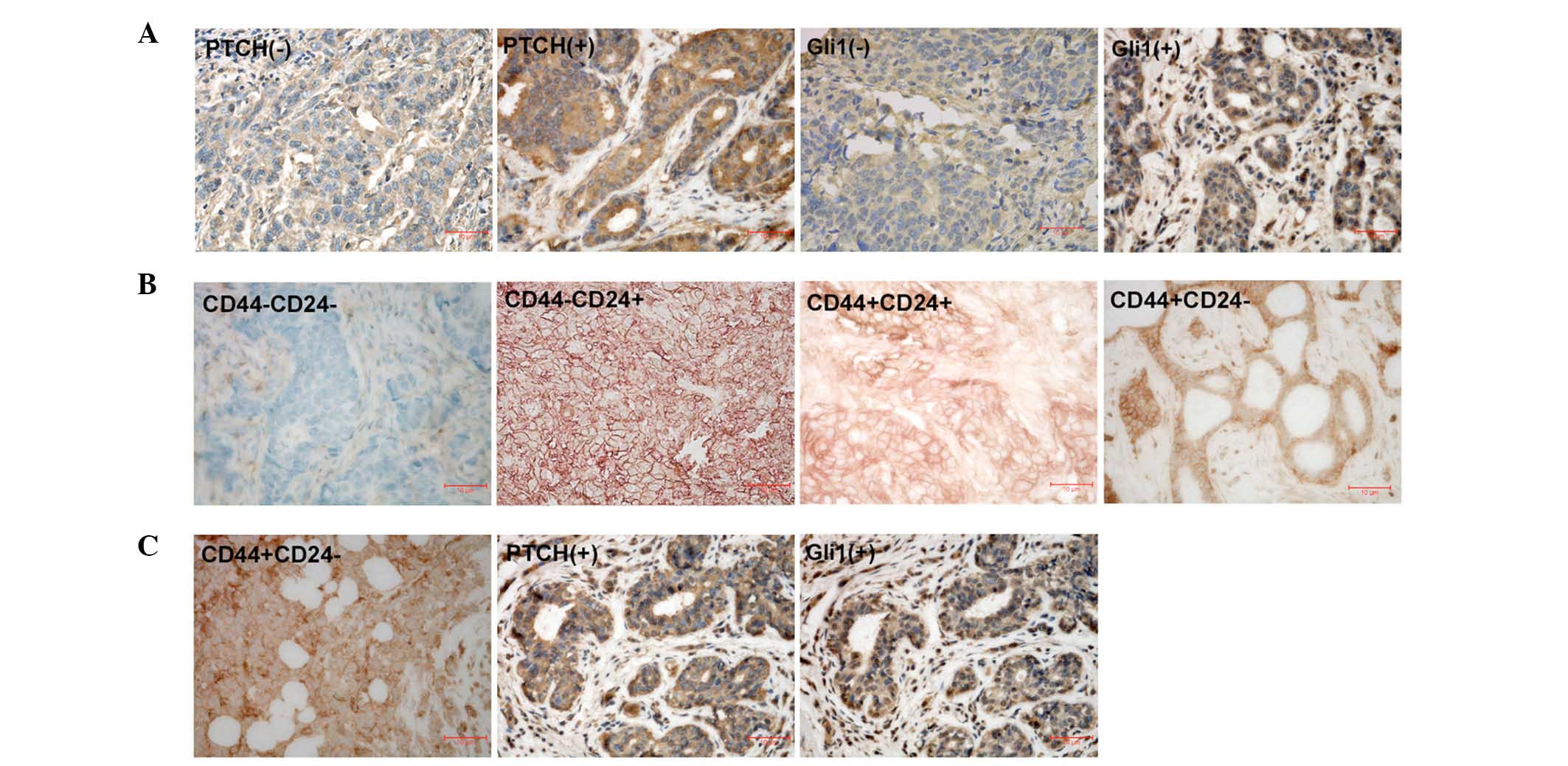
Representative immunohistochemical staining for
PTCH, Gli1 and CD44/CD24 in breast cancer samples is presented in
Fig. 2. PTCH-positive
immunoreactivity was observed in 157 (59.0%) out of 266 breast
cancer samples, and Gli1-postive immunoreactivity was detected in
140 (52.6%) out of 266 breast cancer samples (P<0.001). The
CD44+/CD24− phenotype was observed in 99
(37.2%) out of 266 breast cancer samples.
