Introduction
Acute pancreatitis (AP) is an acute, reversible
inflammatory disease of the pancreas, which may affect several
organs (1). Necrotizing
pancreatitis is characterized by the presence of necrosis in
pancreatic and/or peripancreatic tissues (2). The incidence of AP varies from 17.5
to 109 per 100,000 admissions to emergency departments (3,4). The
risk factors for AP remain unknown; however, smoking, gallstones,
alcohol and drugs are likely to increase the risk of AP and chronic
pancreatitis (5). AP leads to
local and systemic complications, with an overall mortality rate of
4%. In addition, the mortality rate for acute necrotizing
pancreatitis (ANP) is 10–25% (1,6).
Ultrastructural alterations to the pancreatic tissues of patients
with ANP were observed >30 years ago, including morphological
changes in zymogen granules and increased autophagy in the
pancreatic acinar cells (7).
Watanabe et al (8)
described the morphological features of autophagy in
cerulein-induced AP, leading to the understanding of the
association between autophagy and AP. In the pathogenesis of
pancreatitis, intracellular trypsinogen activation has an important
role in its development (9).
Morphological changes in zymogen granules and activation of
plasminogen are associated with the formation of cytoplasmic
vacuoles, which have an important role in autophagy (10).
In cell nuclei, high-motility group box protein 1
(HMGB1) has a role in the transcription of numerous genes, which
are crucial to the autophagy process (11–14).
In the cytoplasm, HMBG1 binds to Beclin 1, thus sustaining the
Beclin 1-Ptdlns3KC3 complex during the activation of autophagy
(15). Extracellular HMGB1 is
involved in the inflammatory response of ANP by triggering
secretion of cytokines, including tumor necrosis factor (TNF)-α and
interleukin (IL)-1β (16,17). Furthermore, as a late inflammatory
factor, HMGB1 has been reported to have a key role in the
development of AP (18).
The exact expression pattern of HMGB1 during ANP
development remains unknown. Therefore, the present study aimed to
investigate the involvement of HMGB1 in the development of ANP, in
order to explore the role of HMGB1 and the autophagy signal Beclin
1 in the development of ANP. The results of the present study may
provide evidence regarding the treatment of ANP.
Materials and methods
Materials and reagents
A total of 48 healthy male Sprague Dawley rats
(weight, 350±30 g; age, 12–14 weeks), were provided by the
Experimental Animal Center of the Third Xiangya Hospital (Changsha,
China). Sodium taurocholate was purchased from Sigma-Aldrich (Merck
Millipore, Darmstadt, Germany). Amylase polyclonal antibody (cat.
no. YT5164) and enzyme-linked immunosorbent assay (ELISA) kit for
HMGB1 (Total HMG-1 Cell-Based Colorimetric ELISA kit) were obtained
from ImmunoWay Biotechnology Company (Plano, TX, USA). Rabbit
anti-HMGB1 (cat. no. ab79823) and anti-Beclin 1 (cat. no. ab55878)
primary antibodies were from Abcam (Cambridge, MA, USA). Goat
anti-rabbit secondary antibodies were purchased from Jackson
ImmunoResearch (West Grove, PA, USA; cat. no. 111-005-144). The
color pre-stained protein marker was purchased from Fermentas
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). The Bradford
protein concentration assay kit was obtained from Beyotime
Institute of Biotechnology (Haimen, China). TRIzol®
reagent was purchased from Invitrogen (Thermo Fisher Scientific,
Inc.). The SYBR Green quantitative polymerase chain reaction (qPCR)
Mix was purchased from Toyobo Co., Ltd. (Osaka, Japan).
Establishment of animal models and
experimental groups
The present study was approved by the ethics
committee of the Experimental Animal Center of the Third Xiangya
Hospital. The rats were housed at a constant temperature of 22°C
under a 12-h light/dark cycle. The rats had access to standard food
and water ad libitum. The ANP rat model was created
according to the method described by Aho et al (19). The rats were fasted for 24 h, with
ad libitum access to water. Subsequently, the rats were
anesthetized by intraperitoneal injection with 10% chloral hydrate
(0.3 ml/100 g). An incision was made into the ventral midline for
entry into the abdominal cavity, in order to expose the common bile
duct and the pancreatic duct. The biliopancreatic duct was occluded
at the distal duodenum using a vascular clip. For the experimental
group, the canal was infused slowly with 5% sodium taurocholate
(0.1 ml/100 g) using a 1-ml syringe at the proximal end. For the
control group, the canal was infused with an equal quantity of
normal saline. After injection, the needle remained motionless for
10 min to prevent drug reflux. After modeling, the skin was
disinfected, the peritoneum was closed with a continuous
single-layer suture, and the skin was sutured with a full-layer
simple-interval suture. After the abdomen had been closed, the rats
were injected with 5 ml saline at the inside position of the lower
limb muscle to supplement water lost during the operation.
Experimental (ANP) rats were grouped according to
the following time points after modeling: 0 h (n=4), 3 h (n=8), 6 h
(n=8), 12 h (n=8) and 24 h (n=8). The control rats were grouped
according to the following time points: 3 h (n=3), 6 h (n=3), 12 h
(n=3) and 24 h (n=3). Rats were sacrificed, at the aforementioned
time points, by cardiac puncture following anesthetization by
intraperiotneal injection with 10% chloral hydrate (0.3 ml/100 g
body weight).
Specimen collection and
preservation
Rats were anesthetized and sacrificed. Blood samples
(~5 ml) were collected from the heart, which were maintained at 4°C
for 30 min and centrifuged at 1,000 × g for 15 min at normal
temperature. The supernatant was then collected and preserved at
−80°C, avoiding repeated freezing and thawing. For electron
microscopy, pancreatic specimens were maintained in 4%
paraformaldehyde for 24 h at 4°C, and were then transferred to
tissue preservation solution (0.2% sodium azide) for preservation.
