Introduction
Nasopharyngeal carcinoma (NPC) is a specific type of
head and neck squamous cell carcinoma, which is derived from
epithelial cells in the nasopharynx (1). It is a relatively common malignancy
in Southeast Asia compared with other regions, and is particularly
common in the Cantonese region of Guangdong, China (2). More than 84,000 novel cases of NPC
are diagnosed each year, according to global cancer statistics
reported by the International Agency for Research on Cancer. Among
these cases, 80% are located in Asia and 5% are located in Europe
(3). There are various factors
that are associated with the initiation and progression of NPC,
including genetic susceptibility, environmental factors and
Epstein-Barr virus latent infection (4). At present, the main therapeutic
strategies for the treatment of patients with NPC are radiotherapy
and comprehensive chemotherapy (5). However, despite advances in the
development of therapeutic strategies for the treatment of patients
with primary NPC, the 5-year survival rate increased from 50% in
the 1980s to 70% in the 1990s (6).
Distant metastasis is the predominant cause of treatment failure,
and 30–60% of patients with NPC will eventually develop distant
metastasis and succumb to disseminated disease (7). Therefore, a better understanding
regarding the molecular mechanisms underlying NPC metastasis is
essential for determining novel therapeutic targets for the
suppression of cancer metastasis.
MicroRNAs (miRNAs/miRs) belong to a group of highly
conserved, endogenous, non-protein-coding small RNA molecules,
which are 19–25 nucleotides in length. miRNAs negatively regulate
gene expression at the post-transcriptional level by binding to the
3′-untranslated region (3′-UTR) of target mRNAs, thus resulting in
mRNA degradation or translational inhibition (8–10).
Previous studies have demonstrated that miRNAs serve important
functions in various biological processes, including cell cycle
progression, cell proliferation, migration, invasion, apoptosis,
differentiation and development (11,12).
Therefore, aberrant expression of miRNAs may be involved in the
pathogenesis of a wide range of human diseases, including cancer
(13). It has previously been
indicated that miRNAs can function as oncogenes by suppressing the
expression of target tumor suppressor genes, or as tumor
suppressors by inhibiting the expression of target oncogenes in
tumor progression (14).
Therefore, targeting miRNAs may be investigated as a novel
therapeutic strategy for the treatment of patients with NPC.
The expression and functions of miR-152 have been
studied in several types of cancer; however, to the best of our
knowledge, no studies have elucidated the expression, functions and
mechanisms of miR-152 in NPC. The present study demonstrated that
miR-152 was significantly downregulated in NPC tissue samples and
cell lines. The present study further explored the functions of
miR-152 in cell proliferation, migration and invasion. In addition,
V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B
(MAFB) was identified as a direct target gene of miR-152. The newly
identified miR-152/MAFB pathway provided information regarding the
functions of miRNAs in NPC initiation and progression, and may
provide a novel therapeutic strategy for the treatment of NPC.
Materials and methods
Clinical specimens
A total of 17 primary NPC specimens were obtained
from patients who had undergone surgery at the Affiliated Sixth
People's Hospital of Shanghai Jiao Tong University (Shanghai,
China) between January 2012 and November 2014 (8 male and 9 female;
age range, 15–63 years). A total of 8 normal nasopharyngeal
epithelial specimens were obtained from biopsy-negative cases at
the Affiliated Sixth People's Hospital of Shanghai Jiao Tong
University (6 male and 2 female; age range, 18–45 years). Tissue
specimens were immediately frozen in liquid nitrogen and stored at
−80°C. None of the patients with NPC had received radiotherapy or
chemotherapy prior to surgery. The present study was approved by
the Affiliated Sixth People's Hospital's Protection of Human
Subjects Committee. Written informed consent was obtained from each
patient involved in the present study.
Cell culture
Four human NPC cell lines: CNE1, SUNE1, 5-8F and
CNE2, and the NP69 normal nasopharyngeal epithelial cell line were
purchased from American Type Culture Collection (Manassas, VA,
USA). The human embryonic kidney (HEK)293T cell line was purchased
from the Chinese Academy of Sciences (Shanghai, China). NP69 cells
were cultured in keratinocyte-serum-free medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 30 µg/ml
bovine pituitary extract (BD Biosciences, San Diego, CA, USA).
CNE1, CNE2, 5-8F and SUNE1 cell lines were cultured in RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml
penicillin and 100 mg/ml streptomycin (all Gibco; Thermo Fisher
Scientific, Inc.). HEK293T cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.).
All cells were cultured at 37°C in a humidified incubator
containing 5% CO2.