Association of PTCH and Gli1
expression with CD44 and CD24 expression
Spearman rank correlation analysis was used to
analyze the association between PTCH and Gli1 expression in breast
cancer. The expression levels of PTCH were positively correlated
with those of Gli1 (r=0.235, P<0.001). In addition, the
expression levels of CD44+/CD24− were
positively correlated with those of PTCH (r=0.167, P=0.006) and
Gli1 (r=0.185, P=0.003) (Table
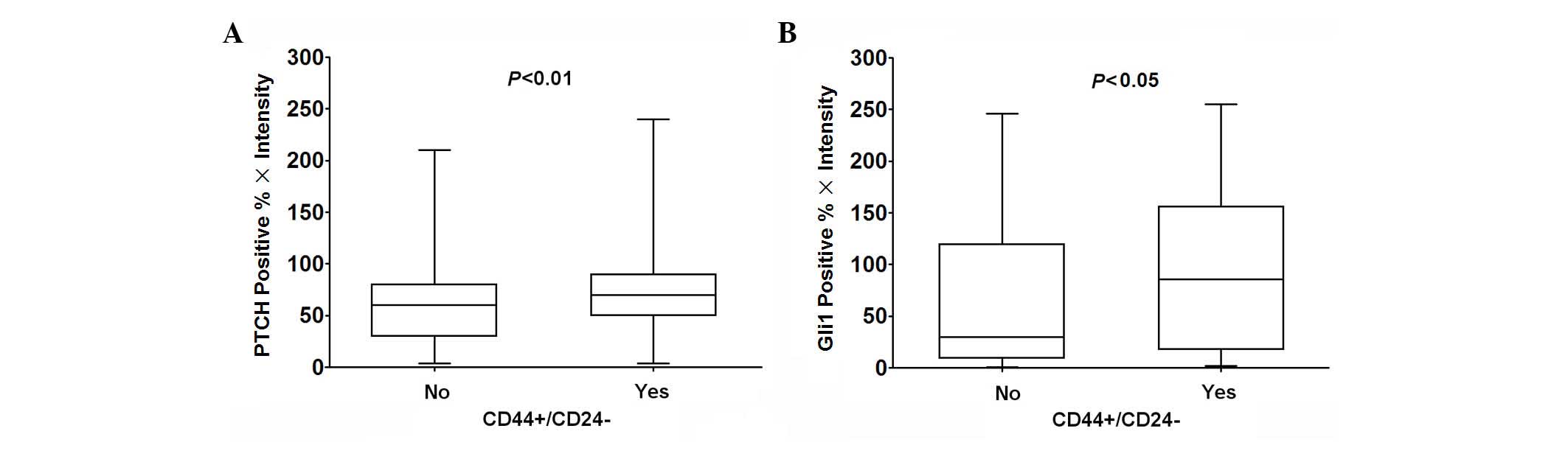
II). Compared with breast cancer with a
non-CD44+/CD24− phenotype, PTCH and Gli1
expression was significantly increased in breast cancer tissues
with the CD44+/CD24− phenotype (Mann-Whitney
U test; P<0.01, 0.05; Fig.
3).
 | Table II.Association of the expression of PTCH
or Gli1 with the expression of CD44+/CD24− in
266 breast cancer tissues. |
Table II.
Association of the expression of PTCH
or Gli1 with the expression of CD44+/CD24− in
266 breast cancer tissues.
|
|
|
CD44+/CD24− |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | No. of cases
(%) | Yes (%) | No (%) | P-value |
|---|
| PTCH |
|
|
|
|
|
Negative | 109 (41.0) | 30 (30.3) | 79 (47.3) | 0.006 |
|
Positive | 157 (59.0) | 69 (69.7) | 88 (52.7) |
|
| Gli1 |
|
|
|
|
|
Negative | 126 (47.4) | 35 (35.4) | 91 (54.5) | 0.003 |
|
Positive | 140 (52.6) | 64 (64.6) | 76 (45.5) |
|
Association of the expression of PTCH,
Gli1, and CD44/CD24 with clinicopathological characteristics of
breast cancer patients
The present study subsequently examined the
association of PTCH, Gli1 and CD44/CD24 expression with the
clinicopathological characteristics of patients with breast cancer
(Table III). PTCH expression was
associated with larger tumors (>2.0 cm; P=0.002), lymph node
metastasis (P=0.003) and Grade II–III tumors (P=0.012); Gli1
expression was associated with larger tumors (>2.0 cm; P=0.028),
lymph node metastasis (P=0.024), invasive lobular carcinoma
(P=0.003) and Grade II–III tumors (P=0.001). Combined expression of
PTCH and Gli1 was associated with larger tumors (>2.0 cm;
P=0.001), lymph node metastasis (P=0.003), invasive lobular
carcinoma (P=0.016) and Grade II–III tumors (P<0.001).
CD44+/CD24− expression was associated with
age (≤50 years old; P=0.014), premenopausal state (P=0.011) and
lymph node metastasis (P=0.032).
 | Table III.Association of the expression of Gli1
and PTCH with the clinicopathological features in breast cancer
patients with the CD44+/CD24− phenotype. |
Table III.