Other pancreatic tissues were collected, placed in cryogenic tubes
and maintained in liquid nitrogen.
Detection of serum amylase and HMGB1
levels by ELISA assay
Serum amylase and HMGB1 levels were used to
determine if ANP modeling was successful. Serum amylase and HMGB1
levels were assessed using ELISA, according to the manufacturer's
protocol with the amylase antibody used at a dilution of
1:20,000.
Electron microscopy
The pancreatic tissues were trimmed to 1×1×1 mm3,
fixed with 2.5% pentanediol for 24 h, and post-fixed with 2% osmium
tetroxide for 2 h; dehydrated with a gradient of 50%, 70%, 90%, and
100% dehydrated acetone, in each solution three times with each
time 10 min. The samples were then placed in an epoxy resin and
pure acetone mixture (1:1) at 37°C for 24 h, prior to embedding in
a mixture of Epon812 resin, methyl nadic anhydride,
dodecenylsuccinic anhydride, and dimethylaminomethyt phenol, at
60°C for 24 h. The blocks were trimmed with semi-thin glasscutter,
stained with toluidine blue, and observed under light microscopy to
select areas with pancreatic acinar structure. Ultrathin sections
with a thickness of 500 Å were obtained with an ultrathin microtome
and stained with uranyl acetate and lead nitrate. The sections were
observed under transmission electron microscopy (Nissan HT7700;
Nissan Corporation, Tokyo, Japan) to examine changes of the
pancreatic acinar autophagosomes.
Immunohistochemistry of HMGB1 and
Beclin 1 in pancreatic tissue
Sections (4 µm) of paraffin-embedded pancreatic
tissues were dewaxed and hydrated, and were washed three times with
phosphate-buffered saline (PBS; pH 7.4; 5 min/wash). Subsequently,
the samples were boiled under high-pressure in 10 mM citric acid
buffer (pH 6.0) for 30 min for antigen retrieval, cooled to room
temperature, and washed with distilled water and PBS (2×5 min).
Sections were incubated with 3% hydrogen peroxide at room
temperature for 1 min, and were washed three times with PBS (5
min/wash). Sections were then blocked with goat antiserum (Abcam)
for 7 min at 37°C, and were washed with PBS (5 min). Subsequently,
the samples were incubated with the following primary antibodies at
4°C overnight: Anti-HMGB1 or anti-Beclin 1 (1:200). Sections were
then rinsed three times with PBS (5 min/wash), and a biotinylated
secondary antibody (1:100) was then added to the sections, which
were incubated at room temperature for 1 h and rinsed in a
humidified chamber. Subsequently, enzyme-labeled streptavidin (50
µl; Wuhan Boster Biological Technology, Co., Ltd., Wuhan, China)
was added, incubated at room temperature for 30 min and rinsed in a
humidified chamber. Sections were stained with DAB (Sigma-Aldrich;
Merck Millipore), observed under a BX53 microscope (Olympus
Corporation, Tokyo, Japan), and when the color was suitable, the
staining reaction was terminated by rinsing. Sections were then
counterstained with hematoxylin, rinsed with water, dehydrated in
gradient alcohol, cleared with xylene, and finally sealed with
neutral resin. Images were captured using a camera system (Motic
B5; Motic, Xiamen, China). Yellow-brown staining of the nuclei or
cytoplasm was recognized as positive staining. Densitometric
quantitative analysis of the expression levels was carried out
using the Image-Pro Plus 6.0 image system (Media Cybernetics, Inc.,
Rockville, MD, USA).
Nuclear extraction
Tissue samples were washed with saline to remove
blood. Subsequently, 400 µl solution A [200 µl KCl (10 mM), 2 ml
HEPES (10 mM, pH 7.9), 20 µl EDTA (0.1 mM), 1.2 ml MgCl2
(1.5 mM), 200 µl DTT (1 mM), 100 µl PMSF (0.5 mM) and 16.28 ml of
dH2O] was added. Tissue samples were homogenized and
placed on ice for 15 min. Samples were then centrifuged at 15,000 ×
g for 1 min at 4°C. The supernatants (cytoplasmic proteins) were
transferred to a new tube, and the pellet (nuclei) was resuspended
in 400 µl solution B [5 ml glycerol (25%), 4 ml HEPES (20 mM; pH
7.9), 4 ml NaCl (400 mM), 200 µl EDTA (1 mM), 200 µl DTT (1 mM),
200 µl PMSF (1 mM) and 6.4 ml dH2O). Samples were placed
on ice for 20 min, and were then centrifuged at 15,000 × g for 10
min at 4°C. The supernatants (nuclear proteins) were transferred to
new tubes and were stored at −80°C until further use.
Western blot analysis for HMGB1 and
Beclin 1
Following extraction of the cytoplasmic and nuclear
proteins, 5% SDS-polyacrylamide gel electrophoresis was used to
separate the proteins (10 µg samples), which were then transferred
to polyvinylidene fluoride (PVDF) membranes. After blocking with 5%
skim milk at room temperature for 2 h, the PVDF membranes were
agitated overnight at 4°C with anti-HMGB1 or anti-Beclin 1
(1:1,000). Subsequently, the membranes were agitated with
horseradish peroxidase-labeled goat anti-rabbit secondary antibody
(1:1,000) at room temperature for 1 h. The blots were visualized
following treatment with enhanced chemiluminescence solution in a
dark room for light exposure. The PVDF membrane was then cleaned
with eluent to remove antibodies, and was hybridized with rabbit
polyclonal anti-β-actin antibodies (1:1,000; Abcam; cat. no.
