Introduction
Bartter syndrome (BS) is a group of autosomal kidney
disorders characterized by hypokalemic metabolic alkalosis,
hypercalciuria and salt wasting (1). BS has five distinct genetic subtypes.
BS types 1, 2, 3 and 4A are all autosomal recessive and are caused
by loss-of-function mutations in the solute carrier family 12,
member 1 (SLC12A1), potassium inwardly-rectifying channel
subfamily J, member 1 (KCNJ1), chloride channel Kb
(CLCNKB), and Barttin CLCNK type accessory β-subunit
(BSND) genes, respectively (1). BS type 4B is caused by simultaneous
heterozygous mutations in the CLCNKA and CLCNKB genes
(2). BS type 5 demonstrates
autosomal dominant inheritance, and is caused by a gain-of-function
mutation in the CASR gene (3). Clinically, BS is classified into
three types based primarily on phenotype and age of onset,
including antenatal BS (BS type 1 and type 2), classic BS (BS type
3), and antenatal BS with sensorineural deafness (BS type 4A, type
4B, and type 5) (2,4). Of these subtypes, BS type 1 (Online
Mendelian Inheritance in Man; no. 600839; www.omim.org/entry/600839) is clinically categorized
as antenatal BS and is usually associated with life-threatening
phenotypes (4,5). The SLC12A1 gene is currently
the only known gene that leads to BS type 1 (6,7).
This gene is located at 15q21.1 and contains 27 exons. Of these 27
exons, 26 of them are protein-encoding exons. The SLC12A1
gene has kidney-specific transcription (NM_000338.2; National
Center for Biotechnology Information; www.ncbi.nlm.nih.gov) and encodes the Na-K-2Cl
co-transporter. The carrier-frequency was estimated to be 1 in 360
(8). To date, 68 different BS type
1-causing mutations have been documented in the Human Gene Mutation
Database (HGMD; www.hgmd.cf.ac.uk) and in the literature (7–21).
The mutation types included missense/nonsense mutations, splicing
mutations, small insertions and small deletions. Deletions
encompassing an entire exon have not yet been reported.
In the current study of a single patient suspected
to have BS type I, a novel homozygous frameshift mutation
(c.1833delT) in SLC12A1 was initially detected by Sanger
sequencing. However, in the parental study, the c.1833delT
frameshift mutation was detected only the father but not in the
mother. Upon further investigation, and after paternal segmental
isodisomy 15 was ruled out, it was discovered that the proband
harbored a 3.16 kb deletion spanning the region where the
c.1833delT mutation was detected. Clearly, this deletion was missed
by Sanger sequencing. To the best of our knowledge, the present
study is the first to report an extensive deletion in the
SLC12A1 gene associated with BS type 1.
Materials and methods
DNA isolation
Genomic DNA was extracted from the peripheral white
blood cells of the proband and their parents using an automatic
nucleic acid isolation system (QuickGene-610L; Fujifilm Holdings
Corporation, Tokyo, Japan) according to the manufacturer's
protocols. The DNA concentration and quality were measured using
the Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific,
Inc., Wilmington, DE, USA) according to manufacturer's
instructions.
Sanger sequencing
Polymerase chain reaction (PCR) was performed using
specific primers designed and generated by Primer3 v0.4.0 software,
(http://bioinfo.ut.ee/primer3/) in our
laboratory (University of Oklahoma Health Sciences Center, Oklahoma
City, OK, USA; Table I) targeting
the coding region and the intron-exon junctions of the
SLC12A1 gene. Sanger sequencing was performed using the
BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and an ABI 3500xL genetic
analyzer (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Sequences were analyzed using Mutation
Surveyor software v4.0.9 (SoftGenetics LLC., State College, PA,
USA).
 | Table I.Primers used for Sanger sequencing of
the SLC12A1 gene. |
Table I.
Primers used for Sanger sequencing of
the SLC12A1 gene.
| Exon | Forward primer | Reverse primer | Product size
(bp) |
|---|
| 2 |
5′-AGCTCCCTAATGGAAGCACA-3′ |
5′-GTAAGTAAGATGGATAGTGTTT-3′ | 531 |
| 3 and 4 |
5′-TCAATTGTTTTGATTTGCTTTGA-3′ |
5′-TGCTTATTGAATTTATGATTTACATGC-3′ | 507 |
| 5 |
5′-GAGGCATGGACCTGAAAACT-3′ |
5′-GCATCCAGCTCCTCCAAATA-3′ | 337 |
| 6 |
5′-TCAGATAGTCACAATCGTTTGGTT-3′ |
5′-TCCCTTAGTGCCCTGAGAAG-3′ | 289 |
| 7 |
5′-CCTATATGGCCCCAGGTGTA-3′ |
5′-TGCCTGCTCATTTCACCATA-3′ | 351 |
| 8 |
5′-TCTGGGTAGCAGAGACTTAACTGA-3′ |
5′-TGATGGGGATGGTGATGAG-3′ | 311 |
| 9 |
5′-GGACTAGGGAAGCCAATGGT-3′ |
5′-TTTGAATCTGTAGGGTAATATGGTCA-3′ | 318 |
| 10 |
5′-CATCAACTTGCTGTTTGCTTG-3′ |
5′-TGCTGCATTGAAAGCTCACT-3′ | 313 |
| 11 |
5′-CAGAGGCTAAGAAATGGACCTT-3′ |
5′-GCCAATAATCAATCAGTTGCAG-3′ | 363 |
| 12 |
5′-CTGGGCGATAGAGCGAGACT-3′ |
