Introduction
Gliomas are the most frequent type of primary tumor
in the brain and account for 50–60% of all brain tumors worldwide
(1). Despite advances in cancer
treatment, this statistic has not changed significantly (2,3).
Previous reports have suggested that several genes contribute to
the pathogenesis of glioma, including protein-coding genes
(4,5), microRNAs (miRNAs) (6) and long non-coding RNAs (lncRNAs)
(7,8). However, the mechanisms of the
majority of genes in glioma remain to be elucidated. Therefore, an
improved understanding of the molecular mechanisms involved in the
development, progression and metastasis of glioma is essential for
identifying novel prognostic molecular markers and designing more
individualized and effective therapeutic strategies.
lncRNAs, which are a subgroup of non-coding RNAs,
are non-protein coding transcripts with a length of >200
nucleotides (9,10). Previous reports have shown that
lncRNAs have multifunctional roles in modulating embryonic
pluripotency, differentiation, development and various diseases,
particularly in cancer (11–13).
lncRNAs may be classified according to their mode of action and
their functions in cells, including genetic imprinting (14,15),
modulating the cancer epigenome (9), serving as molecular decoys (16) and post-transcriptional regulation
(10). Accumulating evidence has
shown that lncRNA expression profiling may facilitate the diagnosis
and prognosis of human cancer, including bladder cancer (17), leukemia (18), breast cancer (9) and rectal cancer (19), suggesting that lncRNAs may serve as
effective therapeutic targets for intervention. Previous evidence
indicates that lncRNAs are important in the pathogenesis of glioma,
including HOTAIR (20), H19
(21) and Linc-POU3F3 (8). Although thousands of lncRNAs have
been annotated, only a few lncRNAs have been functionally
characterized in glioma.
Nuclear factor IA (NFIA), a member of the NFI
family, is essential in glial development in the central nervous
system; it specifies glial identity, maintains glial progenitors
and regulates astrocyte differentiation (22,23).
Increasing evidence has shown that NFIA is involved in and may be
central in a variety of biological processes through complicated
mechanisms in glioma (24,25). For example, Glasgow et al
(26) first characterized the
miR-223/NFIA axis in glioma. The RP5-833A20.1 lncRNA is located in
intron 2 of the NFIA gene and has an opposite transcription
direction to NFIA. Our previous investigations showed that NFIA is
important in the progression of atherosclerosis, which is regulated
by RP5-833A20.1 (27). However,
whether NFIA is targeted by RP5-833A20.1 in glioma remains to be
elucidated.
The present study aimed to examine the role of
RP5-833A20.1 in glioma and to investigate whether NFIA is targeted
by RP5-833A20.1. It was found that the expression of RP5-833A20.1
was decreased, whereas the expression of NFIA was increased in
glioma tissues, compared with adjacent normal tissues. Furthermore,
the present study investigated the effects of the expression of
RP5-833A20.1 on U251 cell phenotype in vitro via a
gain-of-function experiment. It was shown that the overexpression
of RP5-833A20.1 suppressed the expression of NFIA, promoted the
expression of miR-382-5p and enhanced the methylation level of NFIA
in the U251 cells. These results indicated that RP5-833A20.1 may be
a novel therapeutic target for glioma.
Materials and methods
Human samples
Samples of human glioma and adjacent healthy tissues
were obtained from 20 patients (11 male and 9 female, age,
44.6±18.4) undergoing surgery at the Department of Neurosurgery,
Nanfang Hospital of Southern Medical University (Guangzhou, China).
None of the patients had received prior chemotherapy or biotherapy,
and diagnoses were confirmed pathologically in all cases. Central
glioma tissues and adjacent nontumor tissues, located 2 cm from the
center in the same patient were collected. All specimens were snap
frozen at the time of surgery and stored in liquid nitrogen. The
samples were collected with informed consent from patients and
ethical consent was granted from the Committees for Ethical Review
of Research involving Human Subjects of Southern Medical
University.
RNA extraction and RT-qPCR assays
Total RNA from the cultured cells or human tissues
was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in accordance with the
manufacturer's protocol. Prior to the addition of TRIzol, the
tissues were ground using liquid nitrogen. The miRNAs involved in
putatively regulating NFIA were detected using the All-in-One™
miRNA RT-qPCR kit (GeneCopoeia, Rockville, MD, USA) according to
the manufacturer's protocol in 20 µl reaction volumes. Prior to the
addition of TRIzol, the tissues were ground with liquid nitrogen.
cDNA was generated from 1000 ng of total RNA and 2 µl cDNA were
used for PCR analysis. The amplification conditions were as
follows: Stage 1, 95°C for 30 sec; stage 2, 95°C for 3 sec and then
60°C for 30 sec, 40 cycles; stage 3, 95°C for 15 sec, 60°C for 1
min, 95°C for 15 sec, 60°C for 15 sec. The RT-qPCR was performed on
an ABI 7500 Fast Real Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The expression of U6 RNA was used as an
endogenous control (28). The mRNA
levels were evaluated using the ABI 7500 Fast Real-Time PCR system
with SYBR green detection chemistry (Takara Bio, Inc., Shiga,
Japan). The expression of GAPDH was used as an internal control
(29). Quantitative measurements
were determined using the ΔΔCq method (30). All samples were measured in
triplicate and the mean value was considered for comparative
analysis. The sequences of the primers used are listed in Table I.
 | Table I.Oligonucleotide sequences used in the
present study. |
Table I.