ab8227) or rabbit anti-histone H3 (1:1,000; RayBiotech, Inc.,
Norcross, GA, USA; cat. no. 168-10463) and a goat anti-rabbit
horseradish-peroxidase secondary antibody (1:1,000; Abcam; cat. no.
ab6721). The films were scanned, the optical density values of
HMGB1, Beclin 1 and β-actin (internal control) bands were measured
using an image analysis system (Quantity One software version 4.62;
Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the ratios were
obtained for semi-quantitative analysis.
qPCR for HMGB1 and Beclin 1
Total RNA was extracted from the pancreatic tissues
using TRIzol® at various time points (0, 3, 6, 12 and 24
h) following ANP modeling. For each sample, 2.0 µg total RNA was
reverse transcribed to cDNA using 1 µl oligo (dT) (0.5 µg/µl)
primer according to the protocol of the First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.). PCR primers were designed
according to the gene sequences downloaded from National Center for
Biotechnology Information (http://www.ncbi.nlm.nih.gov/genbank/) using Primer 5.0
software (Premier Biosoft, Palo Alto, CA, USA). The PCR primer
sequences were sequenced by Sangon Biotech., Co., Ltd. (Shanghai,
China) as follows: HMGB1, sense 5′-GCTCAGAGAGGTGGAAGAC-3′,
antisense 5′-CCAATGGATAAGCCAGGAT-3′; Beclin 1, sense
5′-TGTGGAATGGAATGAAATCAA-3′, antisense 5′-CCCCCAGAACAGTACAACGGC-3′;
and β-actin (internal reference), sense 5′-CCCATCTATGAGGGTTACGC-3′
and antisense 5′-TTTAATGTCACGCACGATTTC-3′. The PCR cycling
conditions were as follows: Pre-denaturation at 95°C for 3 min;
denaturation at 95°C for 10 sec, and annealing and extension at
58°C (HMGB1 and β-actin) or 60°C (Beclin 1) for 30 sec, for a total
of 35 cycles. The PCR reaction was performed in a total of 20 ml,
including 10 µl 2X SYBR Green qPCR mix, 1 µl primers (10 µM), 1 µl
cDNA and 8 µl DEPC-water, the reaction was conducted in an
Eppendorf MasterCycler Realplex qPCR machine. Fluorescence was read
at each extension phase. The Cq values were obtained and calculated
by 2-ΔΔCq formula (20) relative
to the mean of 0 h values, to get the relative expression
values.
Statistical analysis
Data are presented as the mean ± standard deviation,
and were analyzed using Student's t-test and one-way analysis of
variance followed by the Student-Newman-Keuls post-hoc test. SPSS
13.0 (SPSS Inc., Chicago, IL, USA) was used for statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Successful model generation
As presented in Table
I, amylase levels were increased 3 h after modeling. No rats
succumbed within the 24 h experimental period. This is consistent
with our previous modeling approach (21).
 | Table I.Serum amylase levels in a rat ANP
model, as determined by enzyme-linked immunosorbent assay. |
Table I.
Serum amylase levels in a rat ANP
model, as determined by enzyme-linked immunosorbent assay.
| Group | 3 h | 6 h | 12 h | 24 h |
|---|
| Control | 62.81±8.05 | 65.81±6.31 | 70.81±8.57 | 78.81±6.46 |
| ANP | 734.13±65.30a |
1206.76±83.27a,b |
2006.24±119.81a,b |
1825.90±105.14a,b |
TEM
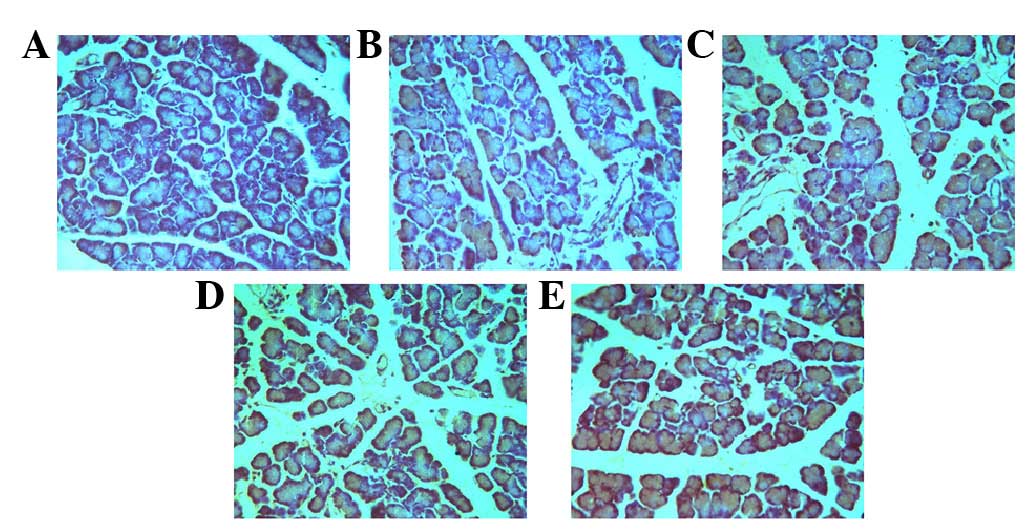
As presented in Fig.
1, phagocytic vesicles appeared in some pancreatic cells at 3
h. The number of phagocytic vesicles increased in a time-dependent
manner. These vesicles exhibited typical autophagy-like morphology
(Fig. 1A-E).