Association of the expression of Gli1
and PTCH with the clinicopathological features in breast cancer
patients with the CD44+/CD24− phenotype.
|
|
| PTCH(+) | Gli1(+) |
CD44+/CD24− |
PTCH(+)/Gli1(+) |
PTCH(+)/CD44+/CD24− |
Gli1(+)/CD44+/CD24− |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Clinicopathological
feature | No. of cases
(%) | Positive n
(%)a | Pb | Positive n
(%)a | Pb | Positive n
(%)a | Pb | Positive n
(%)a | Pb | Positive n
(%)a | Pb | Positive n
(%)a | Pb |
|---|
| Age |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ≤50
years | 138 (51.9) | 82 (59.4) | 0.891 | 70 (50.7) | 0.518 | 61 (44.2) | 0.014 | 51 (37.0) | 0.927 | 40 (29.0) | 0.184 | 36 (26.1) | 0.422 |
| >50
years | 128 (48.1) | 75 (58.6) |
| 70 (54.7) |
| 38 (29.7) |
| 48 (37.5) |
| 28 (21.9) |
| 28 (21.9) |
|
| Menopausal
state |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Premenopausal | 148 (55.6) | 85 (57.4) | 0.555 | 79 (53.4) | 0.785 | 65 (43.9) | 0.011 | 52 (35.1) | 0.431 | 40 (27.0) | 0.540 | 40 (27.0) | 0.205 |
|
Postmenopausal | 118 (44.4) | 72 (61.0) |
| 61 (51.7) |
| 34 (28.8) |
| 47 (39.8) |
| 28 (23.7) |
| 24 (20.3) |
|
| Tumor size |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ≤2.0
cm | 145 (61.4) | 77 (53.1) | 0.002 | 68 (46.9) | 0.028 | 52 (35.9) | 0.816 | 43 (29.7) | 0.001 | 33 (22.8) | 0.122 | 31 (21.4) | 0.209 |
| >2.0
cm | 91 (38.6) | 67 (73.6) |
| 56 (61.5) |
| 34 (37.4) |
| 46 (50.5) |
| 29 (31.9) |
| 26 (28.6) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Negative | 167 (62.8) | 87 (52.1) | 0.003 | 79 (47.3) | 0.024 | 54 (32.3) | 0.032 | 51 (30.5) | 0.003 | 33 (19.8) | 0.005 | 29 (17.4) | 0.001 |
|
Positive | 99 (37.2) | 70 (70.7) |
| 61 (61.6) |
| 45 (45.5) |
| 48 (48.5) |
| 35 (35.4) |
| 35 (35.4) |
|
| Histologic
type |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Invasive ductal carcinoma | 232 (87.2) | 134 (57.8) | 0.274 | 114 (49.1) | 0.003 | 84 (36.2) | 0.373 | 80 (34.5) | 0.016 | 56 (24.1) | 0.164 | 51 (22.0) | 0.038 |
|
Invasive lobular
carcinoma | 34 (12.8) | 23 (67.6) |
| 26 (76.5) |
| 15 (44.1) |
| 19 (55.9) |
| 12 (35.3) |
| 13 (38.2) |
|
| Histological
grade |
|
|
|
|
|
|
|
|
|
|
|
|
|
| I | 42 (21.2) | 18 (42.9) | 0.012 | 14 (33.3) | 0.001 | 13 (31.0) | 0.300 | 7 (16.7) |
<0.001 | 5 (11.9) | 0.020 | 5 (11.9) | 0.033 |
| II | 124 (62.6) | 82 (66.1) |
| 66 (53.2) |
| 55 (44.4) |
| 51 (41.1) |
| 42 (33.9) |
| 35 (28.2) |
|
|
III | 32 (16.2) | 23 (71.9) |
| 25 (78.1) |
| 14 (43.8) |
| 20 (62.5) |
| 11 (34.4) |
| 12 (37.5) |
|
Table III
summarizes the association of PTCH and Gli1 expression with the
clinicopathological features in breast cancer patients with the
CD44+/CD24− phenotype. In tumors with the
CD44+/CD24− phenotype, PTCH expression was
associated with lymph node metastasis (P=0.005) and Grade II–III
tumors (P=0.020) (Table III);
and Gli1 expression was associated with invasive lobular carcinoma
(P=0.038), lymph node metastasis (P=0.001) and Grade II–III tumors
(P=0.033).
