Introduction
Epilepsy is a common chronic neurological disease,
which is characterized by recurring seizures. It has been reported
that the morbidity rate of epilepsy is ~1% worldwide (1). Epilepsy affects >65,000,000
individuals in the world and >100,000 new cases are diagnosed
every year (2).
Electroencephalography (EEG) and magnetic resonance imaging (MRI)
are the most widely used techniques for the diagnosis of epilepsy.
EEG and MRI can detect and locate the epileptic seizures and zones
(3). Usually, a typical EEG of
epilepsy ranges between minutes and hours (4). However the exact etiology of epilepsy
remains to be fully elucidated.
MicroRNAs (miRNAs) are small non-coding RNAs
(ncRNAs) and are the most well-researched family of ncRNAs to date.
Mature miRNA sequences are ~22 nucleotides in length (5). They are involved in depression of the
expression of post-transcriptional target genes via binding to the
3′-untranslated region (3′UTR) (6). miRNAs are abundant in the nervous
system where they serve as effectors of brain development and in
maintenance of the neuronal phenotype (7).
The results of previous expression profiling have
shown unique miRNA expression in acute brain injuries (8). Several studies have demonstrated that
miRNAs affect the pathomechanisms in epileptogenic injuries
(9). Previously, four studies have
profiled the expression of miRNAs in experimental epilepsy, in
which >100 different miRNAs were identified. These results
confirmed that epilepsy is associated with widespread alterations
in the expression of miRNAs (10–13).
The expressed miRNAs were all detected in the brain tissues of
patients or animal models. The present study aimed to examine the
serum expression of miRNAs in patients with epilepsy, and to select
differently expressed miRNA between seizure onset and post-seizure
to explore the function of selected miRNA in epilepsy. This was
performed with the aim to identify the biomarker of epilepsy
treatment.
Materials and methods
Patients and serum samples
Following ethical approval and the provision of
written informed consent, serum samples were collected from three
seizure patients for miRNA microarray analysis and from another 90
patients for confirmation of the miRNA expression. The patients
were evaluated by an epileptologist and underwent a basal EEG and
3T MR brain imaging. Patients experiencing symptoms and EEG
semiology were considered to be at seizure onset. Following
treatment, when the symptoms and EEG semiology tended to be normal,
patients were considered to be post-seizure. All samples were
collected from consenting individuals from the Liaocheng People's
Hospital, Taishan Medical College (Liaocheng, China) and Jinan
University (Shenzhen, China). The sample collection was performed
according to the protocols approved by the Ethics Committee of
Jinan University. The blood samples were collected at seizure onset
and post-seizure.
RNA isolation from human serum
samples
Venous blood (5 ml) was collected and stored at room
temperature for 1 h, followed by centrifugation at 1,600 × g
for 10 min at 4°C. The serum was gently collected and stored at
−80°C or used in the following experiment. For all experiments, 500
µl of human serum was used and total RNA was extracted using TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
concentration and the 260/280 nm absorbance ratio was detected
using a Nanodrop 2000 (Thermo Fisher Scientific, Inc., Pittsburgh,
PA, USA).
MicroRNA expression profiling
Total RNA (100 ng) was used to synthesize first
strand cDNA at 42°C for 2 h using ArrayScript™ reverse
transcriptase, second strand cDNA was at 16°C for 1 h, 65°C for 10
min using reverse transcriptase and DNA polymerase, respectively.