Cell transfection
miR-152 mimics, miR-152 inhibitor, negative control
(NC) mimics, NC inhibitor, MAFB small interfering (si)RNA, NC siRNA
and luciferase reporter plasmid were obtained from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The sequences were as
follows:miR-152 mimics, 5′-UCAGUGCAUGACAGAACUUGG−3′; miR-152
inhibitor, 5′-CCAAGUUCUGUCAUGCACUGA-3′; NC,
5′-UUCUCCGAACGUGUCACGUTT−3′; and NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAA-3′. Cell transfection and co-transfection
were performed using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Post-transfection, the cells were incubated at 37°C until
further assessment.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from homogenized tissues and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
reverse transcription was performed using M-MLV Reverse
Transcription system (M1701; Promega Corporation, Madison, WI,
USA). The reaction system contained 4 µl 5X RT buffer, 0.75 µl dNTP
(10 mM), 1.2 µl primer (Guangzhou RiboBio Co., Ltd., Guangzhou,
China), 0.2 µl MMLV Reverse Transcriptase, 3 µg RNA and double
distilled water. The cycling conditions for reverse transcription
were as follows: 25°C for 30 min; 42°C for 30 min and 85°C for 5
min. The expression levels of miR-152 were detected using
Hairpin-it™ miRNAs qPCR Quantitation kit (Shanghai GenePharma Co.,
Ltd.). The reaction system for qPCR contained 10 µl Real-Time PCR
Buffer, 0.75 µl dNTP (10 mM), 0.4 µl forward primer, 0.4 µl reverse
primer (Guangzhou RiboBio Co., Ltd.), 2 µl cDNA, 0.2 µl Taq DNA
polymerase and double distilled water. The thermocycling conditions
for qPCR were as follows: 95°C for 3 min; then 40 cycles of 95°C
for 12 sec and 62°C for 1 min. U6 small nuclear RNA was used for
normalization of miRNA expression. To detect MAFB mRNA expression
levels, qPCR was performed with SYBR Green Master Mix in an ABI
7500 detection system (Invitrogen; Thermo Fisher Scientific, Inc.).
The reaction system for qPCR contained 10 µl SYBR Green PCR master
mix, 2 µl forward primer and 2 µl reverse primer (Guangzhou RiboBio
Co., Ltd.), 2 µl cDNA and double distilled water. The thermocycling
conditions of qPCR were as follows: 95°C for 10 min, then 40 cycles
of 95°C for 15 sec and 60°C for 1 min. GADPH was used as an
internal control to normalize mRNA expression. Expression levels
were determined using the 2-ΔΔCq method (15).
Cell proliferation assay
Cell proliferation was determined using the MTT
assay (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). A total
of 24 h post-transfection, cells were harvested and seeded into
96-well plates at a density of 3,000 cells/well and were incubated
at 37°C. The MTT assay was performed every 24 h for 4 days,
according to the manufacturer's protocol. Briefly, 20 µl MTT
solution (5 mg/ml) was added to each well. Following a 4 h
incubation, cell culture medium was carefully removed and the
formazan precipitates were dissolved in 200 µl dimethyl sulfoxide.
Absorbance was measured at 490 nm using an automatic multi-well
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
All experiments were performed in triplicate.
Cell migration and invasion
assays
Cell migration and invasion assays were performed
using Transwell apparatus (Costar; Corning Incorporated, Corning,
NY, USA); pore diameter, 8 µm. For the invasion assay, the
Transwell apparatus was pre-coated with Matrigel (BD Biosciences)
according to the manufacturer's protocol. A total of 48 h
post-transfection, cells were harvested and re-suspended as
single-cell solutions in serum-free medium. A total of 1×105 cells
in 200 µl serum-free RPMI-1640 medium were added to the upper
chamber of the Transwell apparatus. RPMI-1640 medium (500 µl)
containing 20% FBS was added to the lower chamber, thus acting as a
chemoattractant. Following a 24 h incubation at 37°C, the cells
were fixed in 100% methanol for 5 min and stained with 0.5% crystal
violet (Beyotime Institute of Biotechnology, Haimen, China) for 5
min. Subsequently, the cells that had not migrated or invaded
through the pores were carefully removed with a cotton swab. Values
for migration and invasion were obtained by counting five fields
per membrane under a microscope (CKX41; Olympus Corporation, Tokyo,
Japan).
Western blotting
A total of 48 h post-transfection, cells were washed
with ice-cold PBS and lysed with radioimmunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology). Bicinchoninic
Acid Protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) was
used to measure the protein concentration, according to the
manufacturer's protocol. Equal amounts of protein (20 µg) were
separated by 10% SDS-PAGE (Beyotime Institute of Biotechnology) and
transferred onto polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked with 5% non-fat dry
milk at room temperature for 1 h. The membranes were then incubated
with mouse anti-human monoclonal MAFB (1:500 dilution; cat. no.
sc-376387; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and
rabbit anti-human polyclonal anti-β-actin (1:1,000 dilution; cat.
no. sc-1616-R; Santa Cruz Biotechnology, Inc.). Following an
overnight incubation at 4°C, the membranes were washed with PBS
containing 0.5% Tween and incubated with the corresponding
horseradish peroxidase-conjugated secondary antibody (1:5,000
dilution; sc-2005 for MAFB; sc-2301 for β-actin; Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. Following
washing, the bands were visualized using an enhanced
chemiluminescence solution (Pierce; Thermo Fisher Scientific,
Inc.). β-actin was used as a loading control. AlphaEase FC
software, version 4.0.1 (ProteinSimple, San Jose, CA, USA) was used
to analyze the western blotting results.