5′-ACTTGAGCCAGATGCAAACA-3′ | 401 |
| 13 |
5′-TCCCCAAATCTTCTTGTTTGA-3′ |
5′-CAAACTAAAAGGAAAGCCCTATGA-3′ | 286 |
| 14 |
5′-TGACAGATGCTCGCTATGTTTT-3′ |
5′-GGATTACGCTTCCCACTAGG-3′ | 352 |
| 15 |
5′-AGATTCTGGAACTTGGCCTAAA-3′ |
5′-ATCCACCTTACATATGCCTCA-3′ | 401 |
| 16 |
5′-ATTTGGGCACTCATCTTTGC-3′ |
5′-TTTGTATGACTGCTTATTTTTAGAATG-3′ | 354 |
| 17 |
5′-GAGAGGTTGCCCCATTTTTC-3′ |
5′-GCATGTACCTTTTCTCCTCACC-3′ | 306 |
| 18 |
5′-GGTCATCTCCAAAAGGCTGA-3′ |
5′-TGCTTGGCTGCCTAACTACA-3′ | 413 |
| 19 |
5′-TCAGGATCTTCAGAACATTACTTCA-3′ |
5′-AAGTCAGAGTACCAGGGGAATG-3′ | 353 |
| 20 |
5′-AAATGCATCAGCTCTTGGCTA-3′ |
5′-GCCCTGCTGTATCCCATAAC-3′ | 310 |
| 21 |
5′-GGTGATTTTGTCTTCTTTCATCA-3′ |
5′-TGGAGAGAAACCTTTCAGTTCCT-3′ | 301 |
| 22 |
5′-TTGTTTCTGCCCTCAAAAGC-3′ |
5′-TCCCATATACCTTCTCATGCAGT-3′ | 240 |
| 23 |
5′-ACTTAATTAAGAGCTATCAA-3′ |
5′-ATTAAAGCAACAAACCTCTGAAATG-3′ | 270 |
| 24 |
5′-CCAACCAAAAAGCCTCTGTC-3′ |
5′-TCAAGAAGTCGCTGCAGTAAA-3′ | 336 |
| 25 |
5′-GCATGATATTCAGCTCTGATTCC-3′ |
5′-TGAAAAATGCAGAAAGCTTGG-3′ | 385 |
| 26 |
5′-AAACACCATAAGTTTCTAAGCCTGA-3′ |
5′-CCTGAAGAGTCCCAAGCTTTT-3′ | 308 |
| 27 |
5′-GCTCAGAAATACTAGTGCCGTTA-3′ |
5′-TCCTCTCCAGAGGTTTGCAT-3′ | 324 |
Uniparental disomy study (UPD)
Genomic DNA (100 ng) templates from the proband and
the parents were subject to investigation by PCR amplification.
This was performed using the following four pairs of short tandem
repeat (STR) primer sets (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) on different loci of chromosome 15q: D15S1024,
15q21.1; D15S131, 15q23; D15S984, 15q24.3 and D15S115, 15q25.2. The
amplified PCR products were subjected to fragment analysis using
the 3500xL Genetic Analyzer (Thermo Fisher Scientific, Inc.). Data
analysis was performed using GeneMapper Software 4.1 (Thermo Fisher
Scientific, Inc.).
Ion Personal Genome Machine (PGM) next
generation target sequencing
All products used for Ion PGM sequencing were
supplied by Thermo Fisher Scientific, Inc., unless stated
otherwise. In order to determine the mutant allele distribution and
exclude the potential heterozygous deletion spanning the c.1833delT
mutation as determined by Sanger sequencing, DNA samples of the
proband, the mother and a normal control cell line (Coriell
Institute for Medical Research, Camden, NJ, USA) were subjected to
Ion PGM target sequencing to detect the possible deletion based on
the method in the previous study by Faiz et al (22). Genomic DNA (10 ng) from each sample
was used to prepare a barcoded library of specific amplicons using
the Ion AmpliSeq Library kit 2.0 and the Ion AmpliSeq™ custom
panels for the SLC12A1 gene, designed using the Ion
AmpliSeq™Designer web-hosted software v3.0 (www.ampliseq.com). The libraries generated from the
each DNA sample were clonally amplified with the Ion PGM™ Template
OT2 200 kit and the Ion OneTouch™ 2 System for templating and
enrichment prior to chip loading. Sequencing was performed using
the Ion 314™ Chip with the Ion PGM™ sequencing 200 kit on the Ion
PGM™ system. Conversion of the raw signal to base calls was
performed on the Torrent Server using Torrent Suite Software v3.4
(Thermo Fisher Scientific, Inc.). Base-called files were aligned to
the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/) hg19 reference genome to
calculate run quality metrics. Sequence reads were generated and
mapped to the hg19 sequence using the Torrent Suite Software v3.4.
The Torrent Variant Caller utilized the mapped reads to identify
single nucleotide polymorphisms, and insertion and deletion
variants. The standard output files of the Ion PGM™ system,
including BAM and VCF, were also analyzed with Integrative Genomics
Viewer software v2.3 (www.broadinstitute.org/igv/).
Array comparative genomic
hybridization (CGH)
Array CGH, using the Agilent 2×400 K chip (Agilent
Technologies, Inc., Santa Clara, CA, USA), was performed to confirm
the deletion in the proband identified by sequencing analysis.
Human reference genomic DNA was purchased from Agilent (Agilent
Technologies, Inc.). The DNA (1 µg) of the proband and the
reference DNA were labeled with cyanine 3 and cyanine 5,
respectively, by random priming (Agilent Technologies, Inc.).
Proband DNA was combined with a normal control DNA sample and
hybridized to an Agilent 2×400 K oligo microarray chip (cat. no.
G4448A; Agilent Technologies, Inc.) by incubating in the Agilent
Microarray Hybridization Oven (Agilent Technologies, Inc.) for 40 h
at 67°C. The slide was then washed using wash buffer (Agilent
Technologies, Inc.) and scanned using the NimbleGen MS 200
Microarray Scanner (Roche NimbleGen, Madison, WI, USA). The Agilent
CytoGenomics 2.7 software program (Agilent Technologies, Inc.) was
used for data analysis.