Oligonucleotide sequences used in the
present study.
| Gene | Sequence |
|---|
| NFIA-F |
5′-AGGTCTTTACCCAGCACATCCTC-3′ |
| NFIA-R |
5′-TCCACTTGACTGACTGCCACTTC-3′ |
| GAPDH-F |
5′-GCACCGTCAAGGCTGAGAAC-3′ |
| GAPDH-R |
5′-TGGTGAAGACGCCAGTGGA-3′ |
| RP5-833A20.1-F |
5′-CATGAGCCACAGCAGTAAGC-3′ |
| RP5-833A20.1-R |
5′-GGAGAACATGGCAGAAATCA-3′ |
|
RP5-833A20.1-RT |
5′-GAAAGGAAGGCATCCAACTT-3′ |
| U6-F |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R |
5′-AACGCTTCACGAATTTGCGT-3′ |
| U6-RT |
5′-AACGCTTCACGAATTTGCGT-3′ |
| BSP-NFIA-F |
5′-AGGATTTTTAATTTTTGGTTTATATTAGTG-3′ |
| BSP-NFIA-R |
5′-AAAAAAAACCTTACCCCCTTCT-3′ |
Cell line and culture conditions
U251 cells were obtained from American Type Culture
Collection (Manassas, VA, USA) and maintained according to the
supplier's recommendation. The U251 human glioma cells were
maintained in Dubecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) with high glucose and sodium
pyruvate, supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) and antibiotics (100 U/ml
penicillin and 100 mg/ml streptomycin). The cells were maintained
in a humidified incubator at 37°C with 5% CO2.
Lentiviral construction and cell
transfection
Packed empty lentivirus (LV) vectors (Landbiology,
Guangzhou, China) with green fluorescent protein (GFP; LV-Mock), an
LV-mediated lncRNA RP5-833A20.1-overexpression vector
(LV-RP5-833A20.1), and an LV-mediated lncRNA RP5-833A20.1-knockdown
vector (LV-siRNA-RP5-833A20.1) with GFP were prepared as described
previously (31). The U251 cells
were incubated in 6-well plates for 24 h at a density of
2ⅹ106 cells/well and then were transfected using DMEM
and polybrene reagents (Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) according to the manufacturer's protocol at a multiplicity of
infection of 1. The U251 human glioma cells were maintained in
Dubecco's modified Eagle's medium (DMEM) with high glucose and
sodium pyruvate, supplemented with 10% FBS and antibiotics (100
U/ml penicillin and 100 mg/ml streptomycin), and plus 2 µg/ml
puromycin for 2 weeks to gain stable clones. The cells were
maintained in a humidified incubator at 37°C with 5%
CO2. The expression level of RP5-833A20.1 was determined
using RT-qPCR analysis.
Cell proliferation assays
For the cell proliferation assays, the U251 cells
were seeded in 96-well plates at the density of 2×103
cells/well. Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto,
Japan) solution (10 µl) was added to each well and the cells were
incubated for 1.5 h at 37°C. The absorbance at 450 nm was recorded
using a Thermo Multiskan MK3 reader (Thermo Fisher Scientific,
Inc.). A total of five replicate wells were set up in each group
and five independent experiments were performed.
Cell invasion assays
For the invasion assays, 5×104 cells in
200 ml serum-free medium were seeded into the upper chamber of a
Transwell insert (8-mm pore size; EMD Millipore, Billerica, MA,
USA) coated with Matrigel. The lower chamber was filled with 600
liters medium containing 10% FBS. Following incubation in a
humidified incubator at 37°C with 5% CO2 for 12 h, the
cells remaining on the upper membrane were removed with a cotton
swab. The cells, which had migrated or invaded through the membrane
were stained with methanol and 0.1% crystal violet, images were
captured, and the cells were counted using a BX51 microscope
(Olympus, Tokyo, Japan). Invasion was assessed by counting the
number of stained cell nuclei from 10 randomly selected fields per
filter in each group (magnification, ×400), with the cell counts
expressed as the mean number of cells per field of view. Three
independent experiments were performed in triplicate.
Flow cytometric analysis of
apoptosis
The U251 cells were harvested 48 h following
transfection and resuspended 100 µl of the solution (1 ×
105 cells) to a 5 ml culture tube and add 5 µl of FITC
Annexin V (BD Biosciences, San Jose, CA, USA) and 5 µl PI (BD
Biosciences). Next, the cells were gently vortexed and incubated
for 15 min at 25°C in the dark, 400 µl of 1X Binding Buffer was
added to each tube. The results were analyzed using a flow
cytometer (FACScan; BD Biosciences) within 1 h. Cell Quest version
6.0 (BD Biosciences) was used to analyze the results. All samples
were assayed in triplicate.
Cell cycle distribution
The treated U251 cells were trypsinized, fixed in
70% ethanol, washed twice with phosphate-buffered saline (PBS), and
then labeled with propidium iodide (Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China) in the presence of 30 KU/ml RNase A (KeyGen
Biotech Co., Ltd.) for 30 min in the dark (50 g/ml). The samples
were run on a FACScan flow cytometer (BD Biosciences), and the
percentages of cells within each phase of the cell cycle were
analyzed using Cell Quest version 6.0.
Western blot analysis
The cells were washed twice with ice-cold PBS and
the cell lysates were harvested using a KeyGen Biotech Whole Cell
Lysis Assay kit (KeyGen Biotech Co., Ltd.) according to the
manufacturer's protocol. The proteins were quantified by KeyGEN
Bradford protein quantitation assay (KeyGen Biotech Co., Ltd.) and
50 µg protein per lane was fractionated by 12% SDS-PAGE,
transferred onto a PVDF membrane, blocked with 5% bovine serum
albumin (Beyotime Institute of Biotechnology, Haimen, China) for 1
h at room temperature and immunoblotted with primary antibodies
overnight at 4°C. Following incubation with the corresponding
secondary antibodies conjugated to horseradish peroxidase, the
signals of the membranes were detected by chemiluminescence western
blotting substrate (Thermo Fisher Scientific, Inc.). The band
intensity of western blotting and the normalization were analyzed
using ImageJ version 1.6.0 software (National Institutes of Health,
Bethesda, MD, USA). The primary antibodies used were obtained from
Abcam (Cambridge, UK) as follows: NFIA (cat. no. ab41851; 1:1,000);
p53 (cat. no. ab31333; 1:1,000); p21 (cat. no. ab47452; 1:1,000);
PAI-1 (cat. no. ab66705; 1:1,000) and β-actin (cat. no. ab8227;
1:2,000). The secondary antibody used was goat anti-rabbit IgG
(cat. no. bs-0295G; 1:5,000; BIOSS, Beijing, China).
Bisulfite PCR sequencing (BSP)
To assess the methylation level of the NFIA gene
promoter, U251 cells with knocked down RP5-833A20.1 or transfected
with a short hairpin negative control vectors (LAND) were used.