Dual-luciferase report assay
HEK293T cells were cultured in a 12-well plate until
they reached 80–90% confluence. Luciferase reporter plasmids,
including pGL3-MAFB-3′UTR wild-type (Wt) and pGL3-MAFB-3′UTR
mutant-type (Mut), were synthesized and confirmed by Shanghai
GenePharma Co., Ltd. HEK293T cells were co-transfected with miR-152
mimics or NC mimics and PGL3-MAFB-3′-UTR Wt or PGL3-MAFB-3′-UTR Mut
using Lipofectamine® 2000, according to the
manufacturer's protocol. A total of 48 h post-transfection,
luciferase activity was detected using the Dual-Luciferase Reporter
Assay system according to the manufacturer's instuctions (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity for each transfected well. Each
assay was replicated three times.
Statistical analysis
Each assay was repeated a minimum of 3 times. Data
are presented as the mean ± standard deviation, and were analyzed
using Student's t-test with SPSS 17 software (SPSS, Inc., Chicago,
IL, USA). Two-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-152 expression in NPC tissues and
cell lines
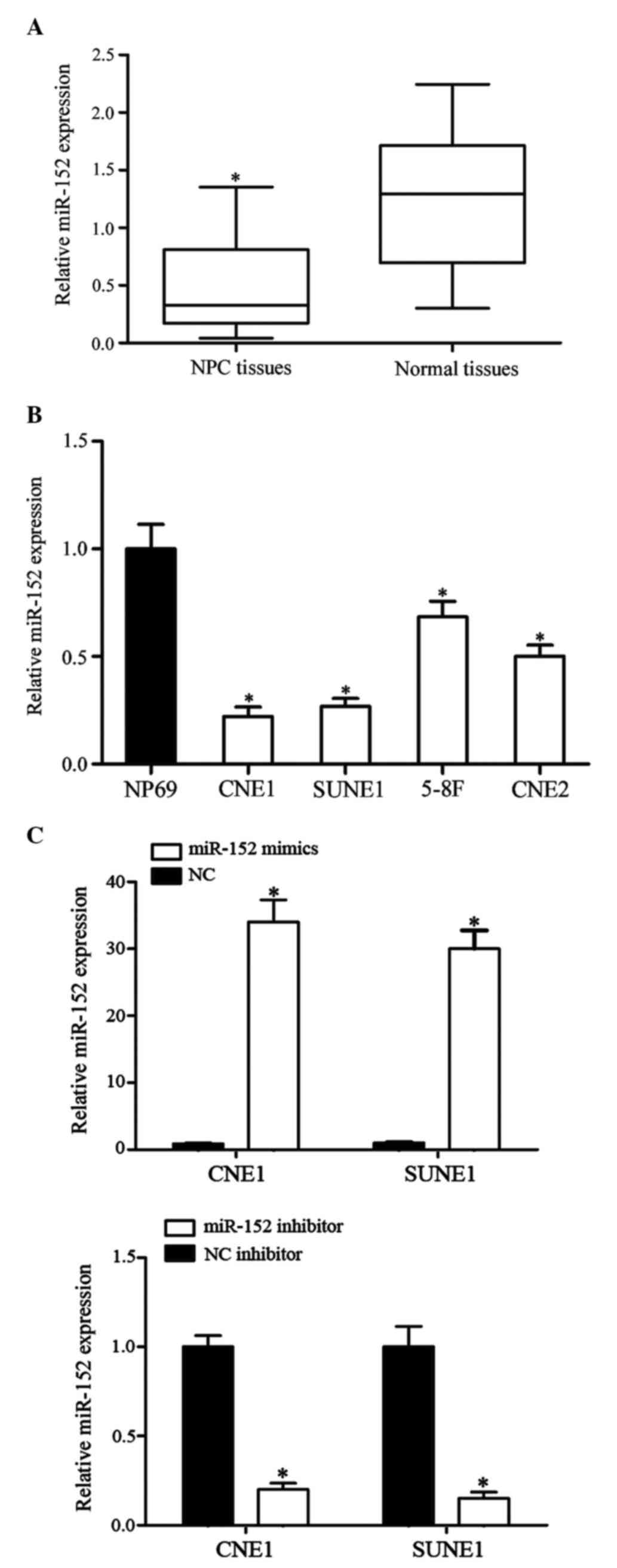
In the present study, the expression levels of
miR-152 were detected in 17 NPC biopsy specimens and eight normal
nasopharyngeal epithelial specimens using RT-qPCR. As shown in
Fig. 1A, miR-152 was downregulated
in NPC tissues compared with in normal nasopharyngeal epithelial
specimens (P<0.05).