Long-range PCR
To confirm the deletion detected by Ion PGM study,
DNA (200 ng) of the proband and the parents was subjected to
long-rang PCR study. Long-range PCR using forward primers targeting
intron 12 and reverse primers targeting intron 16 (Table I) was performed using the Expand
Long Template PCR system (Roche Diagnostics, Indianapolis, IN,
USA). The normal PCR products were expected to be 8784 bp
(GRCh38.p7). The thermal cycling program was provided by the kit
manual with an annealing temperature of 59°C. The PCR products were
loaded on a 1% low electroendosmosis agarose gel (Lonza Rockland,
Rockland, ME, USA) for electrophoresis. A 1 kb DNA (Thermo Fisher
Scientific, Inc.) was used as marker.
Detection of breakpoints
Using the long-range PCR products (0.5 µl) from the
proband as a template, multiple primers (generated by Primer3, v
0.4.0; purchased from Integrated DNA Technologies, Inc.,
Coralville, IA, USA) targeting the deletion of SLC12A1 gene
were designed to determine the breakpoints of the deletion. The
product from the forward primer (5′-AAGGATGACGTCTCTATATT-3′) and
the reverse primer (5′-ACAGAGTTTCGCTCTTGTTG-3′) produced a clean
band in the 1% low electroendosmosis agarose gel, visualized under
UV light, which was subjected to Sanger sequencing (as described
above) for the detection of breakpoints.
Literature review
To investigate the SLC12A1 mutation patterns,
Human Gene Mutation Database (www.hgmd.cf.ac.uk) and previous English-language
studies identified in PubMed (https://www.ncbi.nlm.nih.gov/pubmed) were reviewed
using the search terms ‘Bartter syndrome type 1 OR SLC12A1 OR
NKCC2’.
Results
Following sequencing of the entire coding region and
the intron-exon boundaries of the SLC12A1 gene in the
proband, a novel homozygous frameshift mutation in exon 15
(c.1833delT) was identified (Fig.
1). This deletion causes a change from phenylalanine to leucine
at amino acid position 611 of the SLC12A1 gene, as well as
the generation of a premature stop codon in the 32nd coding frame.
The presence of this stop codon leads to protein truncation and is
considered to be deleterious. In the family history, the proband's
parents stated that they were non-consanguineous. As this mutation
is novel and the proband's parents were unrelated, a parental
mutation assay was suggested. Unexpectedly, this c.1866delT
mutation was only detected in the father's DNA, but not in the
mother's. The genotype of the proband and the parents indicated
that the father did not have the large deletion encompassing exon
15. Therefore, heterozygous deletion encompassing exon 15 was
suspected in the proband and the mother. The purpose of Ion PGM
sequencing was to detect this deletion. Thus, the father was not
subject to Ion PGM analysis. To rule out possible paternal
segmental isodisomy 15, which may lead to the identified homozygous
mutation, a UPD study using four STR markers localized to four
different loci in the chromosome 15q21.1-q25.2 region was
performed. The results excluded the possibility of paternal
isodisomy (data not shown). Based on the genotype of the proband
and the parents, heterozygous deletions encompassing exon 15 were
suspected in the proband and the mother. To exclude a possible
heterozygous deletion that encompassed the c.1833delT mutation, the
same quantity of DNA samples from the proband, the mother, and a
normal control were subjected to Ion PGM target sequencing. The Ion
PGM data clearly demonstrate the presence of the c.1833delT single
nucleotide deletion in the proband but not in the proband's mother.
The read count of exons 14 and 15 from proband and the mother was
markedly lower when compared with normal control, which indicated
the presence of a potentially large deletion in this region
(Fig. 2). To verify the Ion PGM
findings, array CGH was performed using an Agilent 2×400 K chip.
The results demonstrated that one probe located between exons 14
and 15 was deleted, whereas the other probes were in the normal
base line (Fig. 3). This confirmed
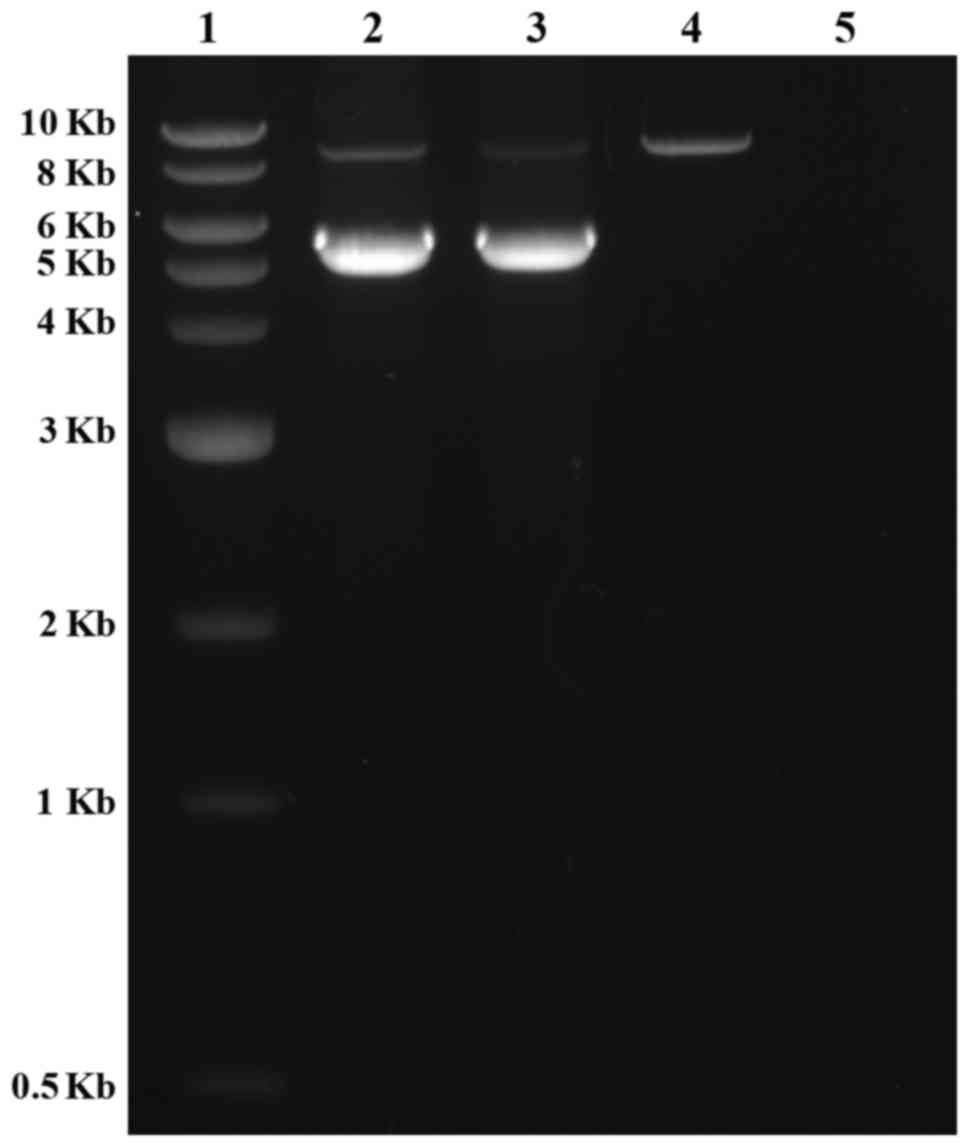
the Ion PGM analysis results. Long-range PCR using forward primers
in intron 12 and reverse primers in intron 16 was applied to
confirm the deletion in exons 14 and 15, as well as determine the
size of the deleted region. The results clearly indicated that the
father had one normal band with an expected size of 8.8 kb, whereas
the mother and the proband had two bands that were 8.8 kb and 5.8
kb in size. This was due to the presence of a 3.0 kb deletion in
one allele (Fig. 4). To determine
the breakpoints of the deletion, the junction fragment was
sequenced. The large deletion was observed to be
c.1685–726_1941+239del, and was 3,160 bp in size (Fig. 5).