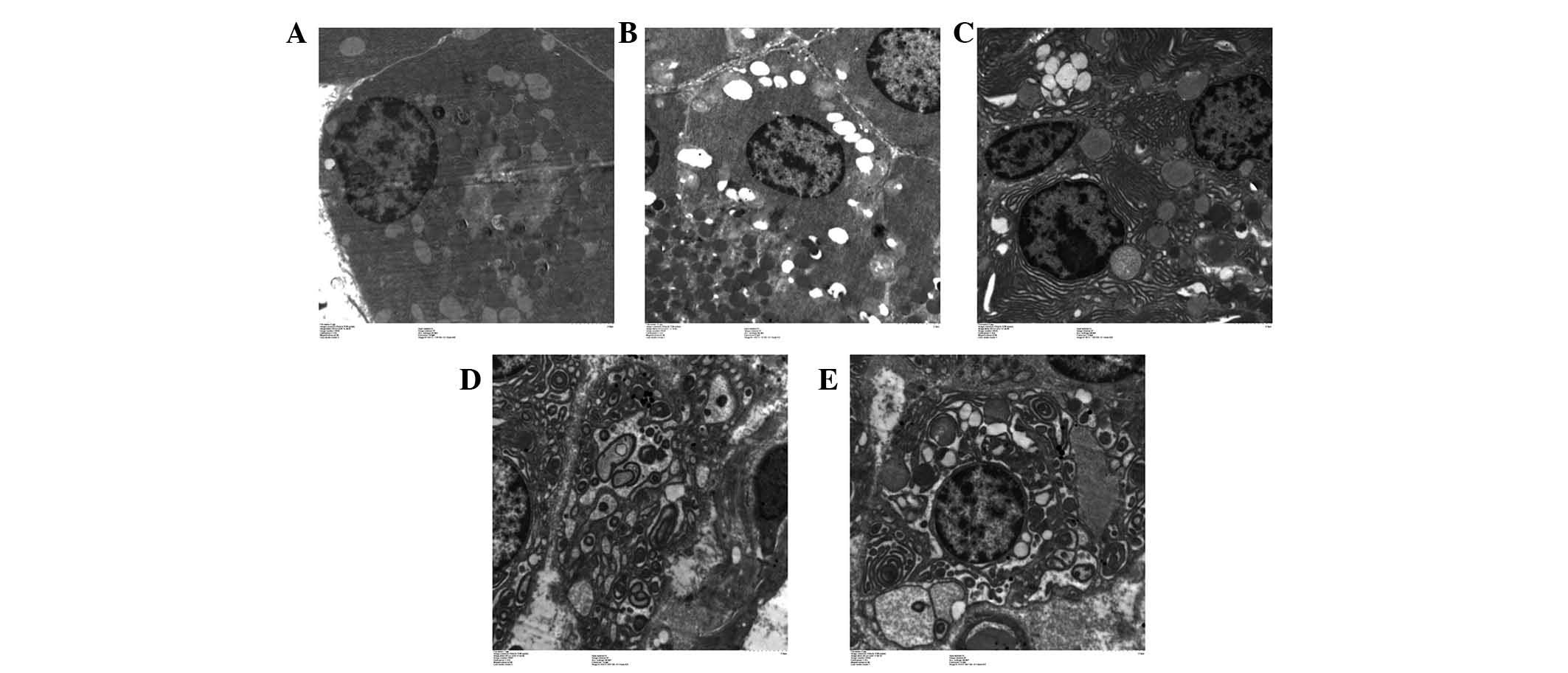 | Figure 1.Autophagy observed under transmission
electron microscopy. (A) Pancreatic acinar cells in the acute
necrotizing pancreatitis (ANP)-0 h group exhibited normal acinar
nucleolus, mitochondria and rough endoplasmic reticulum.
Occasionally, visible phagosomes and lysosomes were generated due
to the normal stress of starvation. (B) In the ANP-3 h group, the
number of phagocytic vesicles was increased, indicating enhanced
autophagy. (C) In the ANP-6 h group, the pancreatic acinar cells
exhibited cystic expansion of the endoplasmic reticulum,
mitochondrial swelling, and margination of nuclear chromatin. (D)
In the ANP-12 h group, phagosomes continued to increase, and the
fusion of phagocytic vesicles with lysosomes increased, resulting
in a large number of double membrane-like structures. (E) In the
ANP-24 h group, the large number of double membrane-like structures