T7 Biotin IVT Mix was used to generate multiple copies of
biotin-modified aRNA from the double stranded cDNA templates,
incubated for 2–14 h at 40°C (MessageAmp™ Premier RNA Amplification
kit; Thermo Fisher Scientific, Inc.). miRNAs were individually
detected using specific oligonucleotides (Takara Bio, Inc.). A
single miRNA-specific oligonucleotide was designed against each
mature miRNA sequence, and miRNA-specific primers were extended
using DNA polymerase. Universal primers were used to amplify the
cDNA templates and the primer complimentary to the array was
fluorescently labeled using miRCURY Hy3/Hy5 Power Labeling kit
(Takara Bio Inc.). The labeled, single-stranded PCR products were
hybridized to a Human v2.0 miRNA Expression BeadChip (Illumina,
Inc., San Diego, CA) with 1,146 human miRNAs (97% coverage in the
miRBase 12.0 database) (14,15).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis of the expression of miRNAs and mRNAs
RT for the individual qPCR analyses was performed
using 500 ng of total RNA and a Reverse Transcription kit (Promega
Corporation, Madison, WI, USA). RT-specific primers for human
miRNAs miR-378, miR-30a, miR-106b and miR-15a (Applied Biosystems;
Thermo Fisher Scientific, Inc.) were used for all miRNA RT
procedures. Individual qPCR analyses were performed on the 7900HT
Fast Realtime system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using miR-378-, miR-30a-, miR-106b- and miR-15a-specific
Taqman miRNA assays (Applied Biosystems Thermo Fisher Scientific,
Inc.). RNU6B was used for the normalization of miRNA expression
levels. The mRNA levels of calcium/calmodulin-dependent protein
kinase type IV (CAMK4), were determined using TaqMan Real-Time PCR
analysis, according to manufacturer's protocols (Applied
Biosystems; Thermo Fisher Scientific, Inc.). To determine mRNA
expression levels, total RNA (10 ng) were reverse transcribed using
iScript Reverse Transcription Supermix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The cDNA templates were amplified with TaqMan
PreAmp Master mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The PCR programs consisted in a hot start of 95°C for 10
min, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. The
primers used were: CAMK4, sense 5′-AGCACATTCAAACCACCACA-3′,
antisense 5′-GGACTCAGAGATCCGTCTGC-3′; miR-30a, sense
5′-TGTAAACATCCTCGAC-3′, antisense 5′-ACATCCAGTGTAGCATA-3′; miR-378,
sense 5′-ACACTCCAGCTGGGACTGGACTTGGAGTC-3′, antisense
5′-TGGTGTCGGAGTCG-3′; miR-106a, sence
5′-CGGAATTCATCTCGAGACGCCAACTTG-3′, antisense
5′-CGGGATCCCTTCATTCAAGGTCAATGAG-3′; miR-15a, sense 5′-
GTCGTATCCAGTGCAGGGTCCGAGGTATTC-3′, antisense
5′-GCACTGGATACGACCACAAA-3′; RNU6B, sense 5′-CTCGCTTCGGCAGCACA-3′,
antisense 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH,
5′-AAGGTCGGAGTCAACGGATT-3′, antisense 5′-CTGGAAGATGGTGATGGGATT-3′.
Each sample was analyzed in triplicate, in three independent
experiments. The level of each mRNA was measured using the
quantification cycle (Cq) and the level of the target was
calculated as described above for the miRNAs. The gene expression
levels were normalized against the expression of GAPDH. The
relative fold change in expression of the target gene transcript
was determined using the comparative quantification method
(2−ΔΔCq) (16). High or
low expression of miRNAs and mRNAs were defined on the basis of the
median expression, determined separately for each cohort.
Construction of luc-UTR vectors
The full-length CAMK4 3′-UTRs were cloned into the
EcoRI and HindIII sites of the pMIR-REPORT luciferase
vector (Ambion; Thermo Fisher Scientific, Inc.) using a
PCR-generated fragment. A Luc-mut vector, in which the first seven
nucleotides complementary to the miR-30a seed-region were mutated,
served as a mutant control. The binding sites of CAMK4 were UGU UUA
CA and these were replaced with the CAC CCG UG in the Luc-mut
vector.
Luciferase reporter assay
The Luc-wild-type (wt), Luc-mut and Luc-control
vectors were co-transfected within miR-30a mimics into 293T cells
(Cell bank of Chinese Academy of Sciences, Shanghai, China) using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The
pMIR-REPORT β-galactosidase control vector (Thermo Fisher
Scientific, Inc.) was transfected into cells to serve as a control.
Luciferase activity was measured in the cell lysates 48 h following
transfection using a dual-light luminescent reporter gene assay kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The results
were normalized against the activity of β-galactosidase (17).
Western blot analysis
Cells was lysed in ice-cold lysis buffer (1% NP-40;
50 mM Tris-HCl, pH 8.0; 100 mM sodium fluoride; 30 mM sodium
pyrophosphate; 2 mM sodium molybdate; 5 mM EDTA and 2 mM sodium
orthovanadate). Lysates were centrifuged at 10,000 × g for
15 min at 4°C. The concentration was determined by Pierce
bicinchoninic acid Protein Assay kit (Thermo Fisher Scientific,
Inc.). Protein lysates (50 µg) were separated on 10%
SDS-polyacrylamide gels and electrophoretically transferred onto
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Membranes were blocked in 5% nonfat milk in TBS-Tween (TBST)
for 1 h at room temperature. The primary rabbit anti-human CaMK4
antibody (cat. no. 4032; Cell Signaling Technology, Inc., Danvers,
MA, USA) was diluted 1:1,000 in 5% milk with TBST and incubated
with the membrane at 4°C overnight. Then blots were washed three
times with TBST. Secondary goat anti-rabbit IgG (cat. no. ab6721;
Abcam, Cambridge, UK) was diluted at 1:3,000 in milk with TBST and
incubated with the membrane at 37°C for 1 h. The hybridization
signals were detected by chemiluminescence (Invitrogen; Thermo
Fisher Scientific, Inc.) and captured using a ChemiDOC (Bio-Rad
Laboratories, Inc.).