Genotype of all documented BS type 1
patients
BS type 1 is a rare autosomal recessive disease
caused by mutations in the SLC12A1 gene. Through examining
previous reports, 72 BS type 1 patients with SLC12A1
mutations were documented (Table
II). Of these, approximately 45% of patients harbored
homozygous mutations, 34% were compound heterozygous mutations, and
following sequencing all exons and intron-exon boundaries of the
gene, and only one mutation was detected in the remaining 21% of
patients. Of these 72 patients, the mutations included
missense/nonsense mutations, splicing mutations, small insertions
and small deletions. To date, no large deletion has been reported.
To investigate SLC12A1 mutation patterns, previous studies
were reviewed alongside the data of the present study which focused
on the following three aspects: i) The mutation distribution
patterns; ii) the possible reasons for the high proportion of
homozygous mutations; iii) the possible reasons why a heterozygous
mutation was identified in the present study.
 | Table II.Summary of mutations in the
SLC12A1 gene (NM_000338.2). |
Table II.
Summary of mutations in the
SLC12A1 gene (NM_000338.2).
| Population
origin | Nucleotide
change | Amino acid
change | Mutation type | Exon | Reference |
|---|
| USA |
c.1833delT/c.1685-726_1941+239del | F611Lfs*32/Exons 14
and 15 deletion | F/D | 15/14,15 | Present case |
| Saudi Arabia |
c.814G>T/c.814G>T |
p.V272F/p.V272F | M homo | 6/6 |
(7) |
| Saudi Arabia |
c.814G>T/c.2467delC |
p.V272F/p.Q823fs | M/F | 6/20 |
(7) |
| Saudi Arabia |
c.1519T>C/c.1519T>C |
p.S507P/p.S507P | M homo | 12/12 |
(7) |
| Saudi Arabia |
c.1942G>T/c.1942G>T |
p.D648N/p.D648N | M homo | 15/15 |
(7) |
| Italy |
c.904delC/c.904delC |
p.R302fs/p.R302fs | F homo | 7/7 |
(7) |
| Saudi Arabia |
c.583-584dupA/c.583-584dupA |
p.M195fs/p.M195fs | F homo | 4/4 |
(7) |
| – |
c.2239delT/c.2239delT |
p.F747fs/p.F747fs | F homo | 18/18 |
(8) |
| – |
c.769G>A/c.769G>A |
p.G257S/p.G257S | M homo | 6/6 |
(8) |
| – |
c.769G>A/c.769G>A |
p.G257S/p.G257S | M homo | 6/6 |
(8) |
| – |
c.1663G>A/c.1663G>A |
p.A555T/p.A555T | M homo | 13/13 |
(8) |
| – |
c.595C>T/c.1966C>T |
p.R199C/p.Q656X | M/N | 4/16 |
(8) |
| – |
c.1576-1578delAAC/c.1576-1578delAAC |
p.N526del/p.N526del | D homo | 13/13 |
(8) |
| – |
c.1432G>A/c.1432G>A |
p.G478R/p.G478R | M homo | 11/11 |
(8) |
| – | c.24-27delTGTA | p.V9fs | F/− | 2/ |
(8) |
| – | c.1954G>A | p.G652S | M/− | 16/ |
(8) |
| – |
c.2495A>C/c.2495A>C |
p.E832A/p.E832A | M homo | 21/21 |
(8) |
| – | c.606G>C | p.W202C | M/− | 4/ |
(8) |
| – |
c.1834G>A/c.1834G>A |
p.G612R/p.G612R | M homo | 15/15 |
(8) |
| – | c.535T>A | p.W179R | M/− | 3/ |
(8) |
| – | c.1966C>T/Intron
25 | p.Q656X/Splice
mutant | N/S | 16/26 |
(8) |
|
| 5′splice
gt>at |
|
|
|
|
| Costa Rican |
c.1875G>A/c.1875G>A |
p.W625X/p.W625X | N homo | 15/15 |
(9) |
| Costa Rican |
c.1875G>A/c.1875G>A |
p.W625X/p.W625X | N homo | 15/15 |
(9) |
| Costa Rican |
c.1875G>A/c.1875G>A |
p.W625X/p.W625X | N homo | 15/15 |
(9) |
| Costa Rican | c.1875G>A | p.W625X | N/− | 15/ |
(9) |
| Costa Rican | c.1875G>A | p.W625X | N/− | 15/ |
(9) |
| German |
c.924G>A/c.924G>A |
p.R302Q/p.R302Q | M homo | 7/7 | (10) |
| Yugoslavia |
c.1195delT/c.1195delT | F stop at 1318/F
stop at 1318 | F homo | 9/9 | (10) |
| Morocco |
c.1541G>A/c.1541G>A |
p.A508T/p.A508T | M homo | 12/12 | (10) |
| German |
c.1548C>A/c.1548C>A |
p.A510D/p.A510D | M homo | 12/12 | (10) |
| Turkey |