Association of the expression of PTCH,
Gli1 and CD44/CD24 with the survival of patients with breast
cancer
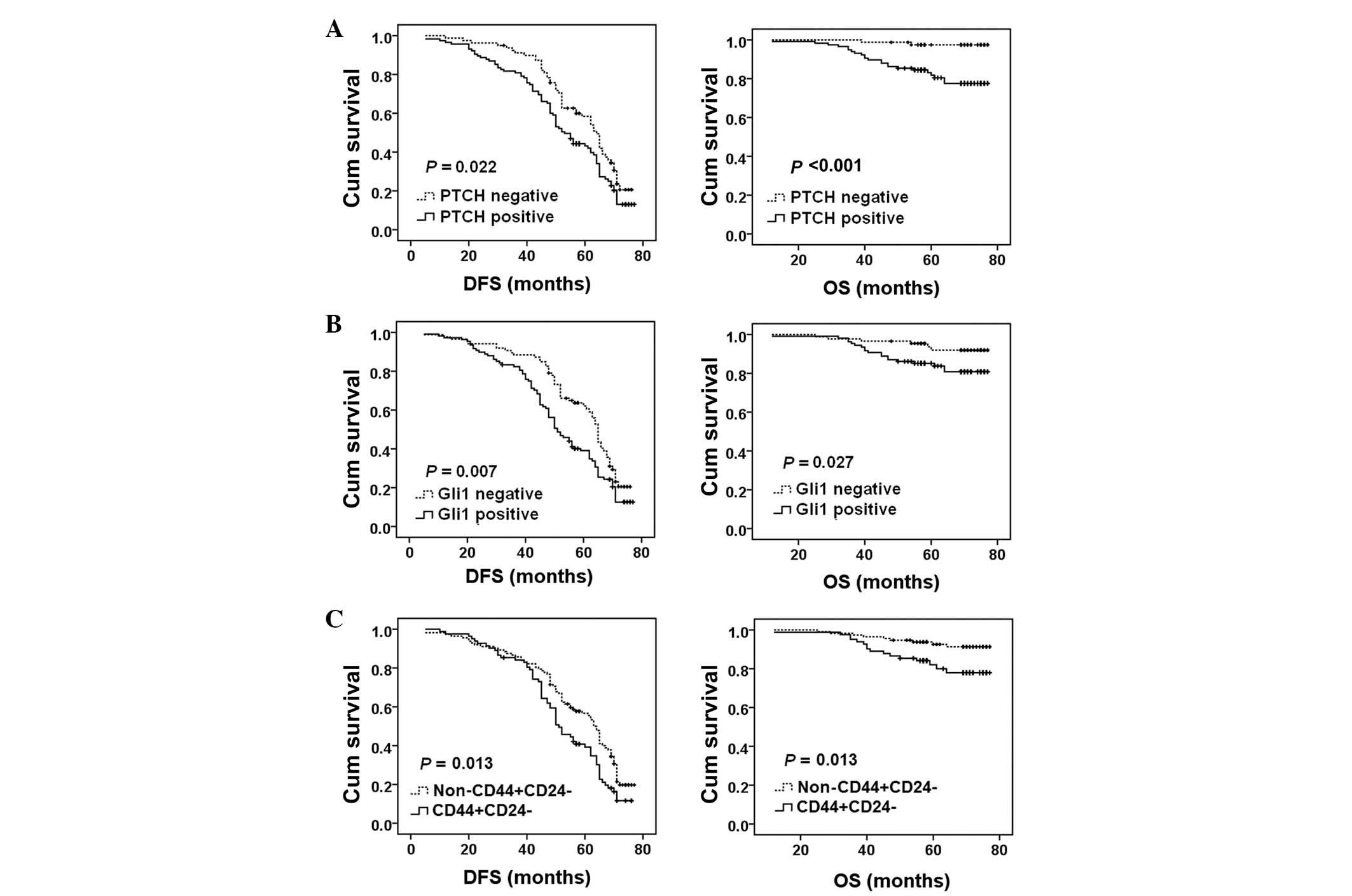
The present study performed a Kaplan-Meier analysis
to evaluate the association between the expression of PTCH, Gli1
and CD44/CD24 and the DFS or OS in 195 patients with breast cancer
that were treated with chemotherapy. PTCH expression was
significantly associated with a shorter DFS (P=0.022) and OS
(P<0.001) (Fig. 4A). Gli1
expression was significantly associated with a shorter DFS
(P=0.007) and OS (P=0.027) (Fig.
4B). CD44+/CD24− expression was
significantly associated with a shorter DFS (P=0.013) and OS
(P=0.013) (Fig. 4C).