Genomic DNA was isolated using a Promega wizard genomic DNA
purification kit (Promega, Madison, MA, USA) according to the
manufacturer's protocol. DNA modification was performed using an
EpiTect Bisulfite kit (Qiagen, Shanghai, China) to convert
unmethylated cytosines to uracils. The PCR was performed using the
aforementioned protocol. Following BSP, the PCR products were used
for cloning and sequencing. The sequences of the BSP primers are
listed in Table I.
Statistical analysis
All experiments were performed three times. Data are
presented as the mean ± standard deviation and analyzed using SPSS
13.0 software (SPSS, Inc., Chicago, IL, USA) with Student's t-test
or one-way analysis of variance. P<0.05 was considered to
indicate a statistically significant difference.
Results
RP5-833A20.1 is downregulated and NFIA
is upregulated in glioma
In our previous study, it was shown that the
RP5-833A20.1/miR-382-5p/NFIA pathway is essential for the
regulation cardiovascular disease (27). Several studies have shown that NFIA
is important in glioma (25,26).
Thus, the present study hypothesized that RP5-833A20.1 may also
function in glioma by regulating the expression of NFIA. To confirm
this, the present study examined the RNA expression levels of
RP5-833A20.1 and NFIA in 20 paired glioma and adjacent nontumor
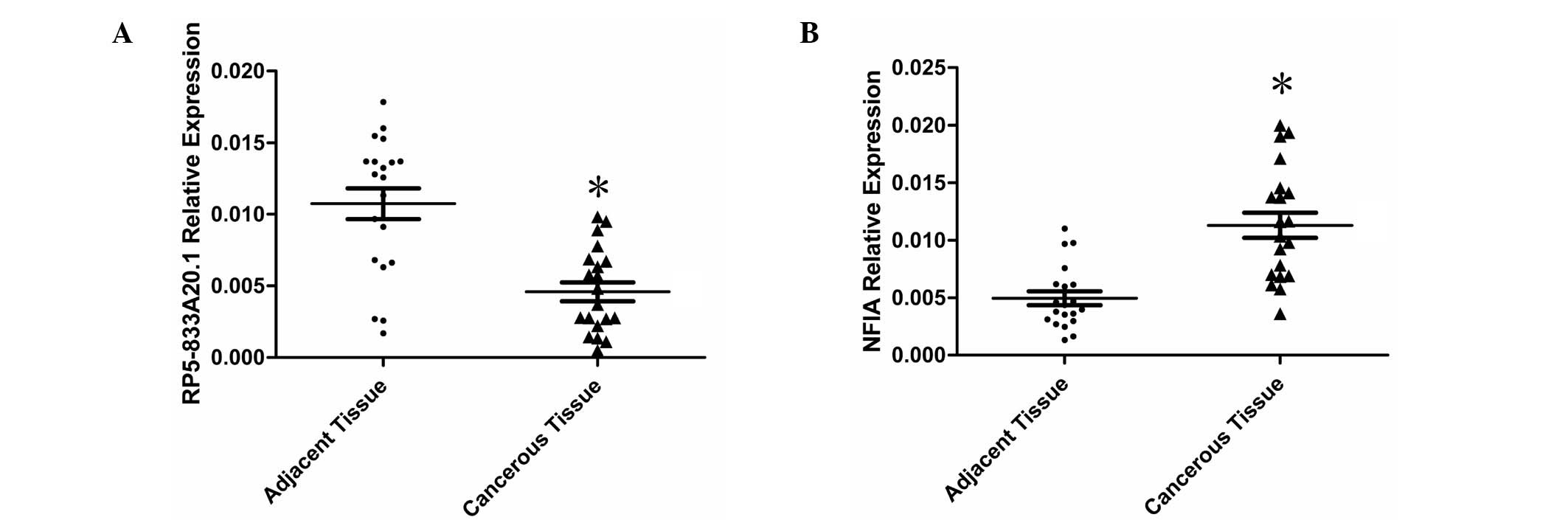
tissue samples. The expression of RP5-833A20.1 was significantly
downregulated (P<0.01) in 80% (16/20) of the cancerous tissues,
compared with the paired adjacent nontumor tissues (Fig. 1A). The expression of NFIA was
significantly upregulated (P<0.01) in 90% (18/20) of the
cancerous tissues, compared with the paired adjacent nontumor
tissues (Fig. 1B).