c.1595-1597del3/c.1595-1597del3 |
p.526delN/p.526delN | D homo | 13/13 | (10) |
| Belgium |
c.747G>A& | p.G243E | M/− | 6/ | (10) |
| German |
c.1326G>A/c.614C>G |
p.C436Y/p.R199G | M/M | 11/4 | (10) |
| Italy | c.1451G>A | p.G478R | M/− | 11/ | (10) |
| German | c.1541G>A | p.A508T | M/− | 12/ | (10) |
| Estonia | c.3013T>G | p.Y998X | N/− | 25/ | (10) |
| Caucasian | c.629–6A>G |
| S/− | 4/ | (11) |
| Caucasian |
c.629-6A>G/c.976–14C>G |
| S/S | 4/8 | (11) |
| Caucasian |
c.1103A>G/c.1103A>G |
p.E368G/p.E368G | M homo | 9/9 | (11) |
| Morocco |
c.1522G>A/c.1522G>A |
p.A508T/p.A508T | M homo | 12/12 | (11) |
| Caucasian |
c.577G>A/c.799G>T |
p.G193R/p.A267S | M/M | 3/6 | (11) |
| Morocco |
c.2281C>T/c.2281C>T |
p.R761X/p.R761X | N homo | 18/18 | (11) |
| Caucasian |
c.337–339dup/c.1041–1045del |
p.E113dup/p.P348QfsX3 | I/F | 3/8 | (11) |
| Caucasian | c.1883C>A | p.A628D | M/− | 15/ | (11) |
| North African |
c.1327G>A/c.1327G>A |
p.G443R/p.G443R | M homo | 11/11 | (11) |
| Caucasian |
c.1010C>T/c.3164+1G>A | p.A337V/Loss splice
donor site | M/S | 8/26 | (11) |
| Mali |
c.955G>A/c.955G>A |
p.G319R/p.G319R | M homo | 7/7 | (11) |
| Algeria |
c.2117delA/c.2117delA |
p.K706RfsX23/p.K706RfsX23 | F homo | 17/17 | (11) |
| Italy |
c.1381T>C/c.1381T>C |
p.C461R/p.C461R | M homo | 11/11 | (12) |
| Italy |
c.1062delG/c.1062delG |
p.K354NfsX73/p.K354NfsX73 | F homo | 8/8 | (12) |
| Italy |
c.1381T>C/c.1630C>T | p.C461R/P544S | M/M | 11/13 | (12) |
| Italy |
c.904delC/c.904delC |
p.R302GfsX2/p.R302GfsX2 | F homo | 7/7 | (12) |
| Italy |
c.1663G>A/c.1663G>A |
p.A555T/p.A555T | M homo | 13/13 | (12) |
| Italy |
c.347G>A/c.1954G>A |
p.R116H/p.G652S | M/M | 2/16 | (12) |
| Italy |
c.551T>A/c.611T>C | p.L184Q/V204A | M/M | 3/4 | (12) |
| Italy |
c.1190G>C/c.3164+1G>A | p.G397A/loss | M/S | 9/26 | (12) |
|
|
| splice donor
site |
|
|
|
| Italy |
c.1493C>T/c.1522G>A | p.A498V/A508T | M/M | 12/12 | (12) |
| Italy |
c.904delC/c.1493C>T |
p.R302GfsX2/A498V | F/M | 7/12 | (12) |
| Italy | – |
p.D12fs/p.R302fs | F/F | 2/7 | (13) |
| Italy | – | p.L522fs/− | F/− | 13/ | (13) |
| Italy | – |
p.R302fs/p.R302fs | F homo | 7/7 | (13) |
| Italy |
c.347G>A/c.1954G>A |
p.R116H/p.G652S | M/M | 2/16 | (13) |
| German |
c.530T>A/c.2751dupT |
p.F177Y/p.D918fs | M/F | 4/22 | (14) |
| – | c.1411C>T | p.R471X | N/− | 11/ | (15) |
| – | c.2095G>A | p.D699N | M/− | 17/ | (15) |
| Japan |
c.1664C>T/c.2426G>T |
p.A555V/p.G809V | M/M | 13/20 | (16) |
| Korea |
c.1277G>A/c.1679T>C |
p.C436Y/p.L560P | M/M | 11/13 | (17) |
| Japan |
c.724+4A>G/c.2095delG | Exon 6
skipping/p.D699fs | S/F | 6/17 | (18) |
| Japan |
c.904C>T/c.2807G>A |
p.R302W/p.W936X | M/N | 7/23 | (19) |
| Japan |
c.577G>A/c.724+1G>A | p.G193R/exon 5
skipping | M/S | 4/6 | (19) |
| Japan |
c.732-734delCTA/c.735-737delCTA |
p.Y245del/p.Y245del | D homo | 6/6 | (20) |
| Japan |
c.348insT/c.788G>A |
p.N117X/p.G257S | N/M | 2/6 | (21) |
| Japan |
c.2393-2394delGA/c.2971-2974delCAAA |
p.D792fsX4/p.N984fsX26 | F/F | 19/24 | (21) |
The mutation distribution pattern of
the SLC12A1 gene
All previous relevant studies demonstrated that a
total of 72 BS type 1 patients (patients from the same family and
with the same genotype were excluded) and 144 SLC12A1
alleles have been characterized to date (Table II). In addition to the 66
mutations documented in the HGMD professional database, the
literature review and the current study contributed four additional
mutations to the mutation database of the SLC12A1 gene.