Statistical analysis
All data are presented as the mean ± standard error
of the mean. Analysis was performed using SPSS software (version
17; SPSS, Inc., Chicago, IL, USA). Comparisons between two groups
were made using Student's t-test, whereas multigroup
comparisons were made using analysis of variance followed by
appropriate post-hoc testing. Pearson's correlation analysis was
used to test the correlation between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinical parameters
Serum from three patients at seizure onset and
post-seizure were used for microarray analysis. The patients
included two men and one woman. Serum from an additional 90
patients at seizure onset and post-seizure were used for RT-qPCR
analysis. The patients had a mean age of 39 years (range, 21–60), a
mean number of years following epilepsy diagnosis of 16 years
(range, 5–31) and a mean seizure frequency of 5 (range, 1–20) times
per month.
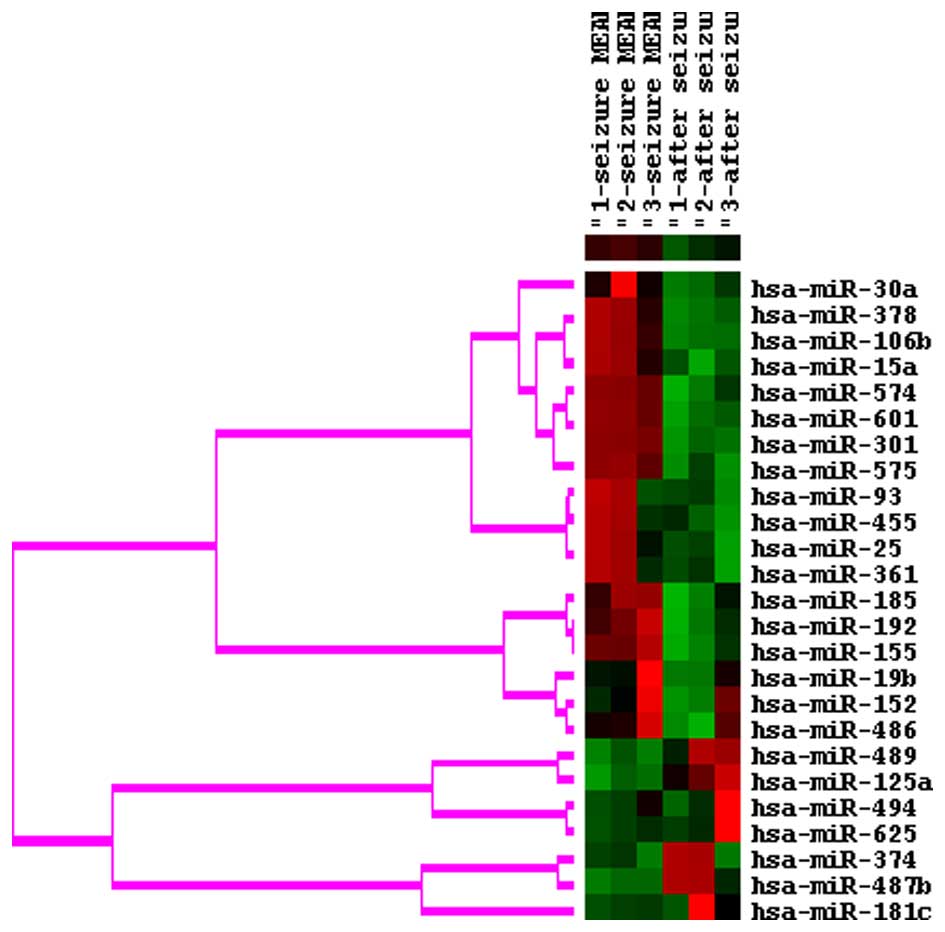
miRNA microarray
The expression levels of 15 miRNAs were increased
and 10 miRNAs were decreased at seizure onset, compared with
post-seizure in the patients with epilepsy, as determined using
miRNA microarray analysis (Fig.