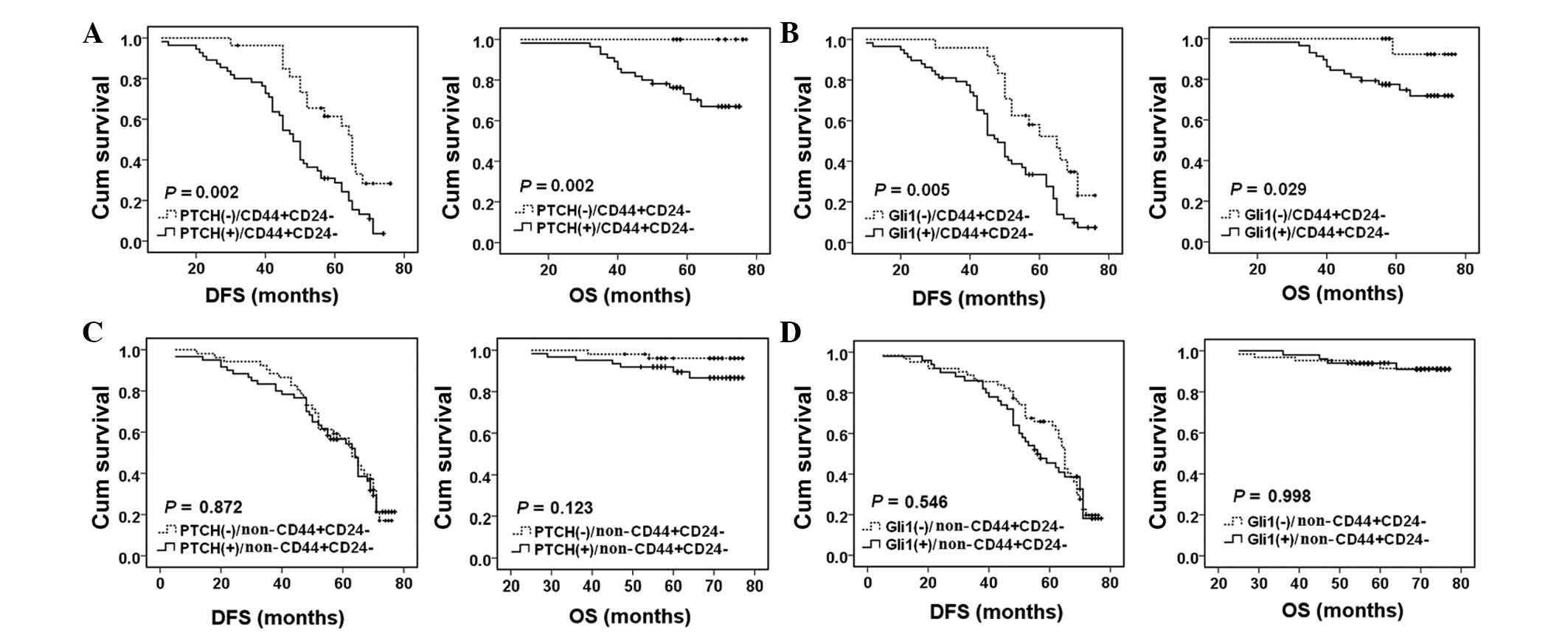
The present study also investigated the association
of the expression of PTCH and Gli1 with the OS or DFS in breast
cancer patients with various CD44/CD24 phenotypes. In patients with
the CD44+/CD24− phenotype, PTCH expression
was significantly associated with a shorter DFS (P=0.002) and OS
(P=0.002) (Fig. 5A). In addition,
Gli1 expression was significantly associated with a shorter DFS
(P=0.005) and OS (P=0.029) (Fig.
5B). However, in patients without the
CD44+/CD24− phenotype, PTCH or Gli1
expression was not significantly associated with OS or DFS
(P>0.05, Fig. 5C and D).
Univariate Cox regression analysis was performed to
evaluate the impact of each clinicopathological variable on the OS
and DFS in 195 patients with breast cancer treated with
chemotherapy (Table IV). The
univariate analysis identified that tumor size and histological
grade were significantly associated with the DFS in patients with
breast cancer. Lymph node metastasis and histological grade were
significantly associated with the OS in patients with breast
cancer. In addition, PTCH, Gli1 and
CD44+/CD24− expression was significantly
associated with a shorter DFS and OS in patients with breast
cancer. Furthermore, the expression of PTCH or Gli1 was
significantly associated with a shorter DFS in breast cancer
patients with the CD44+/CD24− phenotype
(Table IV). Furthermore,
multivariate Cox regression analysis demonstrated that the
expression of PTCH and the CD44+/CD24−
phenotype were independent prognostic factors for a shorter DFS in
patients with breast cancer (Table
V).
 | Table IV.Univariate Cox regression analysis of
the association between clinicopathological features and DFS and OS
in 195 patients with breast cancer treated with chemotherapy. |
Table IV.
Univariate Cox regression analysis of
the association between clinicopathological features and DFS and OS
in 195 patients with breast cancer treated with chemotherapy.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | RR (95% CI) | P | RR (95% CI) | P |
|---|
| Age, years |
|
|
|
|
|
≤50/>50 | 1.046
(0.753~1.453) | 0.788 | 1.623
(0.737~3.576) | 0.229 |
| Menopausal
state |
|
|
|
|
|
Pre-menopause/Post-menopause | 1.151
(0.827~1.603) | 0.404 | 1.902
(0.863~4.191) | 0.111 |
| Tumor size, cm |
|
|
|
|
|
≤2.0/>2.0 | 1.486
(1.045~2.115) | 0.028 | 0.650
(0.254~1.661) | 0.368 |
| Lymph node
metastasis |
|
|
|
|
|
No/yes | 1.248
(0.893~1.744) | 0.194 | 3.081
(1.360~6975) | 0.007 |
| Histologic
type |
|
|
|
|
|
Invasive ductal
carcinoma/Invasive lobular carcinoma | 1.208
(0.767~1.903) | 0.416 | 2.060
(0.822~5.157) | 0.123 |
| Histological
grade |
|
|
|
|
|
I/II/III | 1.529
(1.082~2.162) | 0.016 | 2.117
(1.000~4.478) | 0.046 |
| PTCH
expression |
|
|
|
|
|
Positive/negative | 1.466
(1.046~2.056) | 0.027 | 8.827
(2.080~37.460) | 0.003 |
| Gli1
expression |
|
|
|
|
|
Positive/negative | 1.564
(1.119~2.186) | 0.009 | 2.692
(1.075~6.742) | 0.034 |
|
CD44+/CD24− |
|
|
|
|
|
Yes/no | 1.501
(1.079~2.088) | 0.016 | 2.709
(1.196~6.138) | 0.017 |
|
CD44+/CD24− patient
group |
|
|
|
|
| PTCH
(positive/negative) | 2.286
(1.311~3.984) | 0.004 | 42.080
(0.607~2917) | 0.084 |
| Gli1
(positive/negative) | 2.177
(1.221~3.881) | 0.008 | 6.928
(0.915~52.466) | 0.061 |
 | Table V.Multivariate Cox regression analysis
of clinicopathological features correlated with DFS and OS in 195
patients with breast cancer treated with chemotherapy. |
Table V.