These are D12fs, intron 25 5′- splice gt>at, c.1833delT, and
c.1685–726_1941+239del. Among all 27 exons, the mutations in the
SLC12A1 gene were distributed across the majority of the
exons except for 5, 10, 14 and 27. Among the exons with mutations,
exons 4, 6, 7, 11, 12, 13 and 15 harbored the most common mutated
regions with >10 mutations in each allele. Among all 70
mutations, the R302fs mutation was the most common mutation and
accounted for 5.5% of all the alleles. All of the R302fs mutations
were detected in Italian population studies. At the same amino acid
site, one homozygous mutation of R302Q was detected in a German
population and a heterozygous mutation of R302W was detected in a
Japanese population suggesting a mutation hot spot in this region.
Furthermore, the mutation of W625X was detected in eight alleles
accounting for 5.5% of all alleles, and were all detected in an
isolated Costa Rican population, suggesting a founder effect in
this population (9). The A508T
mutation had been discovered in six alleles, which accounted for
4.1% of all alleles. Less common mutations, including G257S and
A555T, were reported in five alleles. Mutation of N526del was
reported in four alleles. Four mutations, including G625S, C461R,
V272F, and G478R were detected in 3 alleles, 22 mutations were
observed in two alleles, and the remaining mutations were in one
allele. A total of 15 alleles lacked mutations, accounting for
approximately 20.5% of all patients and 10.2% of all alleles,
respectively. The mutation types included missense, nonsense,
splicing mutations, small deletions and small insertions leading to
frameshift mutations, and a large deletions. To the best of our
knowledge, this study is the first report of a large deletion in
the SLC12A1 gene.
The possible reasons for the high
proportion of homozygous mutations in the SLC12A1 gene
In previous reports, homozygous mutations accounted
for 45.2% of all the cases (Table
II). A number of possible reasons for the high percentage of
identified homozygous mutations exist. First, parental
consanguinity may be a potential factor responsible for the
homozygous status. Out of the 72 patients listed in Table II, 33 cases were characterized as
having apparent homozygosity. Of these, approximately 14 cases had
consanguineous parents (7,10,11),
whereas 12 patients did not, and the status of the remaining seven
cases was unknown. Among the patients from non-consanguineous
families, four performed parental analysis and the presence of a
heterozygous deletion was excluded (12). The remaining patients were not used
for further study. In modern society, consanguineous relationships
are likely to be avoided, particularly for individuals from
non-isolated populations. Therefore, it is uncommon to observe a
high proportion of homozygous mutations in a rare disease. The
authors therefore suggest that patients from non-consanguineous
families that harbor homozygous mutations should undergo a deletion
test, if possible. This will provide more information regarding the
potential reasons for the high percentage of homozygous mutations
observed in BS type 1.
Second, the founder effect may serve a role in the
observed mutation distribution. The study by Kurtz et al
(9), involving BS type 1 patients
from an isolated population in Costa Rica, investigated four
familial and three sporadic cases. Of these cases, three of them
harbored homozygous mutations, two of them demonstrated
heterozygous mutations, and the remaining two cases were negative
for mutations. The homozygous and heterozygous mutations identified
were p.W625X, which suggests a founder effect in this Costa Rican
population. In this case, due to the isolated populations, the
incidence was higher when compared with other populations, and the
patients shared the same few mutations (9). Therefore, although parents were not
related to each other, the offspring were more likely to harbor
homozygous mutations.
Regarding the patients in the cohort that were
observed to harbor one heterozygous mutation or no mutation, the
authors of these studies demonstrated that the methods used were
not sensitive enough and may responsible for the undetected
mutations (9). In addition, aside
from those consanguineous parents and parents from isolated
populations, non-related parents carrying the same mutation
demonstrated a 25% chance of producing offspring with a homozygous
mutation. This phenomenon is frequently observed in common
autosomal recessive diseases that have few high-frequency
mutations, such as cystic fibrosis (CF), in which the ΔF508
homozygous mutation is the most frequently observed. This mutation
is present in approximately two-thirds of CF carriers (23). However, for rare autosomal
recessive diseases, homozygous mutations are not commonly observed
in patients from non-consanguineous parents. In the present case of
a single patient with BS type I, Sanger sequencing analysis
detected a frameshift mutation with apparent homozygosity. However,
subsequent analyses demonstrated the presence of a compound large
heterozygous deletion and frameshift mutation. Therefore,
heterozygous deletions encompassing the mutant site should be
excluded as homozygous when the parents are not consanguineous nor
from an isolated population. This will enable clarification of the
mutation pattern and should be applied in clinical practice in the
future.
The possible reasons for identifying
only heterozygous mutations in the SLC12A1 gene
In previous BS type 1 studies (Table II), including our case, 15 out of
73 (20.5%) patients harbored only one mutation as detected by
sequencing all the coding exons and intron-exon boundaries. This is
uncommon in autosomal recessive diseases. There are a number of
potential reasons for this high percentage of patients with
heterozygous mutations. Firstly, the methods used for mutation
detection may not as sensitive enough. Three previous studies used
single-strand conformation polymorphism (SSCP) as a mutation
detection method (7,9,10).
Of the patients reported in these three studies, 7 out of 22
(31.8%) patients harbored one mutation and all lacked mutations in
the KCNJ1 gene (which leads to BS type 2), and was more
challenging to distinguish BS type 1 from BS type 2 by the
phenotype, whilst one patient had a second mutation discovered by
Sanger sequencing in a subsequent study (12). The sensitivity of the SSCP method
was estimated to be 75–98% (24).
Therefore, some mutations may not have been detected due to
limitations of the detection method. However, in the remaining
cases analyzed by direct Sanger sequencing, which demonstrated a
sensitivity of >99.9%, one mutation was detected in 9 out of 52
(17.3%) patients. Of these 9 patients, the second mutation could
not have been a point mutation, small deletion, or small insertion,
as these types of mutation are unlikely to be missed by Sanger
sequencing. In addition, the inaccurate clinical diagnosis may have
resulted in a negative genetic test result. Although it was
difficult to distinguish the subtype of BS in some cases, the
genetic analysis always included SLC12A1, KCNJ1,
CLCNKB and/or BSND if necessary. Therefore, negative
results were less likely due to an incorrect clinical diagnosis.