RP5-833A20.1 suppresses the expression
of NFIA and promotes the expression of miR-382-5p in U251
cells
In our previous study, it was found that
RP5-833A20.1 suppressed the expression of NFIA by promoting the
expression of miR-382-5p in THP-1 macrophages (27). The present study examined whether
RP5-833A20.1 regulates the expression of NFIA by promoting
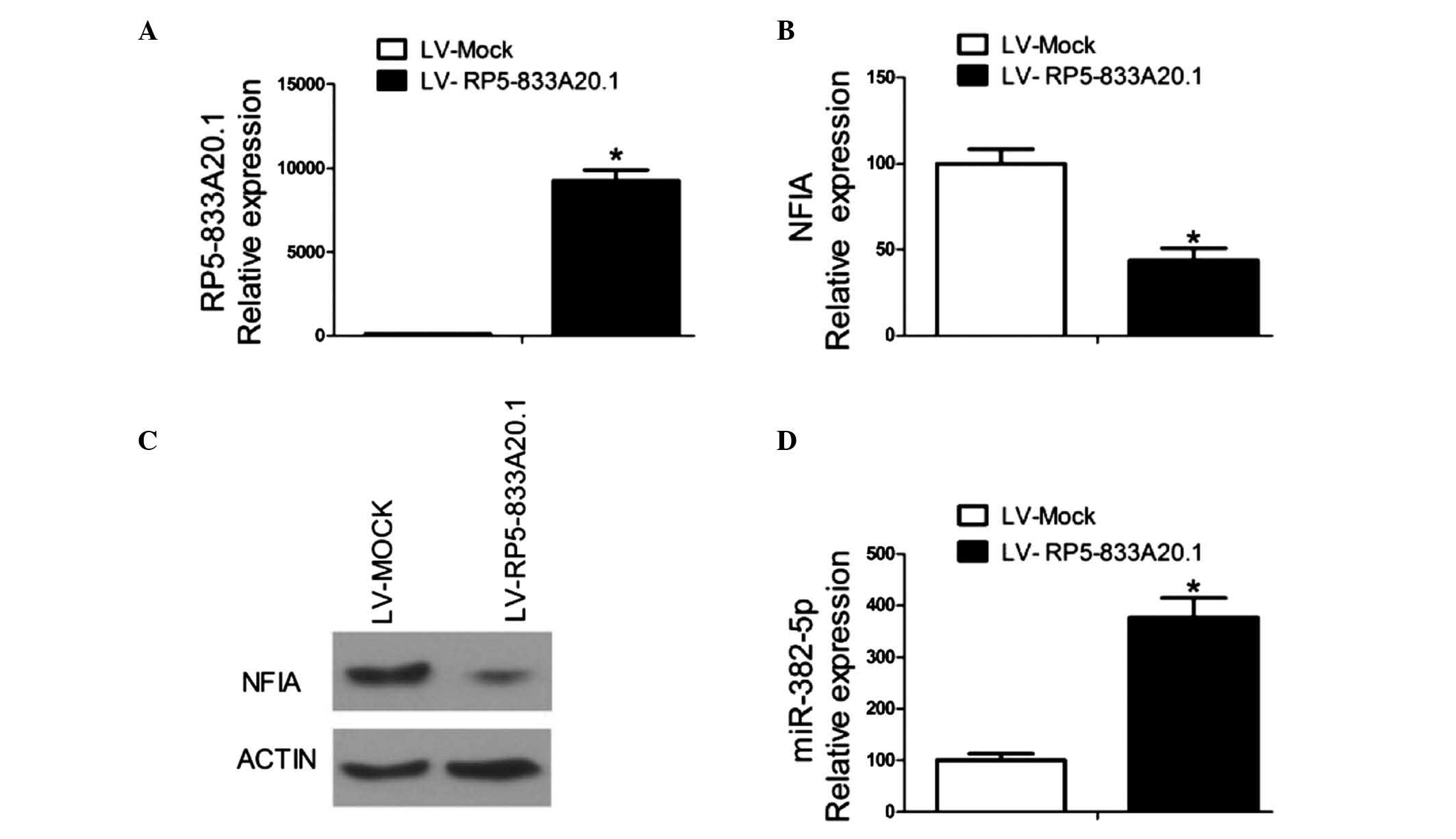
miR-382-5p in U251 cells. Lentivirus-mediated overexpression of
RP5-833A20.1 in the U251 cells(Fig.
2A) markedly inhibited the mRNA and protein expression levels
of NFIA (Fig. 2B and C), whereas
overexpression of RP5-833A20.1 increased the expression of miR-382
in the U251 cells (Fig. 2D). These
results indicated that RP5-833A20.1 suppressed the expression of
NFIA in the U251 cells and that miR-382-5p may be involved in this
process.
RP5-833A20.1 enhances the methylation
level of the NFIA promoter
Increasing evidence has confirmed that lncRNAs can
regulate the DNA methylation of protein-coding genes during the
development of disease (11,32,33).
To assess the role of RP5-833A20.1 in the regulation of DNA
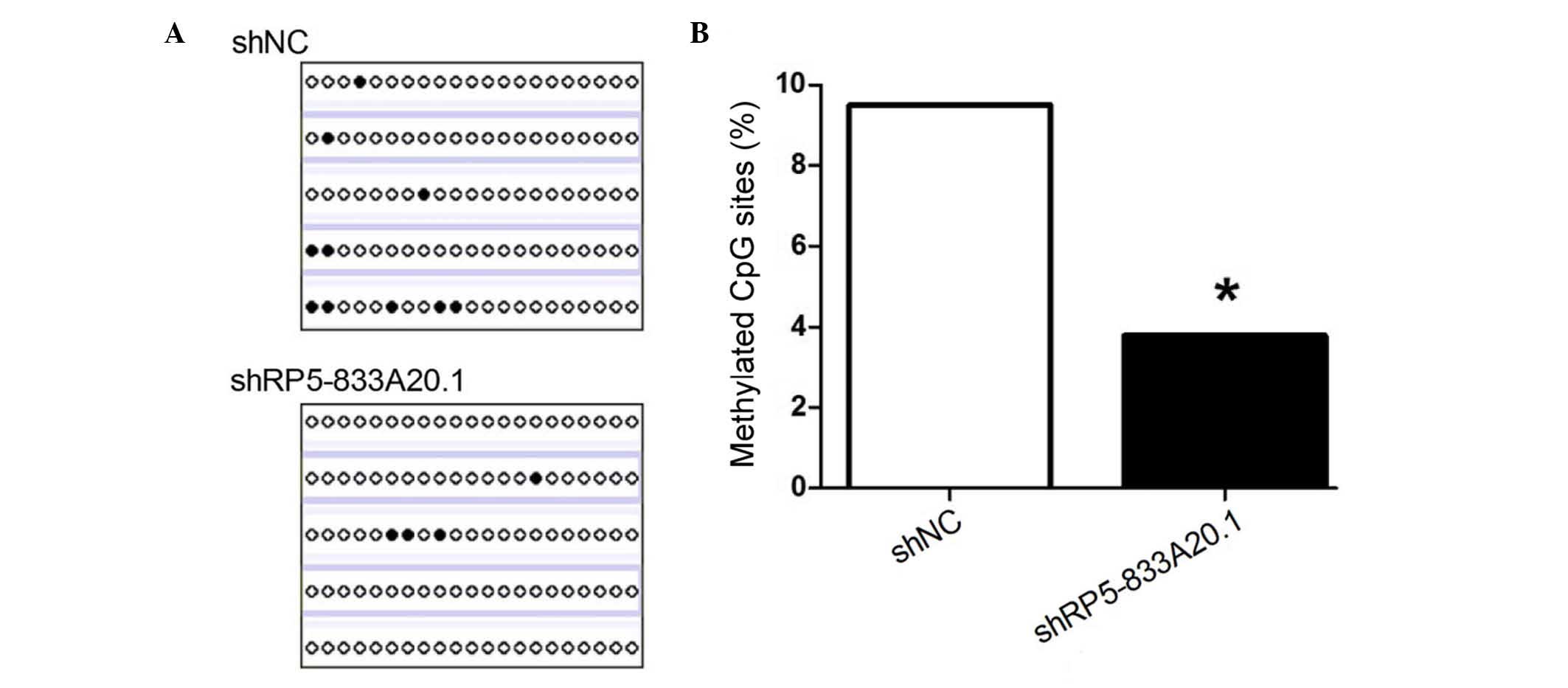
methylation, the present study assessed the methylation level of
the NFIA gene promoter using BSP. As shown in Fig. 3A and B, the knock down of
RP5-833A20.1 decreased the methylation level of the NFIA promoter
in the U251 cells, compared with the control (3.8, vs. 9.5%,
respectively; P<0.05). These results indicated that RP5-833A20.1
repressed the expression of NFIA by enhancing the methylation level
of the NFIA promoter.