increased, providing membrane sources for phagosome formation.
Magnification, ×10,000. |
HMGB1 and Beclin 1 expression in
pancreatic tissue
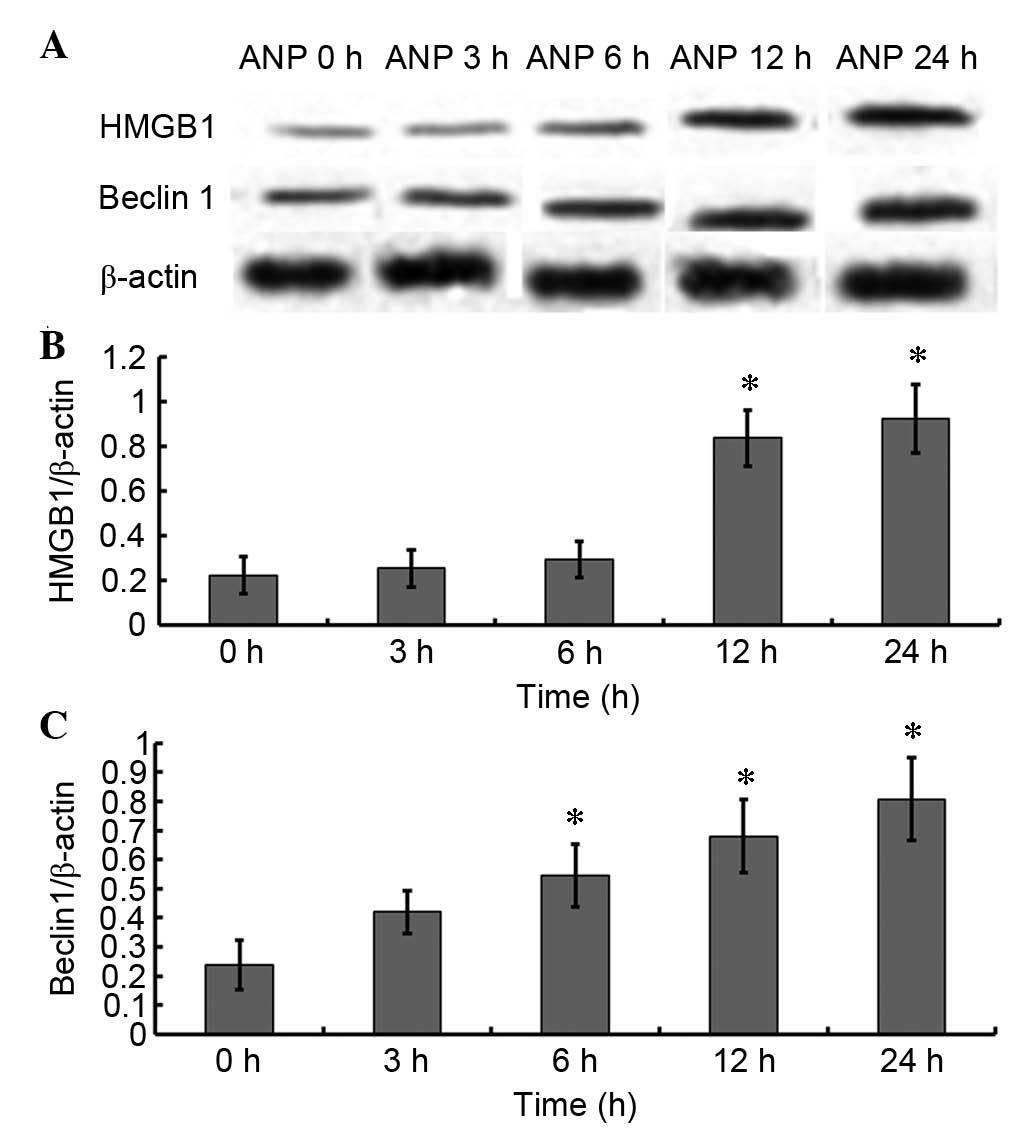
The protein expression levels of HMGB1 and Beclin 1
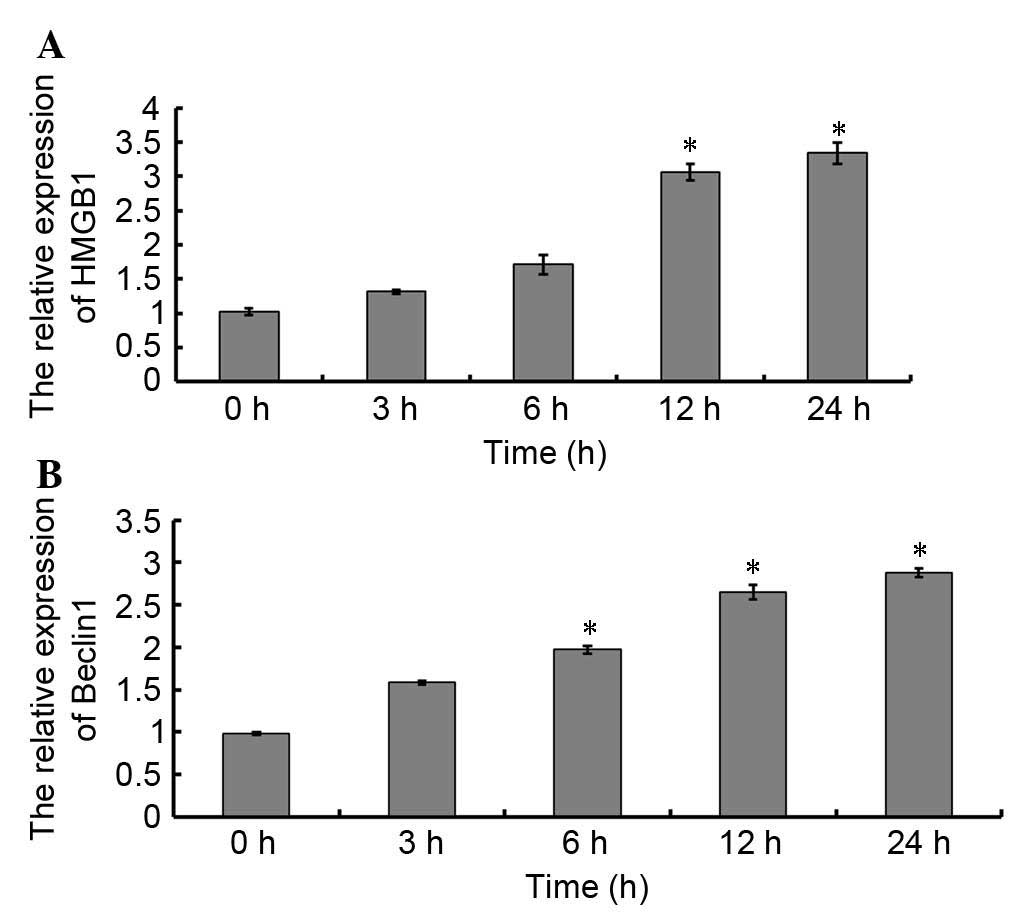
were detected by western blotting (Fig. 2). As presented in Fig. 2A and B, the protein expression
levels of HMGB1 began to increase at 12 h; this finding was
supported by the results of a qPCR (Fig. 3A). However, unlike HMGB1, the
translation (Fig. 2A and C) and
transcription levels (Fig. 3B) of
Beclin 1 began to increase at 3 h. Similar expression patterns of
HMGB1 and Beclin 1 were observed using immunohistochemistry
(Figs. 4 and 5, Table
II).
 | Table II.Quantitative analysis of Beclin 1 and
HMGB1 immunohistochemistry in a rat model of acute necrotizing
pancreatitis. |
Table II.