Additional genes may also contribute to the phenotype. In a
previous study, one patient demonstrating a BS type 4 phenotype,
harbored no detectable mutation in the BSND gene that is
responsible for BS type 4 (2).
Upon further investigation, the patient was observed to harbor
mutations in CLCNKA and CLCNKB genes, and was
characterized as BS type 4B to distinguish from BS type 4A
(2). Based on current evidence for
BS type 1, the presence of mutations in genes other than
SLC12A1 is less likely, however this may need to be
considered for those patients with one or no detectable mutations
in the SLC12A1 gene. Finally, mutations cannot be detected
by the methods used for analysis due to the limitations of the
methods. In the present case, the patient harbored a heterozygous
deletion spanning exons 14 and 15. The deletion was 3.16 kb in
size, which cannot be detected by Sanger sequencing. Therefore, for
patients with heterozygous mutations, the presence of large
deletions should be investigated.
Discussion
In the present study, one patient was suspected of
having BS type 1, and their peripheral blood sample was sent to our
genetics laboratory (University of Oklahoma Health Sciences Center,
Oklahoma City, OK, USA) for SLC12A1 gene sequencing
analysis. The results of Sanger sequencing of the SLC12A1
gene indicated the presence of a novel homozygous frameshift
mutation (c.1833delT) in exon 15 of the proband. Due to the rarity
of this mutation and the lack of a consanguineous relationship
between the parents, a parental study was subsequently performed to
determine the origin of inheritance of this mutation and to exclude
the possible presence of a heterozygous deletion. Notably, a
heterozygous mutation (c.1833delT) was detected in the father but
not in the mother. Three possible reasons were considered to be
responsible for this phenomenon. The first possibility was the
presence of a de novo mutation that has been reported in BS
type 1 previously (21). However,
this possibility was unlikely as the two mutations identified in
the proband were homozygous. The second possibility was segmental
paternal isodisomy of chromosome 15q encompassing the mutant gene.
A similar situation has been reported in studies of Bloom syndrome,
which is caused by a homozygous mutation due to maternal isodisomy
(25). In the current study, as
the proband was not suspected to exhibit Prader-Willi/Angelman
syndromes, four STR markers located between 15q21.1 and 15q25.2
were selected to perform a UPD study. Genotyping of the
microsatellites excluded the possibility that the apparent
homozygous mutation was caused by paternal isodisomy. The final
possibility was the presence of a previously unreported large
deletion encompassing the mutant region on the maternal allele. In
this case, the deleted region should at least include exon 15.
Using cDNA to identify the deletion was not practical, as
SLC12A1 mRNA expression is kidney-specific. To investigate
the deletion in the SLC12A1 gene, parallel sequencing using
Ion PGM was performed on the proband and the mother. It was
revealed that the proband harbored compound heterozygous mutations,
a large deletion on the maternal allele and a frameshift mutation
of F611Lfs*32 on the paternal allele. The deletion was confirmed by
long-range PCR and determined as c.1685-726_1941+239del by
sequencing of the junction fragment.
Sanger sequencing remains as the most popular method
for the detection of mutations in the SLC12A1 gene. The
technique has provided the most accurate results for mutations
including substitutions, small deletions and insertions, however,
it is unable to detect large deletions. Therefore, in the present
case, when the large deletion encompasses the mutant site, the
Sanger sequencing result demonstrated a false homozygous mutation.
Following the genotyping study, a deletion was suspected, therefore
Ion PGM target sequencing was used to exclude this possible
deletion. The results of Ion PGM sequencing indicated the presence
of a large deletion encompassing exons 14 and 15 in the proband and
the mother. To confirm this finding, array CGH and long-range PCR
were performed, which further confirmed the presence of the
deletion in the proband and his mother. By sequencing the junction
fragment, the large deletion was characterized as
c.1685-726_1941+239del. Although this is a single case that
presented a false homozygous mutation result as determined by
Sanger sequencing, in previously reported cases of BS type 1
(Table II), approximately 45% of
patients were observed to harbor homozygous mutations and 21% of
patients harbored only heterozygous mutations. The reasons
underlying this phenomenon remain unclear. Large deletions may
serve an important role in BS type 1, however, due to insufficient
data, no conclusion could be reached. Further deletion analysis of
patients with BS type 1 may enable clarification of this
phenomenon.
In conclusion, the present study was the first to
identify a large deletion encompassing exons 14 and 15 in a patient
with BS type 1, which was not detected by Sanger sequencing. In
addition to the previously reported types of mutations including
missense, nonsense, splicing, frameshift, small deletions and small
insertions, the results of the current study suggest that large
deletions encompassing the whole exon(s) may serve an etiologic
role in BS type 1. These types of mutations may therefore be more
common than originally thought, due to the fact that 21% of
reported cases with only one mutation have been observed.
Acknowledgements
The authors were grateful to the family for
participating in this study and to the University of Oklahoma
Health Sciences Center for supporting this study.
References
|
1
|
Kliegman R and Nelson WE: Nelson textbook
of pediatrics. Elsevier/Saunders; Philadelphia, PA: 2011,
View Article : Google Scholar
|
|
2
|
Nozu K, Inagaki T, Fu XJ, Nozu Y, Kaito H,
Kanda K, Sekine T, Igarashi T, Nakanishi K, Yoshikawa N, et al:
Molecular analysis of digenic inheritance in Bartter syndrome with
sensorineural deafness. J Med Genet. 45:182–186. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vezzoli G, Arcidiacono T, Paloschi V,
Terranegra A, Biasion R, Weber G, Mora S, Syren ML, Coviello D,
Cusi D, et al: Autosomal dominant hypocalcemia with mild type 5
Bartter syndrome. J Nephrol. 19:525–528. 2006.PubMed/NCBI
|
|
4
|
Seyberth HW: An improved terminology and
classification of Bartter-like syndromes. Nat Clin Pract Nephrol.