RP5-833A20.1 inhibits cell
proliferation and invasion in U251 cells
As RP5-833A20.1 was downregulated in glioma tissues
and suppressed the expression of NFIA, the present study then
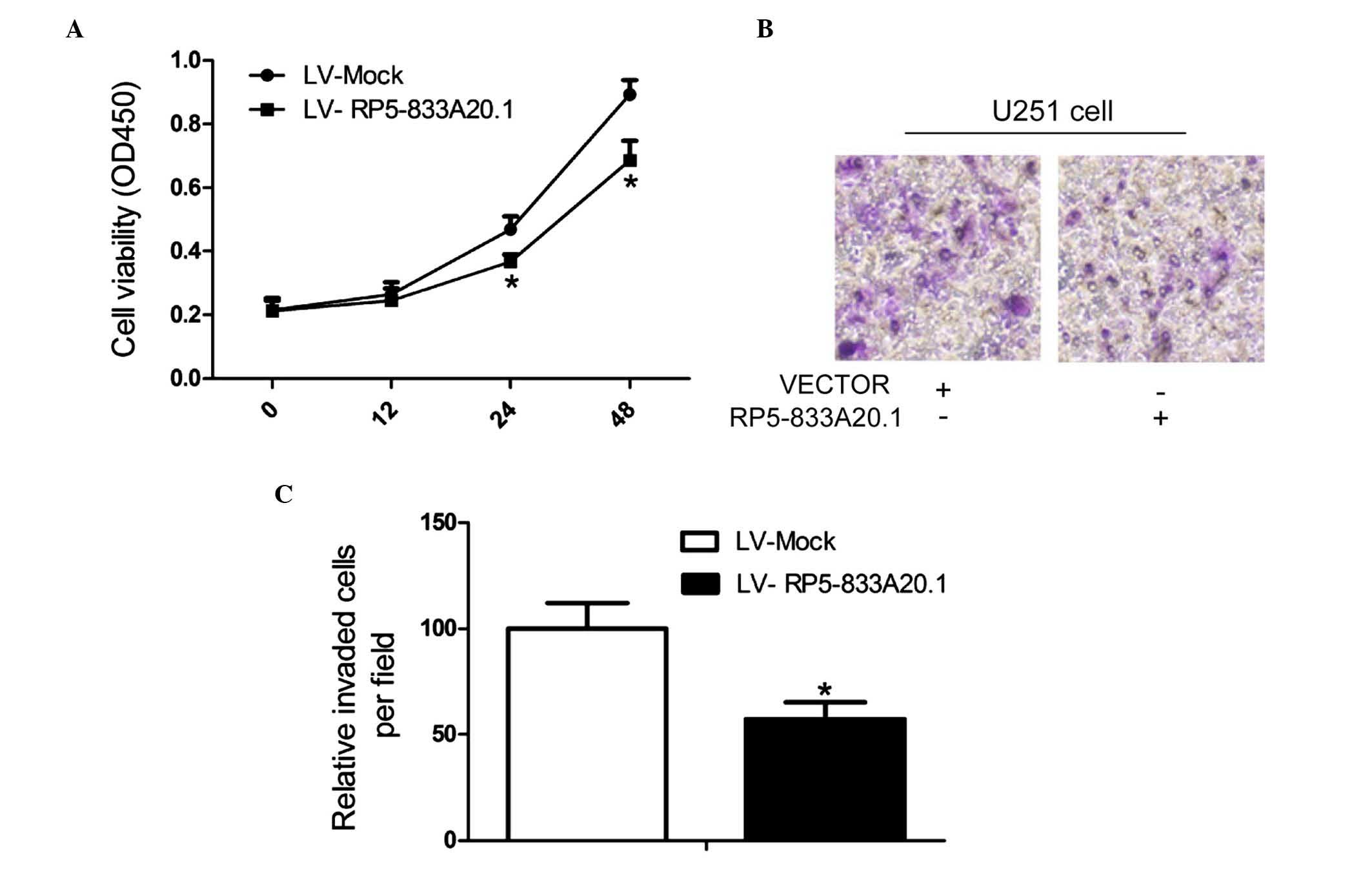
investigated the effect of RP5-833A20.1 on cell proliferation and
invasion. CCK-8 assays revealed that LV-RP5-833A20.1 treatment
significantly decreased the proportion of living U251 cells
(Fig. 4A). The invasive capacity
of the cells was investigated using Transwell assays. Compared with
the control groups, the overexpression of RP5-833A20.1 inhibited
the migratory ability of the U251 cells (Fig. 4B and C).