Quantitative analysis of Beclin 1 and
HMGB1 immunohistochemistry in a rat model of acute necrotizing
pancreatitis.
| Protein | 0 h | 3 h | 6 h | 12 h | 24 h |
|---|
| Beclin 1 | 1.72±0.16 | 4.72±0.62a | 8.86±0.59a | 14.66±0.61a | 19.52±0.77a |
| HMGB1 | 3.72±0.86 | 3.76±0.82 | 4.86±0.99 | 12.46±0.88a | 18.50±0.87a |
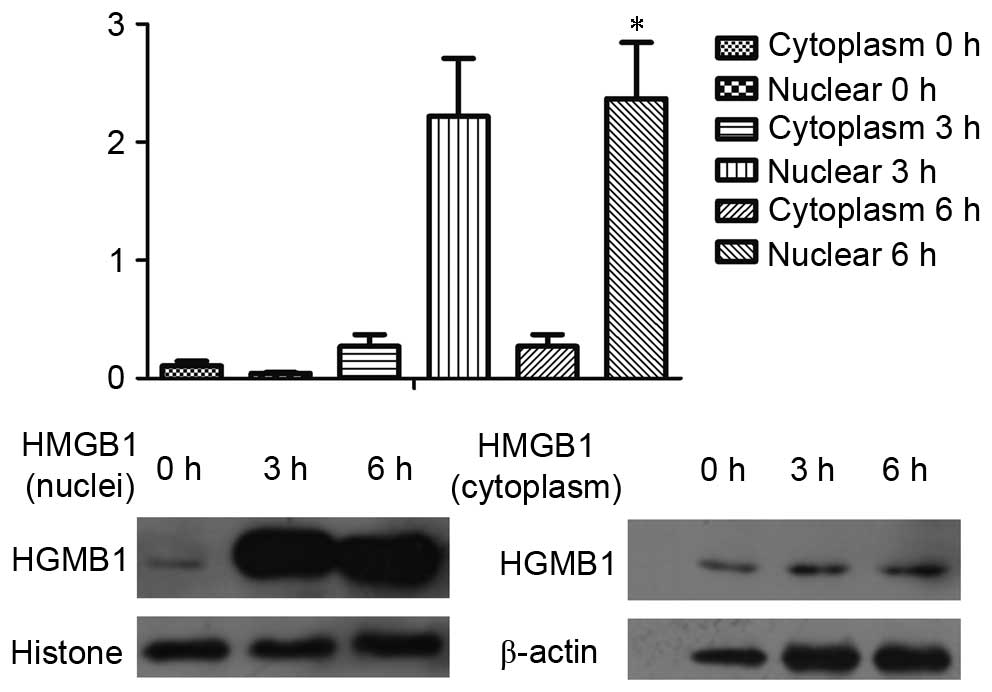
Nuclear protein levels of HMGB1 in
pancreatic cells in the early stage of ANP
Based on the TEM results, autophagy appeared at 3 h,
which corresponded with the increase in amylase levels at 3 h.
However, HMGB1 levels did not increase until 12 h. Since HMGB1 is a
nuclear factor, it may promote autophagy from the nucleus (22). Therefore, the nuclei were isolated
in the present study. Notably, HMGB1 was shown to be accumulated in
the nuclei at the early stage of ANP (Fig. 6).
HMGB1 may be secreted and induce
systemic lesions as an inflammatory factor
It has previously been reported that HMGB1 may be
secreted outside of cells, where is has a role as an inflammatory
cytokine (23). ELISA was used to
evaluate the presence of HMGB1 in the serum at various time points
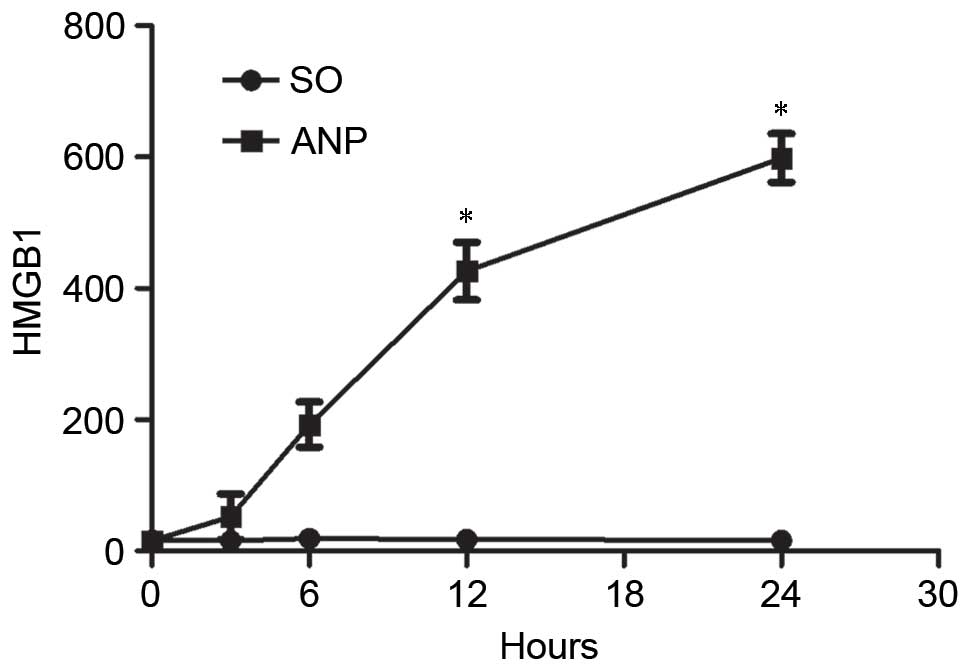
after ANP modeling. As presented in Fig. 7, compared with the control group,
HMGB1 levels in the serum began to increase at 3 h, and were
highest at 24 h after modeling.