Multivariate Cox regression analysis
of clinicopathological features correlated with DFS and OS in 195
patients with breast cancer treated with chemotherapy.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | RR (95% CI) | P | RR (95% CI) | P |
|---|
| Age, years |
|
|
|
|
|
≤50/>50 | 0.895
(0.450~1.780) | 0.751 | 1.796
(0.158~20.473) | 0.637 |
| Menopausal
state |
|
|
|
|
|
Pre-menopause/Post-menopause | 1.659
(0.832~3.306) | 0.150 | 2.280
(0.225~23.130) | 0.486 |
| Tumor size, cm |
|
|
|
|
|
≤2.0/>2.0 | 0.905
(0.528~1.554) | 0.718 | 0.249
(0.050~1.235) | 0.089 |
| Lymph node
metastasis |
|
|
|
|
|
No/yes | 0.615
(0.344~1.101) | 0.102 | 0.706
(0.121~4.107) | 0.698 |
| Histologic
type |
|
|
|
|
|
Invasive ductal
carcinoma/Invasive lobular carcinoma | 1.478
(0.731~2.985) | 0.277 | 1.796
(0.158~20.473) | 0.494 |
| Histological
grade |
|
|
|
|
|
I/II/III | 1.637
(0.958~2.796) | 0.071 | 2.822
(0.700~11.372) | 0.145 |
| PTCH
expression |
|
|
|
|
|
Positive/negative | 2.018
(1.164~3.499) | 0.012 | 4.417
(0.701~27.810) | 0.114 |
| Gli1
expression |
|
|
|
|
|
Positive/negative | 1.061
(0.612~1.842) | 0.832 | 1.783
(0.349~9.123) | 0.487 |
|
CD44+/CD24− |
|
|
|
|
|
Yes/no | 1.888
(1.140~3.126) | 0.014 | 1.909
(0.459~7.942) | 0.374 |
Discussion
It is generally believed that breast CSCs contribute
to chemoresistance, recurrence and metastasis in breast cancer
(4,16,26).
The Hh signaling pathway has been reported to be important for
maintaining the stemness of CSCs (10,13,27).
In addition, the Hh signaling pathway has been demonstrated to be
activated in patients with breast cancer, and inhibition of Hh
signaling reduces the growth of breast cancer cells in vitro
(28). However, it remains to be
elucidated as to whether the Hh signaling pathway affects breast
CSCs in patients with breast cancer. In the present study, PTCH,
Gli1 and CD44/CD24 expression was detected in samples from 266
patients with breast cancer. The results demonstrated that the
expression of PTCH and Gli1, which are the two main components of
the Hh signaling pathway, was higher in breast cancer patients with
the CD44+/CD24− phenotype, as compared with
those with a non-CD44+/CD24− phenotype. The
expression of PTCH and Gli1 was positively correlated with
CD44+/CD24− expression, thus suggesting that
the Hh signaling pathway is activated in breast CSCs. Furthermore,
the expression of PTCH and Gli1 was associated with poor survival
in breast cancer patients with the
CD44+/CD24− phenotype. These findings
suggested that Hh signaling activation in breast CSCs may
contribute to poor outcomes in patients with breast cancer.
It has previously been reported that PTCH expression
is associated with lymph node metastasis and a greater histological
grade in patients with breast cancer (29). Tao et al (30) demonstrated that Gli1 was
significantly upregulated in breast cancer patients with lymph node
metastasis. Furthermore, Xuan et al reported that Gli1
expression was correlated with lymph node metastasis (31). Similarly, the present study
demonstrated that the expression of PTCH and Gli1 was associated
with lymph node metastasis. These findings suggested that the Hh
signaling pathway is important for lymph node metastasis in breast
cancer. Furthermore, the expression of PTCH and Gli1 was more
positively associated with lymph node metastasis in tumors with a
CD44+/CD24− phenotype, further suggesting
that the Hh signaling pathway is important for CSC-mediated
metastasis in breast cancer. It has previously been reported that
breast cancer cells with the CD44+/CD24−
phenotype express high levels of metastasis-associated genes and
exhibit enhanced metastasis (17,32,33).