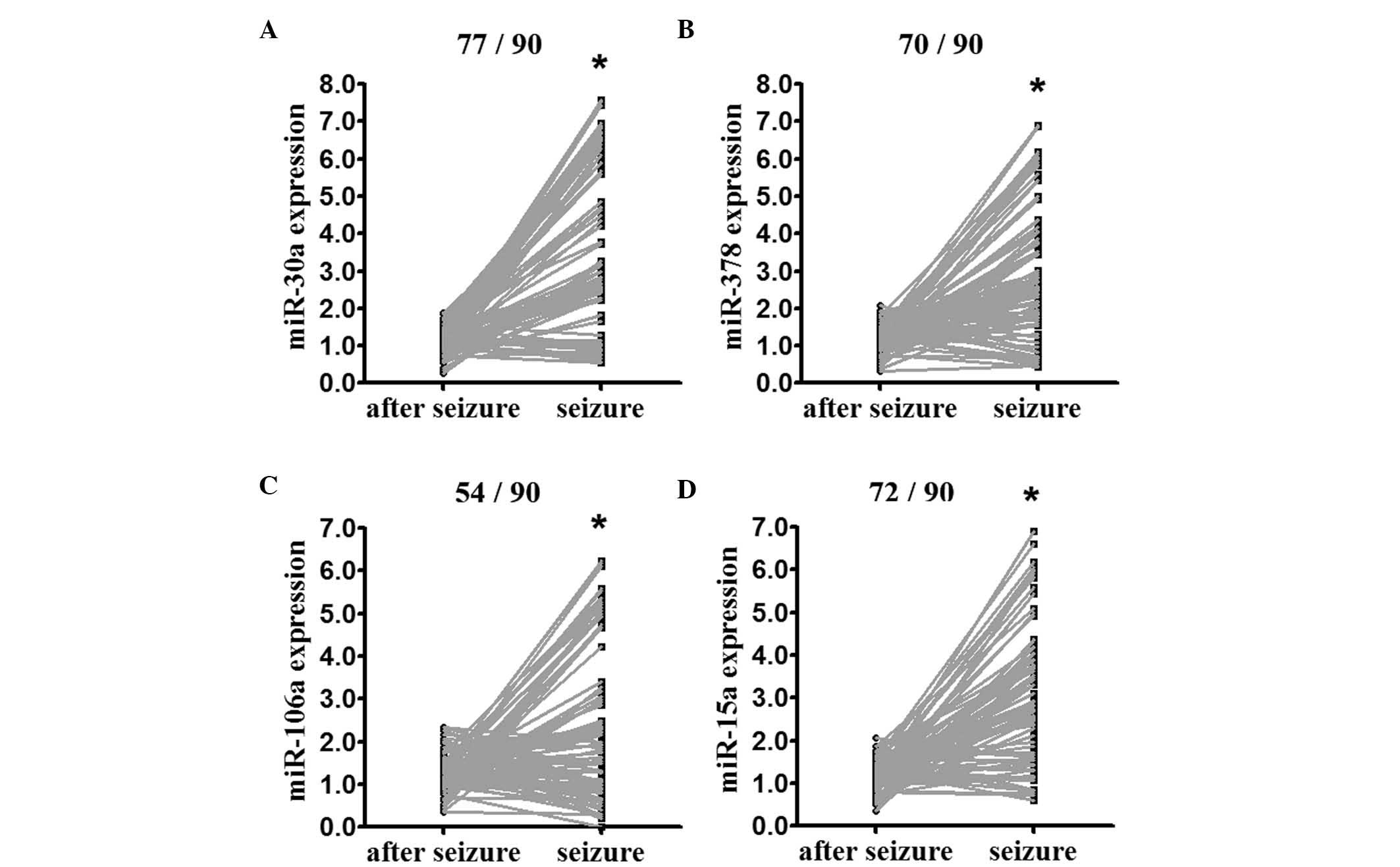
1). The expression of miR-30a, miR-378, miR-106b and miR-15a
was confirmed in the extended cohort of the three original patients
and the additional 90 patients (indicated in Table I), using RT-qPCR analysis. The
expression levels of miR-30a, miR-378, miR-106b and miR-15a, the
top four overexpressed miRNAs identified, were enhanced at seizure
onset, compared with post-seizure (Fig. 2).
 | Table I.Association between expression level
of miRNAs and clinical parameters of epilepsy. |
Table I.
Association between expression level
of miRNAs and clinical parameters of epilepsy.
|
| miR-30a | miR-378 | miR-106b | miR-15a |
|---|
|
|
|
|
|
|
|---|
| Parameter | High | Low | High | Low | High | Low | High | Low |
|---|
| Gender |
|
|
|
|
|
|
|
|
| Male
(52) | 36 | 16 | 28 | 24 | 27 | 25 | 37 | 15 |
| Female
(38) | 25 | 13 | 26 | 12 | 21 | 17 | 20 | 18 |
| Age |
|
|
|
|
|
|
|
|
| ≥39
years (41) | 28 | 13 | 26 | 15 | 25 | 16 | 26 | 15 |
| <39
years (49) | 33 | 16 | 28 | 21 | 23 | 26 | 31 | 18 |
| Years
diagnosed |
|
|
|
|
|
|
|
|
| ≥16
(32) | 20 | 11 | 17 | 15 | 18 | 14 | 19 | 13 |
| <16
(58) | 41 | 18 | 37 | 21 | 30 | 28 | 38 | 20 |
| Seizure
frequency |
|
|
|
|
|
|
|
|
|
≥5/month (37) | 31 |
6a | 22 | 14 | 21 | 15 | 25 | 12 |
|
<5/month (53) | 30 | 23 | 32 | 21 | 26 | 27 | 32 | 21 |
Correlation between upregulated miRNAs
and clinical parameters in patients experiencing seizure
The correlation between upregulated miRNAs and
clinical parameters was statistically analyzed. The expression
levels of miR-378, miR-106b and miR-15a were not associated with
the age, gender, years following diagnosis or seizure frequency.