To further explore the functions of miR-152 in NPC
carcinogenesis and progression, the expression levels of miR-152
were detected in NPC cell lines and the NP69 normal nasopharyngeal
epithelial cell line. Compared with in NP69 cells, miR-152 was
significantly downregulated in CNE1, SUNE1, 5-8F and CNE2 cells
(Fig. 1B). Among these NPC cell
lines, miR-152 expression was lowest in CNE1 and SUNE1 cells
compared with in 5-8F and CNE2 cells. Therefore, CNE1 and SUNE1
cells were used to perform further cell function experiments.
To investigate the functions of miR-152 in CNE1 and
SUNE1 cells, miR-152 mimics, NC mimics, miR-152 inhibitor and NC
inhibitor were transfected into CNE1 and SUNE1 cells. A total of 48
h post-transfection, RT-qPCR was performed to detect the expression
levels of miR-152. As shown in Fig.
1C, miR-152 expression was significantly upregulated in CNE1
and SUNE1 cells transfected with miR-152 mimics, whereas miR-152
was downregulated in CNE1 and SUNE1 cells transfected with the
miR-152 inhibitor (P<0.05).
miR-152 inhibits NPC cell
proliferation
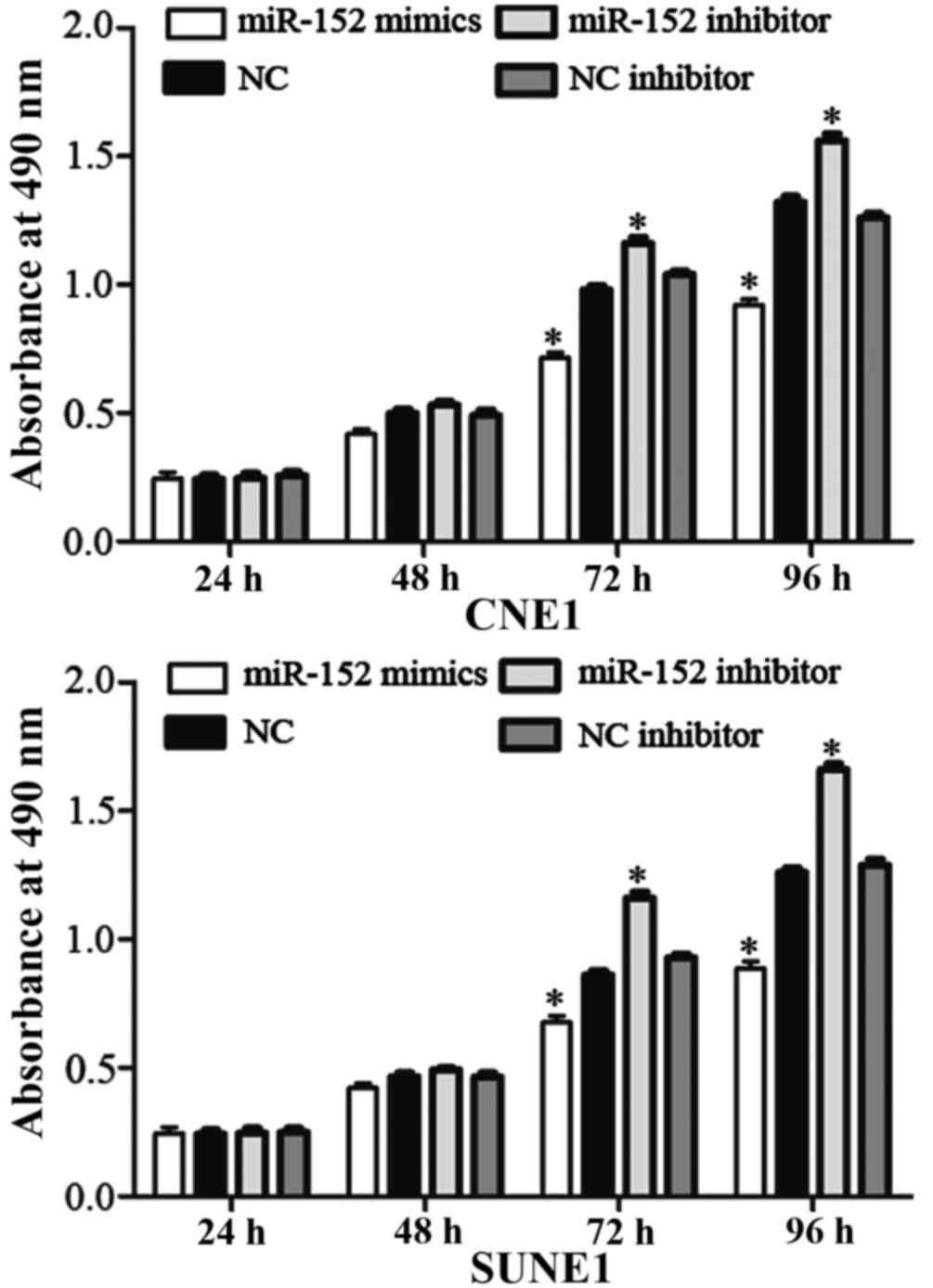
An MTT assay was performed to determine the effects
of miR-152 on cell proliferation. Transfection with miR-152 mimics
inhibited NPC cell proliferation, whereas miR-152 inhibitor
transfection enhanced NPC cell proliferation (Fig. 2; P<0.05). These results indicate
that miR-152 may function as a tumor growth suppressor in NPC.