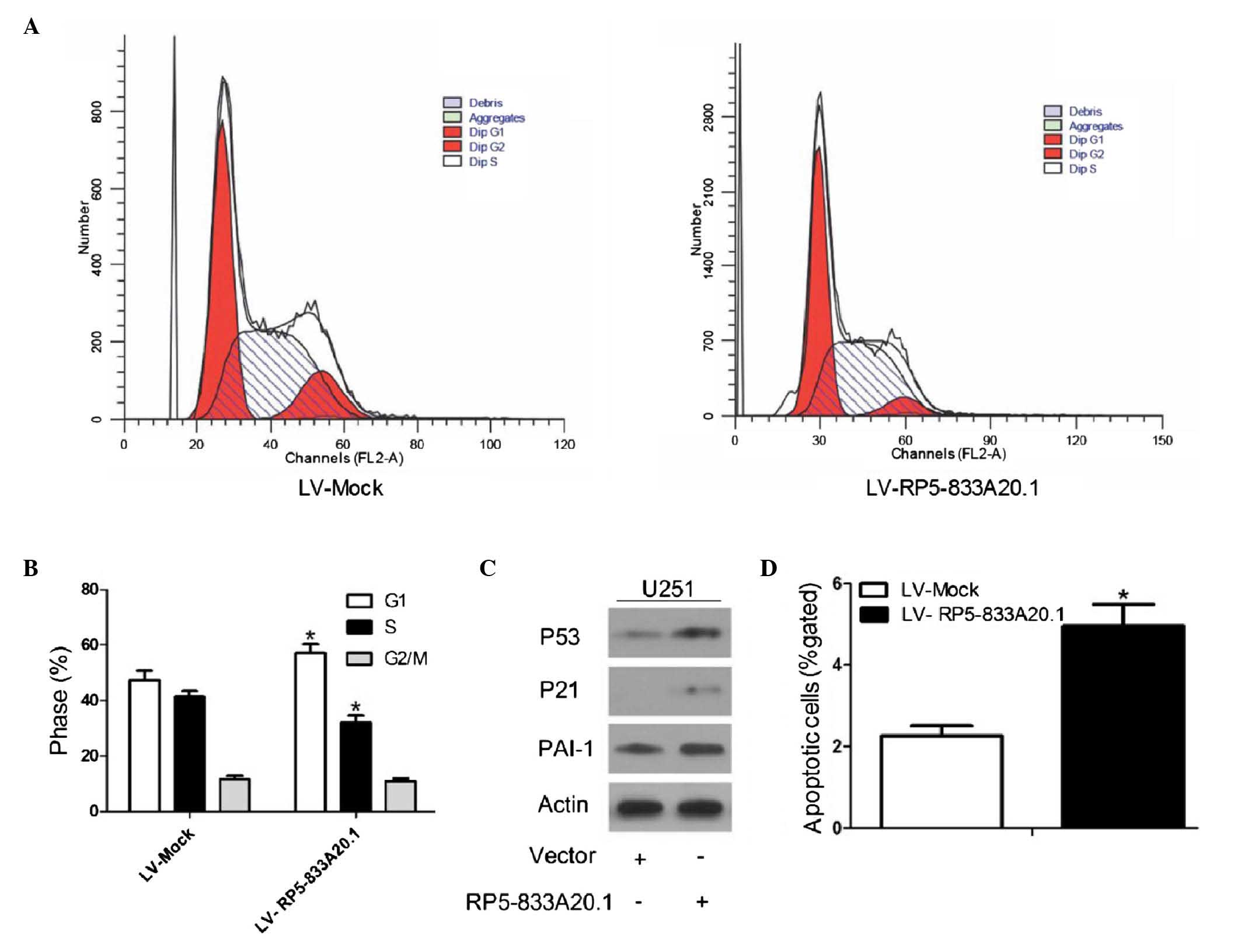
RP5-833A20.1 inhibits cell cycle
progression and induces cellular apoptosis
The present study also investigated whether
RP5-833A20.1 is involved in cell cycle distribution and apoptosis.
FACS analysis showed a significant decrease and increase in the
numbers of cells in the S and G1 phases, respectively, in the U251
cells infected with LV-RP5-833A20.1, compared with the control
(Fig. 5A and B). Consistent with
the FACS data, the expression of G1/S phase checkpoint proteins,
including tumor suppressor p53 (P53) (34), cell cycle regulator p21 (P21)
(35) and plasminogen activator
inhibitor 1 (PAI-1) (36) were
markedly upregulated in the cells infected with LV- RP5-833A20.1
(Fig. 5C). To determine whether
RP5-833A20.1 has a function in cell apoptosis, flow cytometry was
performed. As shown in Fig. 5D,
LV- RP5-833A20.1 significantly induced the apoptotic rate of the
U251 cells.
Discussion
Glioma is the most common form of primary brain
tumor in adults with varying grades of malignancy and histological
subtypes (37). Based on their
likely cellular origins, gliomas can broadly be subclassified into
astrocytomas, oligodendrogliomas, ependymomas and mixed tumors
(38). Despite aggressive
treatment approaches, and an improved understanding of its biology
and underlying molecular mechanisms, successful therapeutic
strategies are limited and long-term survival rates remain
unsatisfactory (39–41). The key finding of the present study
was that the expression of RP5-833A20.1 was decreased in glioma
tissues, compared with corresponding adjacent nontumor tissues from
20 patients with glioma. The results of the present study showed
the pathological roles of RP5-833A20.1 in repressing cell
proliferation, inhibiting cell cycle progression and inducing
apoptosis. Thus, the results of the present study indicated that
RP5-833A20.1 acts as an anti-oncogene in glioma and can be
considered as a potential prognostic indicator for glioma.
Previous studies on novel lncRNAs have suggested
that lncRNAs are characterized and important in the pathogenesis of
cancer, rather than transcriptional noise (42–44).
Accumulating evidence has shown that lncRNAs can drive
carcinogenesis by promoting cell proliferation through the
regulation of cell cycle and apoptosis (17,19,45).
Studies have found that lncRNAs can be used in predicting survival
rates in patients with glioma (46), and are potential biomarkers and
therapeutic targets for glioma (47,48).
However, the roles of lncRNAs in glioma remain to be fully
elucidated. In the present study, the expression of RP5-833A20.1
was downregulated in glioma tissues, compared with corresponding
adjacent nontumor tissues. Furthermore, the overexpression of
RP5-833A20.1 inhibited proliferation and cell cycle progression,
and induced apoptosis in U251 cells. Studies have indicated that
NFIA is involved in and may be central to a variety of biological
processes through complicated mechanisms in glioma (22,23).