The expression of miR-30a was positively associated with seizure
frequency (Table I).
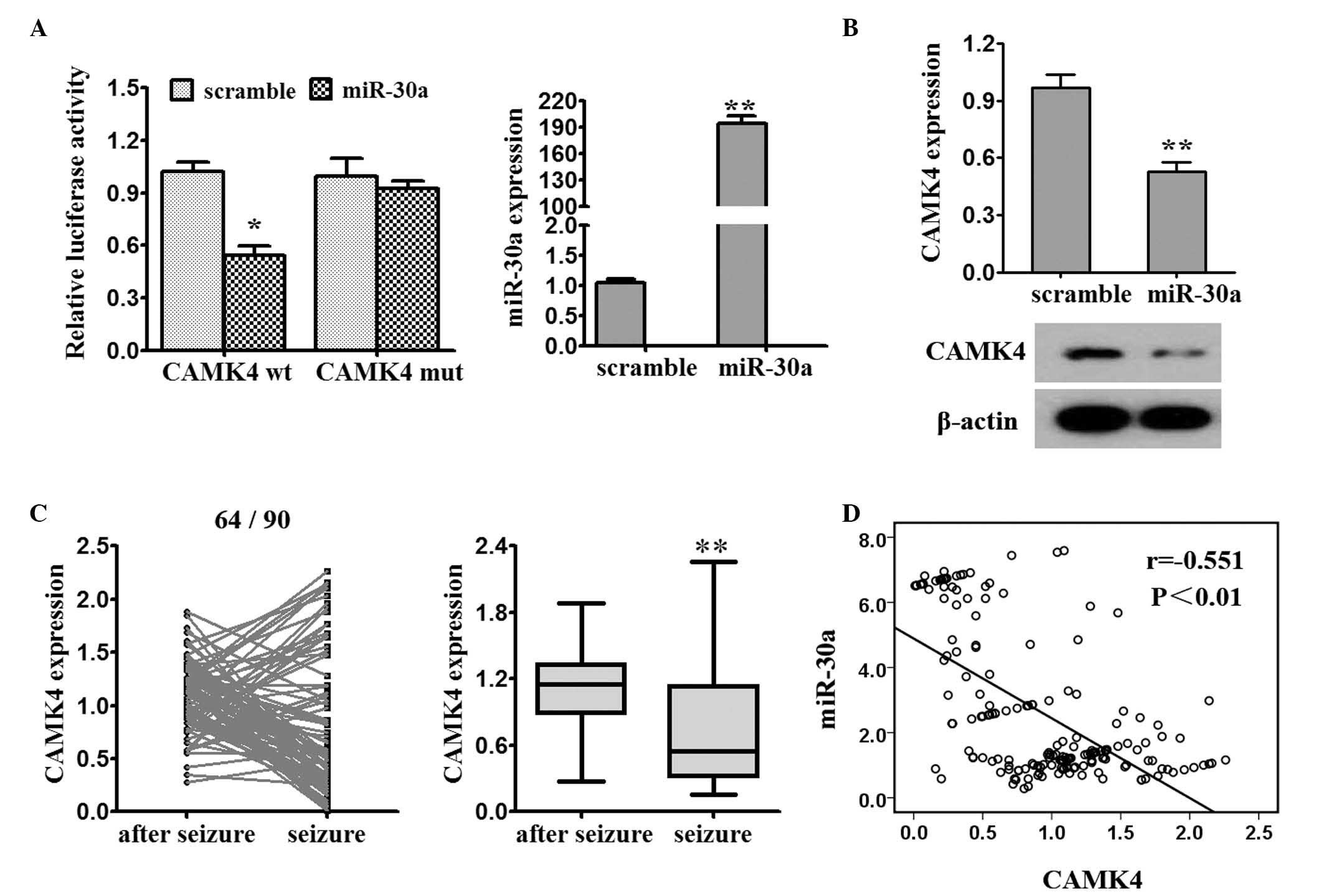
CAMK4 is a target of miR-30a
The online software program, Targetscan 6.0
(www.targetscan.org/vert_71/), was
used to assist in identifying miR-30a targets. The miR-30a mimics
decreased the luciferase activity of the CAMK4-3′-UTR-wt reporter
(Fig. 3A). The results from the
RT-qPCR and western blot analyses showed that the enhanced
expression of miR-30a by the miR-30a mimics in the 293T cells led
to a decrease in endogenous mRNA and protein levels of CaMK4
(Fig. 3B). The mRNA expression of
CAMK4 was significantly reduced post-seizure, compared with at
seizure onset (64/90; Fig. 3C).