miR-152 inhibits NPC cell migration
and invasion
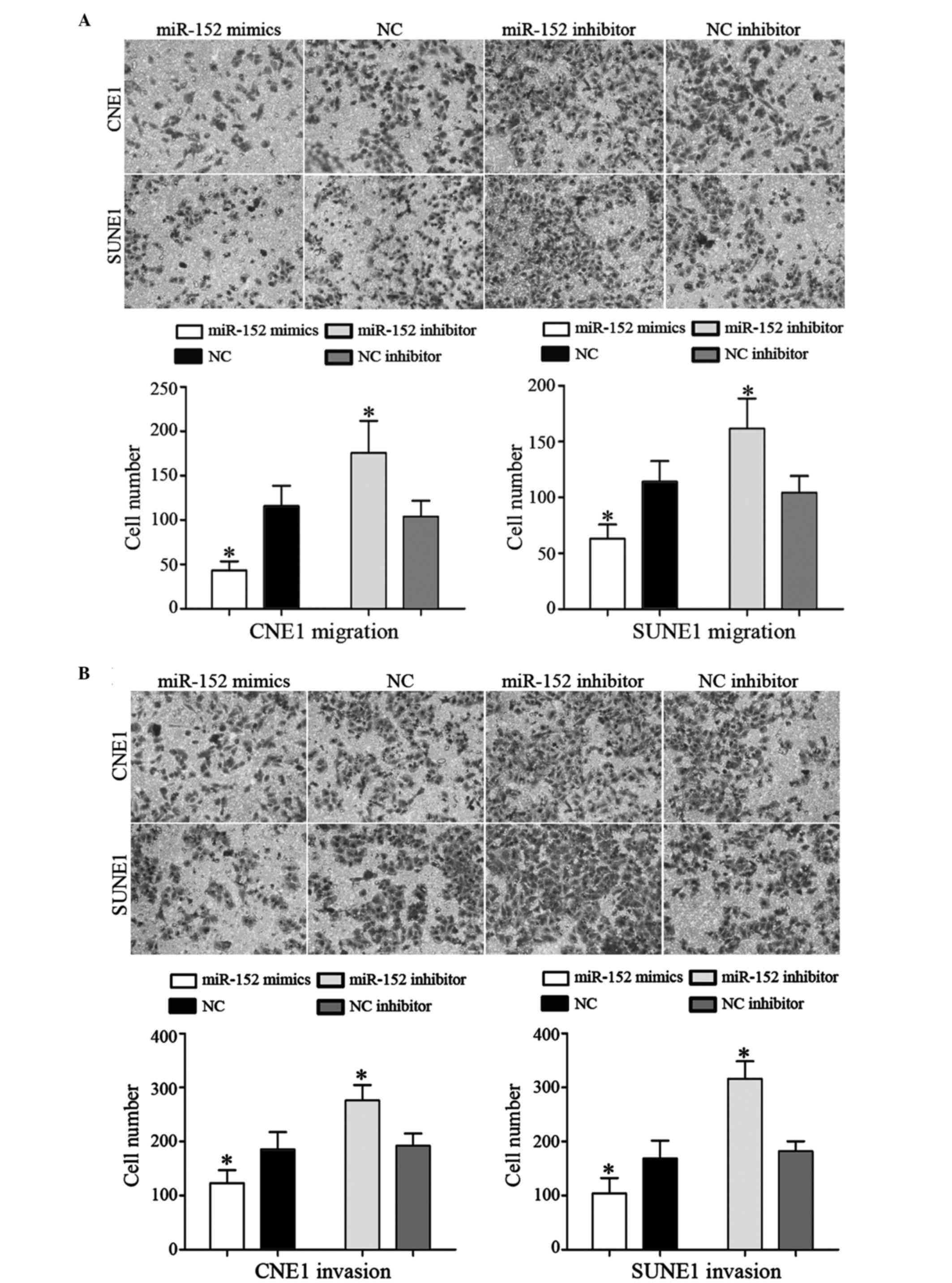
Transwell chambers were used to determine whether
miR-152 inhibited cell migration and invasion. The number of
migrated CNE1 and SUNE1 cells was significantly decreased following
transfection with miR-152 mimics, as compared with cells
transfected with NC mimics. Conversely, transfection with the
miR-152 inhibitor had the opposite effect on migration (Fig. 3A; P<0.05). Furthermore,
transfection with miR-152 mimics decreased the invasive ability of
NPC cells compared with those transfected with NC mimics.