In the present study, it was found that the expression of NFIA was
downregulated in glioma tissues, compared with corresponding
adjacent nontumor tissues. In addition, the overexpression of
RP5-833A20.1 suppressed the expression of NFIA in U251 cells. Lee
et al (25) identified a
novel, tumor-promoting role for NFIA in glioblastoma, which was
mediated via the transcriptional repression of p53, p21 and PAI1
through specific NFIA-recognition sequences in their promoters. In
the present study, the results revealed that P53, P21 and PAI-1
were markedly upregulated in U251 cells overexpressing
RP5-833A20.1. Therefore, RP5-833A20.1 may enhance the levels of
P53, P21 and PAI-1 by suppressing the expression of NFIA.
Previous reports have demonstrated that lncRNAs
exert regulatory control on gene function through their interaction
with miRNAs, a family of small non-coding RNAs, which are important
post-transcriptional regulators of gene expression (49–51).
Based on these findings, the present study examine the potential
roles of lncRNAs in binding to and regulating miRNAs. In our
previous study, it was confirmed that hsa-miR-382-5p is involved in
regulating the expression of NFIA by RP5-833A20.1 (27). The results of the present study
showed that the overexpression of RP5-833A20.1 suppressed the
expression of NFIA in U251 cells. In addition, the level of
miR-382-5p was increased by the overexpression of RP5-833A20.1 in
the U251 cells. Taken together, these results indicated that
RP5-833A20.1 may suppress the expression of NFIA by promoting the
expression of miR-382-5p in U251 cells. However, whether other
miRNAs can be regulated by RP5-833A20.1, and whether glioma can be
affected by miR-382-5p through other targets remain to be
elucidate, requiring further investigation. lncRNAs have dynamic
effects in transcriptional regulation, and are involved in several
human diseases, particularly cancer (52). For example, lincRNA-p21 prevents
reprogramming by sustaining CpG methylation of pluripotency gene
promoters (32). The present study
investigated the potential roles of RP5-833A20.1 in methylation.
The results revealed that the knockdown of RP5-833A20.1 decreased
the level of NFIA promoter methylation. Taken together, these
results indicated that RP5-833A20.1 may repress the expression of
NFIA by enhancing the methylation level of the NFIA promoter.
However, the detailed mechanisms of this process require further
investigation.
In conclusion, the results of the present study
demonstrated RP5-833A20.1 as an anti-oncogene, which inhibited
tumor cell proliferation, induced apoptosis and inhibited cell
cycle progression, and may involve the NFIA pathway. In addition,
RP5-833A20.1 may suppress the expression of NFIA by promoting the
expression of miR-382-5p or enhancing the methylation level of the
NFIA promoter. These findings indicated RP5-833A20.1 as a tumor
suppressor, the downregulation of which may promote glioma
metastasis, and suggested that RP5-833A20.1 may be an effective
target for glioma therapy.
Acknowledgements
This study was financially supported by the National
Natural Sciences Foundation of China (grant no. 81301489), the
President Foundation of Nanfang Hospital, Southern Medical
University (grant nos. 2012B002 and 2014C016), the Guangdong
Provincial Medical research foundation (B2014245) and the Guangdong
Provincial Department of Education Science and Technology
Innovation Project (grant no. 2012KJCX0029). The funding bodies
were not involved in the study design, data collection and
analysis, decision to publish or preparation of the manuscript.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trabelsi S, Brahim DH, Ladib M, Mama N,
Harrabi I, Tlili K, Yacoubi MT, Krifa H, Hmissa S, Saad A and Mokni
M: Glioma epidemiology in the central Tunisian population:
1993–2012. Asian Pac J Cancer Prev. 15:8753–8757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Gittleman H, Stetson L, Virk SM
and Barnholtz-Sloan JS: Epidemiology of gliomas. Cancer Treat Res.
163:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Masamha CP, Xia Z, Yang J, Albrecht TR, Li
M, Shyu AB, Li W and Wagner EJ: CFIm25 links alternative
polyadenylation to glioblastoma tumour suppression. Nature.
510:412–416. 2014.PubMed/NCBI
|
|
5
|
Liang WZ, Chou CT, Chang HT, Cheng JS, Kuo
DH, Ko KC, Chiang NN, Wu RF, Shieh P and Jan CR: The mechanism of
honokiol-induced intracellular Ca(2+) rises and apoptosis in human
glioblastoma cells. Chem Biol Interac. 221:13–23. 2014. View Article : Google Scholar
|
|
6
|
Que T, Song Y, Liu Z, Zheng S, Long H, Li
Z, Liu Y, Wang G, Liu Y, Zhou J, et al: Decreased miRNA-637 is an
unfavorable prognosis marker and promotes glioma cell growth,
migration and invasion via direct targeting Akt1. Oncogene.
34:4952–4963. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bian EB, Li J, Xie YS, Zong G, Li J and
Zhao B: LncRNAs: New players in gliomas, with special emphasis on
the interaction of lncRNAs With EZH2. J Cell Physiol. 230:496–503.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo H, Wu L, Yang Q, Ye M and Zhu X:
Functional linc-POU3F3 is overexpressed and contributes to
tumorigenesis in glioma. Gene. 554:114–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arab K, Park YJ, Lindroth AM, Schäfer A,
Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et
al: Long noncoding RNA TARID directs demethylation and activation
of the tumor suppressor TCF21 via GADD45A. Mol Cell. 55:604–614.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng S, Chen H, Wang Y, Gao W, Fu Z, Zhou
Q, Jiang Y, Lin Q, Tan L, Ye H, et al: Long non-coding RNA
LOC389641 promotes progression of pancreatic ductal adenocarcinoma
and increases cell invasion by regulating E-cadherin in a
TNFRSF10A-related manner. Cancer Lett. 371:354–365. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schoenherr CJ, Levorse JM and Tilghman SM:
Nat Genet. 33:66–69. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JT: The X as model for RNA's niche in
epigenomic regulation. Cold Spring Harb Perspect Biol.
2:a0037492010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-beta-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garding A, Bhattacharya N, Claus R, Ruppel
M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M,
Oakes C, et al: Epigenetic upregulation of lncRNAs at 13q is linked
to the In Cis downregulation of a gene cluster that targets NF-kB.