The expression of miR-30a and CAMK4 were demonstrated to be
negatively correlated (Fig.
3D).
Discussion
The pathogenesis of different types of epilepsy
involves several important biological pathways, a number of which
have been shown to be regulated by miRNAs. In the present study,
the expression of miRNAs was compared at seizure onset with
expression post-seizure in patients with epilepsy. The miRNA
microarray revealed that the expression levels of 15 miRNAs were
increased and 10 miRNAs were decreased at seizure onset, compared
with at post-seizure in patients with epilepsy. The present study
then confirmed that the expression levels of miR-30a, miR-378,
miR-106b and miR-15a were higher at seizure onset, compared with
levels post-seizure in the serum of patients with epilepsy.
miR-30a is involved in tumorigenesis, inflammation
and myoblast differentiation and miR-30a is known to function as a
tumor suppressor in breast cancer, small cell lung cancer and
colorectal carcinoma (18).
miR-30a has been found to be increased in cerebral arteries
following subarachnoid hemorrhage (SAH), compared with the levels
in rats subjected to sham surgery, and may be involved in the
vascular wall changes observed following SAH (19). Wen et al (20) demonstrated that miR-30a-5p is
significantly overexpressed in hepatitis B virus (HBV)-positive
patients with hepatocellular cancer, compared with HBV-positive
controls without cancer. This study revealed that plasma miR-30a
offers potential as an early biomarker for detecting hepatocellular
carcinoma. Overexpression of miR-30a-5p promotes the
differentiation of myoblasts, whereas its inhibition restricts the
differentiation of myoblasts in vitro (21). Circulating levels of miR-30a have
been found to be markedly downregulated in patients with ischemic
stroke until 24 weeks (22). As
all four miRNAs detected in the present study were decreased
following seizures, the present study analyzed the association
between the expression of miRNAs in patients with epilepsy with
seizures and the clinical parameters. The results of the present
study revealed that the expression of miR-30a was higher in the
sera of patients at seizure onset, with that post-seizure, and was
associated with seizures frequency. Several studies have indicated
that miR-30a functions in multiple biological processes via
targeting a number of genes. For example, miR-30a can target
insulin receptor substrate 2 in colorectal tumorigenesis (23). The overexpression of miR-30a
upregulates the expression levels of B cell lymphoma 2-related
protein A1 immediate early response 3 and cyclin D2 by inhibiting
forkhead transcription factor ligand 2 (24). The downregulation of miRNA-30a
alleviates cerebral ischemic injury through enhancing beclin
1-mediated autophagy (25). In the
present study, the hypothesis that CAMK4 was a target of miR-30a
was confirmed. The inhibition of CAMK4 is detrimental in cerebral
ischemia (26). The present study
analyzed the correlation between the expression levels of miR-30a
and CAMK4. The expression of CAMK4 was negatively associated with
that of miR-30a in patients with epilepsy. However, the function of
CAMK4 in epilepsy was not investigated, and CAMK4 may be involved
in the miR-30a-mediated pathway in epilepsy.
miR-378 promotes the migration of liver cancer cells
by downregulating the expression of Fus (27). miR-378 is also considered to be a
diagnostic biomarker in cancer, including renal cell carcinoma
(28) and colorectal cancer
(29). miR-378 may be a potential
biomarker for characterizing non-small cell lung cancer brain
metastasis (30). The
overexpression of miR-378 attenuates high glucose-suppressed
osteogenic differentiation through targeting caspase 3 and
activating the phosphoinositide 3-kinase/Akt signaling pathway
(31). Circulating levels of
miR-378 predicts left ventricular hypertrophy in patients with
aortic stenosis (32). miRNA-378
controls classical brown fat expansion to counteract obesity
(33). miR-106b promotes
hematopoietic cell expansion by targeting sequestosome 1-regulated
pathways in mice (34). miR-106b
is decreased in patients with chronic myeloid leukemia in the
chronic phase (35). A previous
study found that the relative levels of miR-106b prior to and
following Helicobacter pylori eradication were significantly
higher in the high-risk group, compared with the control (36). miR-15a enhances the
radiosensitivity of breast cancer cells by targeting G2 checkpoints
(37). Maudet et al
(38) identified the miR-15a miRNA
family as cellular restriction factors for Salmonella infection
using functional high-throughput screening. The miR-15 family is a
regulator of cardiac hypertrophy and fibrosis, acting by inhibiting
of the transforming growth factor β-pathway (39). However, the functions of miR-378,
miR-106b and miR-15a in the nervous system have not been reported.