In addition, Lin et al (19) revealed that the expression of
CD44+/CD24− was associated with lymph node
metastasis in breast cancer patients with invasive ductal
carcinoma. Since the present study demonstrated that the expression
of PTCH and Gli1 was significantly associated with lymph node
metastasis in tumors with a CD44+/CD24−
phenotype, it may be suggested that the Hh signaling pathway is
essential for CSC-induced lymph node metastasis in patients with
breast cancer.
The present study also demonstrated that the
expression of PTCH and Gli1 was associated with a shorter DFS and
OS in patients with breast cancer, thus suggesting that the Hh
signaling pathway contributes to poor outcomes in patients with
breast cancer. Consistent with these findings, Ramaswamy et
al (34) reported that Gli1
overexpression was associated with a shorter DFS and OS in patients
with breast cancer. Notably, the present study revealed that the
expression of PTCH and Gli1 was significantly associated with a
shorter DFS and OS in breast cancer patients with the
CD44+/CD24− phenotype, but not in patients
without the CD44+/CD24− phenotype. It has
been reported that the CD44+/CD24− phenotype
contributes to poor prognosis in patients with breast cancer
(19,20). The present findings indicated that
the Hh signaling pathway in CSCs contributes to poor prognosis of
breast cancer. Furthermore, a univariate Cox regression analysis
identified that the expression of PTCH, Gli1 and
CD44+/CD24− was associated with poor
prognosis of patients with breast cancer. The multivariate analysis
identified that PTCH expression and the
CD44+/CD24− phenotype were independent
prognostic factors for poor outcome in patients with breast
cancer.
In conclusion, the present study investigated PTCH
and Gli1 expression, and the CD44+/CD24−
phenotype in patients with breast cancer, and analyzed the
association of their expression with clinicopathological
characteristics and prognosis. The results demonstrated that the
expression of PTCH and Gli1 was associated with lymph node
metastasis and a worse clinical outcome in patients with breast
cancer, particularly those with the
CD44+/CD24− phenotype. This study suggests
that the Hh signaling pathway in CSCs may be associated with a poor
prognosis in patients with breast cancer. Therefore, inhibition of
the Hh signaling pathway may be an effective therapeutic strategy
for the inhibition of breast CSCs, thus preventing breast cancer
recurrence and metastasis.
Acknowledgements
The present study was supported by the Program for
Liaoning Innovative Research Team in University, LNIRT, China
(grant no. LT2014016), the Program for Liaoning Excellent Talents
in University, China (grant no. LJQ2014084), and the S&T
Projects in Shenyang, China (grant no. F14-232-6-05).
References
|
1
|
Al-Hajj M and Clarke MF: Self-renewal and
solid tumor stem cells. Oncogene. 23:7274–7282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sampieri K and Fodde R: Cancer stem cells
and metastasis. Semin Cancer Biol. 22:187–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ishii H, Iwatsuki M, Ieta K, Ohta D,
Haraguchi N, Mimori K and Mori M: Cancer stem cells and
chemoradiation resistance. Cancer Sci. 99:1871–1877. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Lewis MT, Huang J, Gutierrez C,
Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC,
et al: Intrinsic resistance of tumorigenic breast cancer cells to
chemotherapy. J Natl Cancer Inst. 100:672–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McDermott SP and Wicha MS: Targeting
breast cancer stem cells. Mol Oncol. 4:404–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: Paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taipale J and Beachy PA: The Hedgehog and
Wnt signalling pathways in cancer. Nature. 411:349–354. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang J and Hui CC: Hedgehog signaling in
development and cancer. Dev Cell. 15:801–812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Briscoe J and Thérond PP: The mechanisms
of Hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu S, Dontu G, Mantle ID, Patel S, Ahn
NS, Jackson KW, Suri P and Wicha MS: Hedgehog signaling and Bmi-1
regulate self-renewal of normal and malignant human mammary stem
cells. Cancer Res. 66:6063–6071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bar EE, Chaudhry A, Lin A, Fan X, Schreck
K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A and
Eberhart CG: Cyclopamine-mediated hedgehog pathway inhibition
depletes stem-like cancer cells in glioblastoma. Stem Cells.
25:2524–2533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clement V, Sanchez P, de Tribolet N,
Radovanovic I and Ruiz i Altaba A: HEDGEHOG-GLI1 signaling
regulates human glioma growth, cancer stem cell self-renewal, and
tumorigenicity. Curr Biol. 17:165–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao C, Chen A, Jamieson CH, Fereshteh M,
Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, et al:
Hedgehog signalling is essential for maintenance of cancer stem
cells in myeloid leukaemia. Nature. 458:776–779. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tanaka H, Nakamura M, Kameda C, Kubo M,
Sato N, Kuroki S, Tanaka M and Katano M: The Hedgehog signaling
pathway plays an essential role in maintaining the
CD44+CD24−/low subpopulation and the side
population of breast cancer cells. Anticancer Res. 29:2147–2157.