Conversely, transfection with the miR-152 inhibitor enhanced cell
invasive ability, as compared with cells transfected with the NC
inhibitor (Fig. 3B; P<0.05).
These results suggest that overexpression of miR-152 may inhibit
NPC cell migration and invasion.
MAFB is a direct target gene of
miR-152
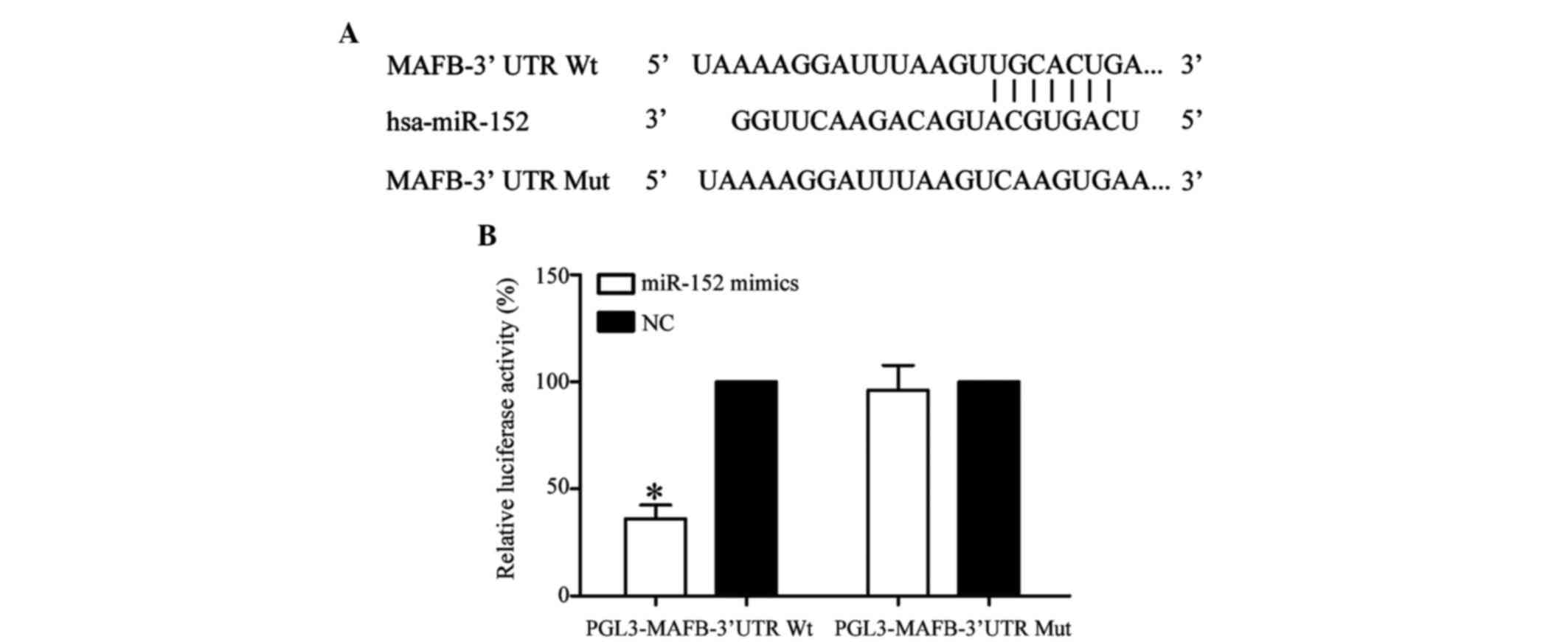
TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/) were used to
predict a set of target genes for miR-152. Among these predicted
targets, MAFB was selected since it was predicted by the two
bioinformatic analyses. As shown in Fig. 4A, MAFB was predicted to be a direct
target gene of miR-152.
To investigate whether MAFB was a direct target gene
of miR-152, a dual-luciferase reporter assay was performed. As
shown in Fig. 4B, miR-152
significantly inhibited luciferase activity in PGL3-MAFB-3′UTR
Wt-transfected HEK293T cells, but not in PGL3-MAFB-3′UTR
Mut-transfected HEK293T cells (P<0.05). These results indicate
that MAFB is a direct target gene of miR-152.