In the present study, it was found that miR-378, miR-106b and
miR-15a were increased in the sera of patients experiencing
seizures. The mechanisms underlying the regulation of the
expression of these miRNAs require further investigation. The
present study suggested that miR-30a may be useful for predicting
prognosis following seizure.
References
|
1
|
van Graan LA, Lemieux L and Chaudhary UJ:
Methods and utility of EEG-fMRI in epilepsy. Quant Imaging Med
Surg. 5:300–312. 2015.PubMed/NCBI
|
|
2
|
Kwan P, Schachter SC and Brodie MJ:
Drug-resistant epilepsy. N Engl J Med. 365:919–926. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahmad MA, Ayaz Y, Jamil M, Gillani S Omer,
Rasheed MB, Imran M, Khan NA, Majeed W and Javaid N: Comparative
analysis of classifiers for developing an adaptive
computer-assisted EEG analysis system for diagnosing epilepsy.
Biomed Res Int. 2015:6380362015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Puttachary S, Sharma S, Stark S and
Thippeswamy T: Seizure-induced oxidative stress in temporal lobe
epilepsy. Biomed Res Int. 2015:7456132015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sano T, Reynolds JP, Jimenez-Mateos EM,
Matsushima S, Taki W and Henshall DC: MicroRNA-34a upregulation
during seizure-induced neuronal death. Cell Death Dis. 3:e2872012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saugstad JA: Non-Coding RNAs in stroke and
neuroprotection. Front Neurol. 6:502015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhalala OG, Srikanth M and Kessler JA: The
emerging roles of microRNAs in CNS injuries. Nat Rev Neurol.
9:328–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu DZ, Tian Y, Ander BP, Xu H, Stamova
BS, Zhan X, Turner RJ, Jickling G and Sharp FR: Brain and blood
microRNA expression profiling of ischemic stroke, intracerebral
hemorrhage, and kainate seizures. J Cereb Blood Flow Metab.
30:92–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henshall DC: MicroRNA and epilepsy:
Profiling, functions and potential clinical applications. Curr Opin
Neurol. 27:199–205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zucchini S, Marucci G, Paradiso B, Lanza
G, Roncon P, Cifelli P, Ferracin M, Giulioni M, Michelucci R,
Rubboli G and Simonato M: Identification of miRNAs differentially
expressed in human epilepsy with or without granule cell pathology.
PLoS One. 9:e1055212014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li MM, Jiang T, Sun Z, Zhang Q, Tan CC, Yu
JT and Tan L: Genome-wide microRNA expression profiles in
hippocampus of rats with chronic temporal lobe epilepsy. Sci Rep.
4:47342014.PubMed/NCBI
|
|
13
|
McKiernan RC, Jimenez-Mateos EM, Sano T,
Bray I, Stallings RL, Simon RP and Henshall DC: Expression
profiling the microRNA response to epileptic preconditioning
identifies miR-184 as a modulator of seizure-induced neuronal
death. Exp Neurol. 2:346–354. 2013.
|
|
14
|
Zeng X, Xiang J, Wu M, Xiong W, Tang H,
Deng M, Li X, Liao Q, Su B, Luo Z, et al: Circulating miR-17,
miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers
in nasopharyngeal carcinoma. PLoS One. 7:e463672012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
You G, Yan W, Zhang W, Wang Y, Bao Z, Li
S, Li S, Li G, Song Y, Kang C and Jiang T: Significance of miR-196b
in tumor-related epilepsy of patients with gliomas. PLoS One.
7:e462182012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao S, Yang Z, Lv R, Zhao J, Wu M, Liao Y
and Liu Q: miR-135b contributes to the radioresistance by targeting
GSK3β in human glioblastoma multiforme cells. PLoS One.
9:e1088102014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang R, Liang L, Luo D, Feng Z, Huang Q,
He R, Gan T, Yang L and Chen G: Downregulation of miR-30a is
associated with poor prognosis in lung cancer. Med Sci Monit.
21:2514–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Müller AH, Povlsen GK, Bang-Berthelsen CH,
Kruse LS, Nielsen J, Warfvinge K and Edvinsson L: Regulation of
microRNAs miR-30a and miR-143 in cerebral vasculature after
experimental subarachnoid hemorrhage in rats. BMC Genomics.
16:1192015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guess MG, Barthel KK, Harrison BC and
Leinwand LA: miR-30 family microRNAs regulate myogenic
differentiation and provide negative feedback on the microRNA
pathway. PLoS One. 10:e01182292015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Long G, Wang F, Li H, Yin Z, Sandip C, Lou
Y, Wang Y, Chen C and Wang DW: Circulating miR-30a, miR-126 and
let-7b as biomarker for ischemic stroke in humans. BMC Neurol.