2009.PubMed/NCBI
|
|
15
|
Jaggupilli A and Elkord E: Significance of
CD44 and CD24 as cancer stem cell markers: An enduring ambiguity.
Clin Dev Immunol. 2012:7080362012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abraham BK, Fritz P, McClellan M,
Hauptvogel P, Athelogou M and Brauch H: Prevalence of
CD44+/CD24−/low cells in breast cancer may
not be associated with clinical outcome but may favor distant
metastasis. Clin Cancer Res. 11:1154–1159. 2005.PubMed/NCBI
|
|
18
|
Sheridan C, Kishimoto H, Fuchs RK,
Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr, Badve S and
Nakshatri H: CD44+/CD24− breast cancer cells
exhibit enhanced invasive properties: An early step necessary for
metastasis. Breast Cancer Res. 8:R592006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin Y, Zhong Y, Guan H, Zhang X and Sun Q:
CD44+/CD24− phenotype contributes to
malignant relapse following surgical resection and chemotherapy in
patients with invasive ductal carcinoma. J Exp Clin Cancer Res.
31:592012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee HE, Kim JH, Kim YJ, Choi SY, Kim SW,
Kang E, Chung IY, Kim IA, Kim EJ, Choi Y, et al: An increase in
cancer stem cell population after primary systemic therapy is a
poor prognostic factor in breast cancer. Br J Cancer.
104:1730–1738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tavassoli FA and Devilee P: World Health
Organization Classification of Tumours Pathology and Genetics
Tumours of the Breast and Female Genital Organs. IARC Press; Lyon:
2003
|
|
22
|
Bai X, Song Z, Fu Y, Yu Z, Zhao L, Zhao H,
Yao W, Huang D, Mi X, Wang E, et al: Clinicopathological
significance and prognostic value of DNA methyltransferase 1, 3a,
and 3b expressions in sporadic epithelial ovarian cancer. PLoS One.
7:e400242012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim TJ, Lee JY, Hwang TK, Kang CS and Choi
YJ: Hedgehog signaling protein expression and its association with
prognostic parameters in prostate cancer: A retrospective study
from the view point of new 2010 anatomic stage/prognostic groups. J
Surg Oncol. 104:472–479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He HC, Chen JH, Chen XB, Qin GQ, Cai C,
Liang YX, Han ZD, Dai QS, Chen YR, Zeng GH, et al: Expression of
hedgehog pathway components is associated with bladder cancer
progression and clinical outcome. Pathol Oncol Res. 18:349–355.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim BW, Cho H, Chung JY, Conway C, Ylaya
K, Kim JH and Hewitt SM: Prognostic assessment of hypoxia and
metabolic markers in cervical cancer using automated digital image
analysis of immunohistochemistry. J Transl Med. 11:1852013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu H, Patel MR, Prescher JA, Patsialou A,
Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, et al:
Cancer stem cells from human breast tumors are involved in
spontaneous metastases in orthotopic mouse models. Proc Natl Acad
Sci USA. 107:18115–18120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song Z, Yue W, Wei B, Wang N, Li T, Guan
L, Shi S, Zeng Q, Pei X and Chen L: Sonic hedgehog pathway is
essential for maintenance of cancer stem-like cells in human
gastric cancer. PLoS One. 6:e176872011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kubo M, Nakamura M, Tasaki A, Yamanaka N,
Nakashima H, Nomura M, Kuroki S and Katano M: Hedgehog signaling
pathway is a new therapeutic target for patients with breast
cancer. Cancer Res. 64:6071–6074. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Im S, Choi HJ, Yoo C, Jung JH, Jeon YW,
Suh YJ and Kang CS: Hedgehog related protein expression in breast
cancer: Gli-2 is associated with poor overall survival. Korean J
Pathol. 47:116–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tao Y, Mao J, Zhang Q and Li L:
Overexpression of Hedgehog signaling molecules and its involvement
in triple-negative breast cancer. Oncol Lett. 2:995–1001.
2011.PubMed/NCBI
|
|
31
|
Xuan Y and Lin Z: Expression of Indian
Hedgehog signaling molecules in breast cancer. J Cancer Res Clin
Oncol. 135:235–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu R, Wang X, Chen GY, Dalerba P, Gurney
A, Hoey T, Sherlock G, Lewicki J, Shedden K and Clarke MF: The
prognostic role of a gene signature from tumorigenic breast-cancer
cells. N Engl J Med. 356:217–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Charafe-Jauffret E, Ginestier C and
Birnbaum D: Breast cancer stem cells: Tools and models to rely on.
BMC Cancer. 9:2022009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X,
Shapiro CL and Majumder S: Hedgehog signaling is a novel
therapeutic target in tamoxifen-resistant breast cancer aberrantly
activated by PI3K/AKT pathway. Cancer Res. 72:5048–5059. 2012.
View Article : Google Scholar : PubMed/NCBI
|