4:560–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhat YR, Vinayaka G and Sreelakshmi K:
Antenatal bartter syndrome: A review. Int J Pediatr.
2012:8571362012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Madrigal G, Saborio P, Mora F, Rincon G
and Guay-Woodford LM: Bartter syndrome in Costa Rica: A description
of 20 cases. Pediatr Nephrol. 11:296–301. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simon DB, Karet FE, Hamdan JM, DiPietro A,
Sanjad SA and Lifton RP: Bartter's syndrome, hypokalaemic alkalosis
with hypercalciuria, is caused by mutations in the Na-K-2Cl
cotransporter NKCC2. Nat Genet. 13:183–188. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji W, Foo JN, O'Roak BJ, Zhao H, Larson
MG, Simon DB, Newton-Cheh C, State MW, Levy D and Lifton RP: Rare
independent mutations in renal salt handling genes contribute to
blood pressure variation. Nat Genet. 40:592–599. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kurtz CL, Karolyi L, Seyberth HW, Koch MC,
Vargas R, Feldmann D, Vollmer M, Knoers NV, Madrigal G and
Guay-Woodford LM: A common NKCC2 mutation in Costa Rican Bartter's
syndrome patients: Evidence for a founder effect. J Am Soc Nephrol.
8:1706–1711. 1997.PubMed/NCBI
|
|
10
|
Vargas-Poussou R, Feldmann D, Vollmer M,
Konrad M, Kelly L, van den Heuvel LP, Tebourbi L, Brandis M,
Karolyi L, Hebert SC, et al: Novel molecular variants of the
Na-K-2Cl cotransporter gene are responsible for antenatal Bartter
syndrome. Am J Hum Genet. 62:1332–1340. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brochard K, Boyer O, Blanchard A, Loirat
C, Niaudet P, Macher MA, Deschenes G, Bensman A, Decramer S, Cochat
P, et al: Phenotype-genotype correlation in antenatal and neonatal
variants of Bartter syndrome. Nephrol Dial Transplant.
24:1455–1464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Puricelli E, Bettinelli A, Borsa N, Sironi
F, Mattiello C, Tammaro F, Tedeschi S and Bianchetti MG: Italian
Collaborative Group for Bartter Syndrome: Long-term follow-up of
patients with Bartter syndrome type I and II. Nephrol Dial
Transplant. 25:2976–2981. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Colussi G, Bettinelli A, Tedeschi S, De
Ferrari ME, Syrén ML, Borsa N, Mattiello C, Casari G and Bianchetti
MG: A thiazide test for the diagnosis of renal tubular hypokalemic
disorders. Clin J Am Soc Nephrol. 2:454–460. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pressler CA, Heinzinger J, Jeck N,
Waldegger P, Pechmann U, Reinalter S, Konrad M, Beetz R, Seyberth
HW and Waldegger S: Late-onset manifestation of antenatal Bartter
syndrome as a result of residual function of the mutated renal
Na+-K+-2Cl- co-transporter. J Am Soc Nephrol. 17:2136–2142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Urbanova M, Reiterová J, Stĕkrová J,
Lnĕnická P and Ryšavá R: DNA analysis of renal electrolyte
transporter genes among patients suffering from Bartter and
Gitelman syndromes: Summary of mutation screening. Folia Biol
(Praha). 57:65–73. 2011.PubMed/NCBI
|
|
16
|
Yamazaki H, Nozu K, Narita I, Nagata M,
Nozu Y, Fu XJ, Matsuo M, Iijima K and Gejyo F: Atypical phenotype
of type I Bartter syndrome accompanied by focal segmental
glomerulosclerosis. Pediatr Nephrol. 24:415–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee EH, Heo JS, Lee HK, Han KH, Kang HG,
Ha IS, Choi Y and Cheong HI: A case of Bartter syndrome type I with
atypical presentations. Korean J Pediatr. 53:809–813. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nozu K, Iijima K, Kawai K, Nozu Y, Nishida
A, Takeshima Y, Fu XJ, Hashimura Y, Kaito H, Nakanishi K, et al: In
vivo and in vitro splicing assay of SLC12A1 in an antenatal
salt-losing tubulopathy patient with an intronic mutation. Hum
Genet. 126:533–538. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nozu K, Iijima K, Kanda K, Nakanishi K,
Yoshikawa N, Satomura K, Kaito H, Hashimura Y, Ninchoji T, Komatsu
H, et al: The pharmacological characteristics of molecular-based
inherited salt-losing tubulopathies. J Clin Endocrinol Metab.
95:E511–E518. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fukuyama S, Okudaira S, Yamazato S,
Yamazato M and Ohta T: Analysis of renal tubular electrolyte
transporter genes in seven patients with hypokalemic metabolic
alkalosis. Kidney Int. 64:808–816. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adachi M, Asakura Y, Sato Y, Tajima T,
Nakajima T, Yamamoto T and Fujieda K: Novel SLC12A1 (NKCC2)
mutations in two families with Bartter syndrome type 1. Endocr J.
54:1003–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Faiz F, Allcock RJ, Hooper AJ and van
Bockxmeer FM: Detection of variations and identifying genomic
breakpoints for large deletions in the LDLR by Ion Torrent
semiconductor sequencing. Atherosclerosis. 230:249–255. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
O'Sullivan BP and Freedman SD: Cystic
fibrosis. Lancet. 373:1891–1904. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Scoggan KA and Bulman DE: Single-strand
conformational polymorphism analysis (SSCP) and sequencing for ion
channel gene mutations. Methods Mol Biol. 217:143–151.
2003.PubMed/NCBI
|
|
25
|
Woodage T, Prasad M, Dixon JW, Selby RE,
Romain DR, Columbano-Green LM, Graham D, Rogan PK, Seip JR, Smith
A, et al: Bloom syndrome and maternal uniparental disomy for
chromosome 15. Am J Hum Genet. 55:74–80. 1994.PubMed/NCBI
|