13:1782013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv
G, Zou Z, Jiang XC, Zou C, Liu W, et al: Role of microRNA 30a
targeting insulin receptor substrate 2 in colorectal tumorigenesis.
Mol Cell Biol. 35:988–1000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang T, Li F and Tang S: MiR-30a
upregulates BCL2A1, IER3 and cyclin D2 expression by targeting
FOXL2. Oncol Lett. 9:967–971. 2015.PubMed/NCBI
|
|
25
|
Wang P, Liang J, Li Y and Li J, Yang X,
Zhang X, Han S, Li S and Li J: Down-regulation of miRNA-30a
alleviates cerebral ischemic injury through enhancing beclin
1-mediated autophagy. Neurochem Res. 39:1279–1291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McCullough LD, Tarabishy S, Liu L,
Benashski S, Xu Y, Ribar T, Means A and Li J: Inhibition of
calcium/calmodulin-dependent protein kinase kinase β and
calcium/calmodulin-dependent protein kinase IV is detrimental in
cerebral ischemia. Stroke. 44:2559–2566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koval AV, Vlasov P, Shichkova P,
Khunderyakova S, Markov Y, Panchenko J, Volodina A, Kondrashov FA
and Katanaev VL: Anti-leprosy drug clofazimine inhibits growth of
triple-negative breast cancer cells via inhibition of canonical Wnt
signaling. Biochem Pharmacol. 87:571–578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang C, Hu J, Lu M, Gu H, Zhou X, Chen X,
Zen K, Zhang CY, Zhang T, Ge J, et al: A panel of five serum miRNAs
as a potential diagnostic tool for early-stage renal cell
carcinoma. Sci Rep. 5:76102015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Clancy C, Joyce MR and Kerin MJ: The use
of circulating microRNAs as diagnostic biomarkers in colorectal
cancer. Cancer Biomark. 15:103–113. 2015.PubMed/NCBI
|
|
30
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
You L, Gu W, Chen L, Pan L, Chen J and
Peng Y: MiR-378 overexpression attenuates high glucose-suppressed
osteogenic differentiation through targeting CASP3 and activating
PI3K/Akt signaling pathway. Int J Clin Exp Pathol. 7:7249–7261.
2014.PubMed/NCBI
|
|
32
|
Chen Z, Li C, Xu Y, Li Y, Yang H and Rao
L: Circulating level of miR-378 predicts left ventricular
hypertrophy in patients with aortic stenosis. PLoS One.
9:e1057022014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pan D, Mao C, Quattrochi B, Friedline RH,
Zhu LJ, Jung DY, Kim JK, Lewis B and Wang YX: MicroRNA-378 controls
classical brown fat expansion to counteract obesity. Nat Commun.
5:47252014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Meenhuis A, van Veelen PA, de Looper H,
van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru
AH, van Adrichem AJ, et al: MiR-17/20/93/106 promote hematopoietic
cell expansion by targeting sequestosome 1-regulated pathways in
mice. Blood. 118:916–925. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fallah P, Amirizadeh N, Poopak B, Toogeh
G, Arefian E, Kohram F, Rad SM Hosseini, Kohram M, Naghadeh H
Teimori and Soleimani M: Expression pattern of key microRNAs in
patients with newly diagnosed chronic myeloid leukemia in chronic
phase. Int J Lab Hematol. 37:560–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shiotani A, Murao T, Kimura Y, Matsumoto
H, Kamada T, Kusunoki H, Inoue K, Uedo N, Iishi H and Haruma K:
Identification of serum miRNAs as novel non-invasive biomarkers for
detection of high risk for early gastric cancer. Br J Cancer.
109:2323–2330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mei Z, Su T, Ye J, Yang C, Zhang S and Xie
C: The miR-15 family enhances the radiosensitivity of breast cancer
cells by targeting G2 checkpoints. Radiat Res. 183:196–207. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Maudet C, Mano M, Sunkavalli U, Sharan M,
Giacca M, Förstner KU and Eulalio A: Functional high-throughput
screening identifies the miR-15 microRNA family as cellular
restriction factors for Salmonella infection. Nat Commun.
5:47182014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tijsen AJ, van der Made I, van den
Hoogenhof MM, Wijnen WJ, van Deel ED, de Groot NE, Alekseev S,
Fluiter K, Schroen B, Goumans MJ, et al: The microRNA-15 family
inhibits the TGFβ-pathway in the heart. Cardiovasc Res. 104:61–71.
2014. View Article : Google Scholar : PubMed/NCBI
|