Discussion
In eukaryotes, autophagy is involved in cell
physiological and pathological processes. Autophagy has a dual
role, and can either protect cells against damage, or promote
pathological processes associated with changes to the cell
environment (24–28). In AP, autophagy is involved in all
pathological processes; it protects the pancreas in the early
stages, but promotes disease progression in the late stages
(10). HMBG1 is involved in
ANP-associated autophagy (18,23,29);
however, the exact involvement of HMGB1 in ANP remains unknown.
The aim of the present study was to determine the
expression pattern of HMGB1 in ANP, and to investigate its
association with autophagy. The results of the present study
demonstrated that autophagy was detected 3 h after generation of an
ANP model; however, HMGB1 expression remained unaltered during the
early stage (0–6 h). HMGB1 was significantly increased at 12 h, and
was shown to be still rising at 24 h. Notably, HMGB1 expression was
significantly increased in the nuclei, rather than in the
cytoplasm, at 3–6 h. In addition, in the serum, HMGB1 levels began
to increase at 3 h, and reached the highest levels at 24 h in the
ANP group.
In the present study, a rat model of ANP was
successfully established by retrogradely injecting sodium
taurocholate into the biliopancreatic duct. Beclin 1, which is a
critical factor in autophagy, continuously increased with the
development of ANP. Furthermore, HMGB1 expression in the pancreatic
tissue was not significantly altered 3 and 6 h after modeling, but
was significantly increased at 12 and 24 h. A previous study
demonstrated time differences in the expression of early
inflammatory factors, including HMGB1, TNF-α, IL-1 and IL-6 in
pancreatic tissue (29). HMGB1 may
act as a key signaling molecule in activating or promoting
autophagy when early inflammatory factors begin to decrease;
therefore, the results of the present study lay a theoretical
foundation to further explore the role of HMGB1 as a key factor in
the development of AP. The results of the present study are
supported by the results of a previous study, which detected
increased serum and pancreatic levels of HMGB1 in humans with
pancreatitis and animal models of pancreatitis (30).
Activation of trypsinogen particles and nuclear
factor (NF)-κB by autophagy is the initiating event of pancreatitis
development, and has been recognized as the main pathological
mechanism underlying the occurrence and development of AP (9). It has previously been demonstrated
that early inflammatory factors, such as NF-κB, are downregulated
~12 h after the onset of AP (18);
however, AP-induced inflammation continues, and other inflammatory
cytokines act to increase inflammation. In autophagy, factors such
as vacuole membrane protein 1, microtubule-associated protein light
chain 3 II and Beclin 1 are involved in the non-caspase-dependent
signaling pathway and have an important role in autophagy (31). However, the exact role of HMGB1 in
ANP-associated autophagy is ill known. In cell nuclei, HMGB1 has a
role in the transcription of numerous genes that are crucial to the
autophagy process (11–14). Indeed, the present study detected
increased nuclear localization of HMGB1 post-ANP induction, which
may be associated with the increased autophagy features observed
using TEM; these results are supported by a previous study
(20). In addition, in AP,
inhibition of Beclin 1 activity can inhibit autophagy and
significantly reduce tissue damage (32). In the cytoplasm, HMBG1 binds to
Beclin 1, sustaining the Beclin 1-Ptdlns3KC3 complex during the
activation of autophagy (15).
Inhibiting HMGB1 action has been shown to suppress
the development of pancreatitis (29). In addition, vitamin K3 is able to
inhibit cerulein-induced AP via the inhibition of autophagy, and
may reduce tissue damage (33).
Therefore, inhibiting autophagy is possible to prevent the
development of AP, whereas inducing autophagy may aggravate the
condition. However, more studies are required to determine if
Beclin 1 or HMGB1 could be considered as possible therapeutic
targets in ANP.
The present study is not without its limitations.
Firstly, it was performed in animals, and the exact involvement of
HMGB1 in ANP may be subtly different in humans. In addition, the
present study very superficially explored the involvement of HMGB1
in the mechanisms underlying ANP. Further studies are required to
explore these mechanisms in detail. For example, Toll-like
receptors and receptor for advanced glycation end products have
been reported to be activated by HMGB1 and mediate the
extracellular effects of HMGB1 (34,35).
In conclusion, in the ANP model generated in the
present study, HMGB1 was initially increased in the nuclei, which
may result in the initiation of autophagy. Subsequently, it was
translocated into the cytoplasm, where it may interact with Beclin
1 to enhance autophagy, and HMGB1 was released into the blood,
leading to deterioration of ANP.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81270545), the
Science and Technology Department of Hunan Province Foundation of
China (grant no. 2013FJ6021), and by the Fundamental Research Funds
for the Central Universities of Central South University (grant no.
2015zzts124).
References
|
1
|
Frossard JL, Steer ML and Pastor CM: Acute
pancreatitis. Lancet. 371:143–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banks PA, Bollen TL, Dervenis C, Gooszen
HG, Johnson CD, Sarr MG, Tsiotos GG and Vege SS: Acute Pancreatitis
Classification Working Group: Classification of acute
pancreatitis-2012: Revision of the atlanta classification and
definitions by international consensus. Gut. 62:102–111. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
O'Farrell A, Allwright S, Toomey D,
Bedford D and Conlon K: Hospital admission for acute pancreatitis
in the Irish population, 1997 2004: Could the increase be due to an
increase in alcohol-related pancreatitis? J Public Health (Oxf).