PLoS Genet. 9:e10033732013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim T, Jeon YJ, Cui R, Lee JH, Peng Y, Kim
SH, Tili E, Alder H and Croce CM: Role of MYC-regulated long
noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl
Cancer Inst. 107:dju5052015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song HR, Gonzalez-Gomez I, Suh GS, Commins
DL, Sposto R, Gilles FH, Deneen B and Erdreich-Epstein A: Uclear
factor IA is expressed in astrocytomas and is associated with
improved survival. Neuro Oncol. 12:122–132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glasgow SM, Zhu W, Stolt CC, Huang TW,
Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C and Deneen B:
Mutual antagonism between Sox10 and NFIA regulates diversification
of glial lineages and glioma subtypes. Nat Neurosci. 17:1322–1329.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brun M, Coles JE, Monckton EA, Glubrecht
DD, Bisgrove D and Godbout R: Nuclear factor I regulates brain
fatty acid-binding protein and glial fibrillary acidic protein gene
expression in malignant glioma cell lines. J Mol Biol. 391:282–300.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JS, Xiao J, Patel P, Schade J, Wan J,
Deneen B, Erdreich-Epstein A and Song HR: A novel tumor-promoting
role for nuclear factor IA in glioblastomas is mediated through
negative regulation of p53, p21 and PAI1. Neuro Oncol. 16:191–203.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Glasgow SM, Laug D, Brawley VS, Zhang Z,
Corder A, Yin Z, Wong ST, Li XN, Foster AE, Ahmed N and Deneen B:
The miR-223/nuclear factor I-A axis regulates glial precursor
proliferation and tumorigenesis in the CNS. J Neurosci.
33:13560–13568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR,
Ma X, Wu SG, Chen ZP, Hu YR, Yang JY, et al:
RP5-833A20.1/miR-382-5p/NFIA-Dependent Signal transduction pathway
contributes to the regulation of cholesterol homeostasis and
inflammatory reaction. Arterioscler Thromb Vasc Biol. 35:87–101.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X,
Wu D and Liu L: The long intergenic noncoding rna ufc1, a target of
MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to
increase levels of β-Catenin in HCC cells. Gastroenterology.
148:415–426 e18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu YW, Ma X, Li XX, Liu XH, Xiao J, Mo ZC,
Xiang J, Liao DF and Tang CK: Eicosapentaenoic acid reduces ABCA1
serine phosphorylation and impairs ABCA1-dependent cholesterol
efflux through cyclic AMP/protein kinase a signaling pathway in
THP-1 macrophage-derived foam cells. Atherosclerosis. 204:e35–e43.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bao X, Wu H, Zhu X, Guo X, Hutchins AP,
Luo Z, Song H, Chen Y, Lai K, Yin M, et al: The p53-induced
lincRNA-p21 derails somatic cell reprogramming by sustaining
H3K9me3 and CpG methylation at pluripotency gene promoters. Cell
Res. 25:80–92. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lai F and Shiekhattar R: Where long
noncoding RNAs meet DNA methylation. Cell Res. 24:263–264. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hao PP, Li H, Lee MJ, Wang YP, Kim JH, Yu
GR, Lee SY, Leem SH, Jang KY and Kim DG: Disruption of a regulatory
loop between DUSP1 and p53 contributes to hepatocellular carcinoma
development and progression. J Hepatol. 62:1278–1286. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ehedego H, Boekschoten MV, Hu W, Doler C,
Haybaeck J, Gaβler N, Müller M, Liedtke C and Trautwein C: p21
ablation in liver enhances DNA damage, cholestasis and
carcinogenesis. Cancer Res. 75:1144–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Giacoia EG, Miyake M, Lawton A, Goodison S
and Rosser CJ: PAI-1 leads to G1-phase cell-cycle progression
through cyclin D3/cdk4/6 upregulation. Mol Cancer Res. 12:322–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang S, Ye Z, Song X, Chen G, Huai C,
Wang Q, Song J, Lu D, Zhao Y and Chen H: Association of EFEMP1 gene
polymorphisms with the risk of glioma: A hospital-based
case-control study in a Chinese Han population. J Neurol Sci.
349:54–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qureshi IA and Mehler MF: Emerging roles
of non-coding RNAs in brain evolution, development, plasticity and
disease. Nat Rev Neurosci. 13:528–541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang XQ and Leung GK: Long non-coding
RNAs in glioma: Functional roles and clinical perspectives.
Neurochem Int. 77:78–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morfouace M, Lalier L, Oliver L, Cheray M,
Pecqueur C, Cartron PF and Vallette FM: Control of glioma cell
death and differentiation by PKM2-Oct4 interaction. Cell Death Dis.
5:e10362014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding rna involved in cancer, neurobiology and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mattick JS and Rinn JL: Discovery and
annotation of long noncoding RNAs. Nat Struct Mol Biol. 22:5–7.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: LncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu H, Xu Q, Liu F, Ye X, Wang J and Meng
X: Identification and validation of long non-coding RNA biomarkers
in human non-small cell lung carcinomas. J Thorac Oncol.
10:645–654. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and Leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun Y, Wang Z and Zhou D: Long non-coding
RNAs as potential biomarkers and therapeutic targets for gliomas.
Med Hypotheses. 81:319–321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu
Y and Zhu W: A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated
by DNMT1 and inhibits migration of glioma cells. Tumour Biol.
35:7935–7944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Imig J, Brunschweiger A, Brümmer A,
Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M
and Hall J: miR-CLIP capture of a miRNA targetome uncovers a
lincRNA H19-miR-106a interaction. Nat Chem Biol. 11:107–114. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC,
Hwang SM and Hu YC: Suppression of hepatocellular carcinoma by
baculovirus-mediated expression of long non-coding RNA PTENP1 and
MicroRNA regulation. Biomaterials. 44:71–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dey BK, Pfeifer K and Dutta A: The H19
long noncoding RNA gives rise to microRNAs miR-675-3p and
miR-675-5p to promote skeletal muscle differentiation and
regeneration. Genes Dev. 28:491–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|