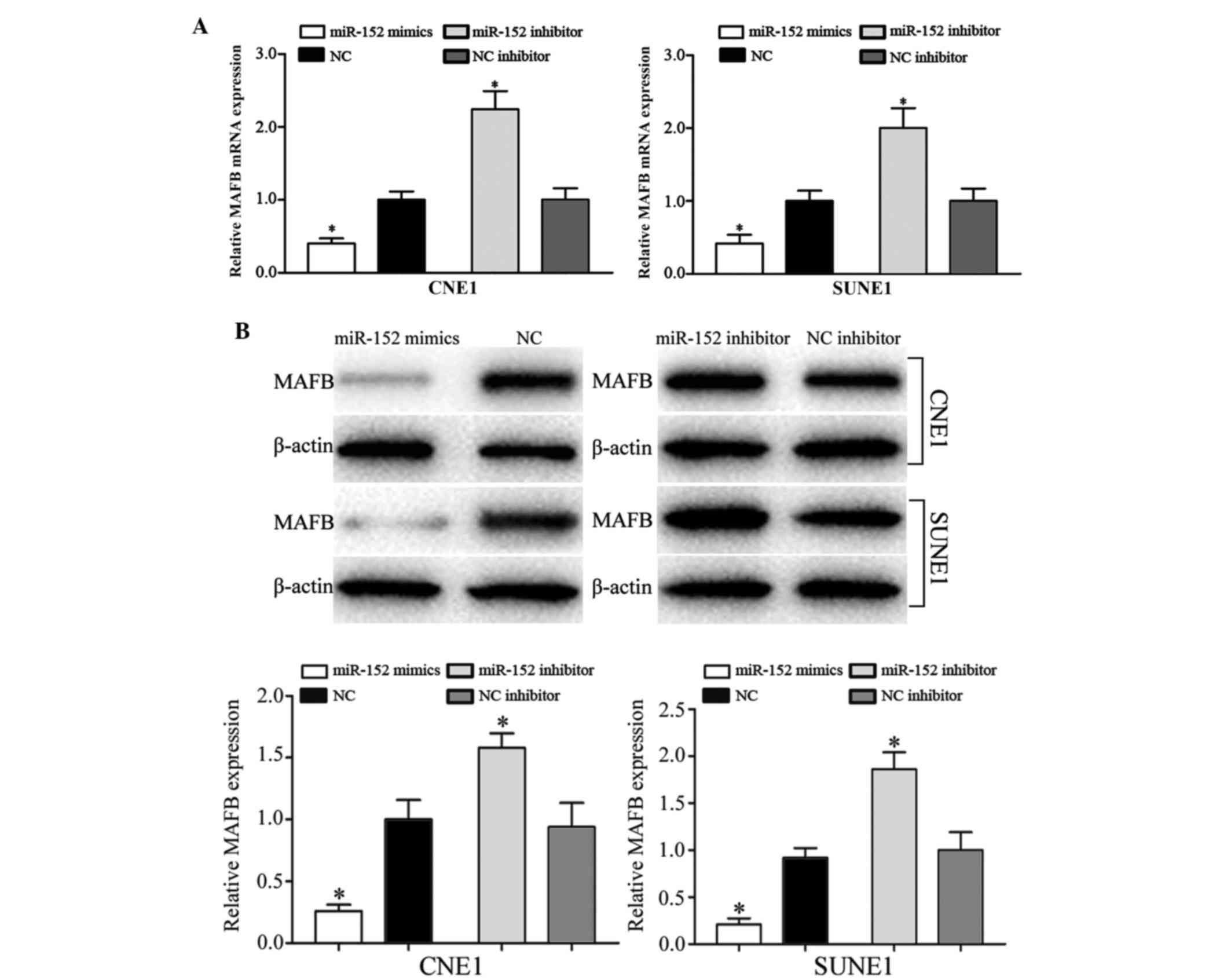
miR-152 negatively regulates MAFB
expression at the mRNA and protein level
To determine the association between miR-152 and
MAFB expression, RT-qPCR and western blotting were performed. As
presented in Fig. 5A, transfection
with miR-152 mimics decreased the mRNA expression levels of MAFB,
whereas the miR-152 inhibitor increased MAFB mRNA expression
(P<0.05). Western blotting revealed that compared with NC
mimics-transfected cells, the protein expression levels of MAFB
were also significantly downregulated in CNE1 and SUNE1 cells
transfected with miR-152 mimics, whereas transfection with the
miR-152 inhibitor enhanced MAFB expression at the protein level
(Fig. 5B; P<0.05).
MAFB is associated with the effects of
miR-152 on NPC cells
The present study explored the endogenous expression
of MAFB in CNE1, SUNE1 and NP69 cells by western blotting. The
expression levels of MAFB were significantly upregulated in CNE1
and SUNE1 cells compared with in NP69 cells (Fig. 6A; P<0.05). It is in accord with
the down-regulation of miR-152 in NPC.