29:398–404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fagenholz PJ, Fernandez-del Castillo C,
Harris NS, Pelletier AJ and Camargo CA Jr: National study of united
states emergency department visits for acute pancreatitis,
1993–2003. BMC Emerg Med. 7:12007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tolstrup JS, Kristiansen L, Becker U and
Grønbaek M: Smoking and risk of acute and chronic pancreatitis
among women and men: A population-based cohort study. Arch Intern
Med. 169:603–609. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
American Gastroenterological Association
(AGA) Institute on ‘Management of Acute Pancreatits’ Clinical
Practice and Economics Committee; AGA Institute Governing Board:
AGA Institute, . Medical position statement on acute pancreatitis.
Gastroenterology. 132:2019–2021. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Helin H, Mero M, Markkula H and Helin M:
Pancreatic acinar ultrastructure in human acute pancreatitis.
Virchows Arch A Pathol Anat Histol. 387:259–270. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Watanabe O, Baccino FM, Steer ML and
Meldolesi J: Supramaximal caerulein stimulation and ultrastructure
of rat pancreatic acinar cell: Early morphological changes during
development of experimental pancreatitis. Am J Physiol.
246:G457–G467. 1984.PubMed/NCBI
|
|
9
|
Sah RP and Saluja A: Molecular mechanisms
of pancreatic injury. Curr Opin Gastroenterol. 27:444–451. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grasso D, Ropolo A, Lo Re A, Boggio V,
Molejon MI, Iovanna JL, Gonzalez CD, Urrutia R and Vaccaro MI:
Zymophagy, a novel selective autophagy pathway mediated by
VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem.
286:8308–8324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang D, Kang R, Cheh CW, Livesey KM, Liang
X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et
al: HMGB1 release and redox regulates autophagy and apoptosis in
cancer cells. Oncogene. 29:5299–5310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang D, Kang R, Livesey KM, Cheh CW,
Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, et al:
Endogenous HMGB1 regulates autophagy. J Cell Biol. 190:881–892.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang D, Kang R, Livesey KM, Kroemer G,
Billiar TR, Van Houten B, Zeh HJ III and Lotze MT: High-mobility
group box 1 is essential for mitochondrial quality control. Cell
Metab. 13:701–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang D, Kang R, Livesey KM, Zeh HJ III and
Lotze MT: High mobility group box 1 (HMGB1) activates an autophagic
response to oxidative stress. Antioxid Redox Signal. 15:2185–2195.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang R, Lotze MT, Zeh HJ, Billiar TR and
Tang D: Cell death and DAMPs in acute pancreatitis. Mol Med.
20:466–477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livesey KM, Kang R, Vernon P, Buchser W,
Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ III, Li L, et
al: p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer
Res. 72:1996–2005. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang ZW, Zhang QY, Zhou MT, Liu NX, Chen
TK, Zhu YF and Wu L: Antioxidant inhibits HMGB1 expression and
reduces pancreas injury in rats with severe acute pancreatitis. Dig
Dis Sci. 55:2529–2536. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aho HJ, Koskensalo SM and Nevalainen TJ:
Experimental pancreatitis in the rat. sodium taurocholate-induced
acute haemorrhagic pancreatitis. Scand J Gastroenterol. 15:411–416.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sailai Y, Yu X, Baiheti P, Tang H, Li Y
and Xu M: Influence of nuclear factor-κB activation on inflammatory
mediators of alveolar macrophages in rats with acute necrotizing
pancreatitis. Investigative Med. 58:38–42. 2010. View Article : Google Scholar
|
|
22
|
Skinner M: Autophagy: In the hands of
HMGB1. Nat Rev Mol Cell Biol. 11:756–757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Scaffidi P, Misteli T and Bianchi ME:
Release of chromatin protein HMGB1 by necrotic cells triggers
inflammation. Nature. 418:191–195. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ogier-Denis E and Codogno P: Autophagy: A
barrier or an adaptive response to cancer. Biochim Biophys Acta.
1603:113–128. 2003.PubMed/NCBI
|
|
25
|
Klionsky DJ: Autophagy. Curr Biol.
15:R282–R283. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: A double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marino G and Lopez-Otin C: Autophagy:
Molecular mechanisms, physiological functions and relevance in
human pathology. Cell Mol Life Sci. 61:1439–1454. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eskelinen EL: Maturation of autophagic
vacuoles in mammalian cells. Autophagy. 1:1–10. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yasuda T, Ueda T, Shinzeki M, Sawa H,
Nakajima T, Takeyama Y and Kuroda Y: Increase of high-mobility
group box chromosomal protein 1 in blood and injured organs in
experimental severe acute pancreatitis. Pancreas. 34:487–488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shen X and Li WQ: High-mobility group box
1 protein and its role in severe acute pancreatitis. World J
Gastroenterol. 21:1424–1435. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pyo JO, Nah J and Jung YK: Molecules and
their functions in autophagy. Exp Mol Med. 44:73–80. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang S, Bing M, Chen F, Sun Y, Chen H and
Chen W: Autophagy regulation by the nuclear factor κB signal axis
in acute pancreatitis. Pancreas. 41:367–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chinzei R, Masuda A, Nishiumi S, Nishida
M, Onoyama M, Sanuki T, Fujita T, Moritoh S, Itoh T, Kutsumi H, et
al: Vitamin K3 attenuates cerulein-induced acute pancreatitis
through inhibition of the autophagic pathway. Pancreas. 40:84–94.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Asavarut P, Zhao H, Gu J and Ma D: The
role of HMGB1 in inflammation-mediated organ injury. Acta
Anaesthesiol Taiwan. 51:28–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schmidt AM, Hofmann M, Taguchi A, Yan SD
and Stern DM: RAGE: A multiligand receptor contributing to the
cellular response in diabetic vasculopathy and inflammation. Semin
Thromb Hemost. 26:485–493. 2000. View Article : Google Scholar : PubMed/NCBI
|