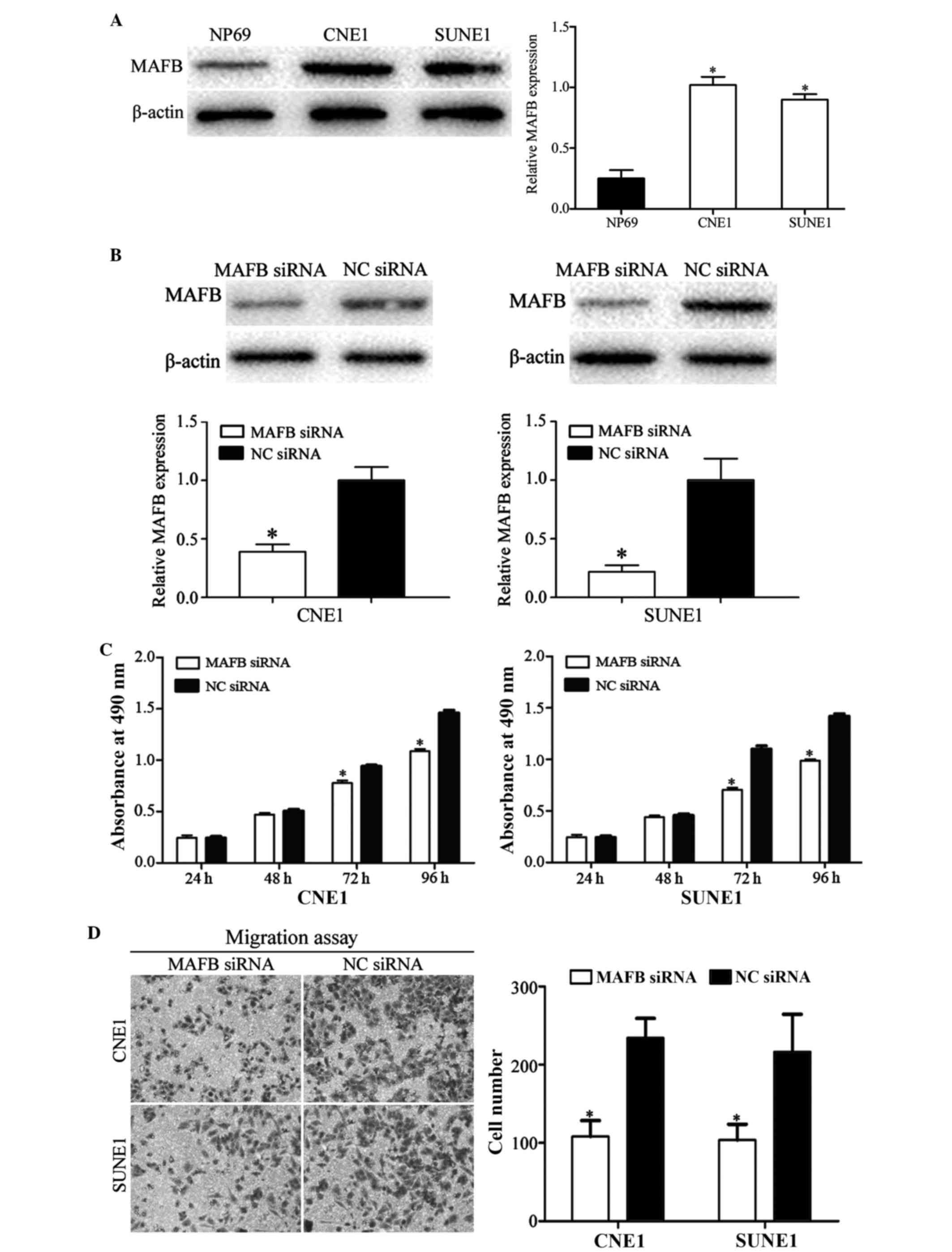 | Figure 6.Effects of MAFB on CNE1 and SUNE1
cells. (A) Western blotting revealed that MAFB was upregulated in
CNE1 and SUNE1 cells compared with NP69 cells. (B) Western blotting
revealed that the protein expression levels of MAFB were
downregulated in CNE1 and SUNE1 cells following transfection with
MAFB siRNA. (C) CNE1 and SUNE1 cells transfected with MAFB siRNA
exhibited significantly decreased cell proliferation. (D) Cell
migration assay indicated that transfection with MAFB siRNA
inhibited CNE1 and SUNE1 cell migration; magnification, ×200.
*P<0.05 vs. respective controls. miR, microRNA; MAFB, V-maf
avian musculoaponeurotic fibrosarcoma oncogene homolog B; siRNA,
small interfering RNA; NC, negative control. (E) Cell invasion
assay indicated that inhibition of MAFB significantly suppressed
CNE1 and SUNE1 cell invasion; magnification, ×200. *P<0.05 vs.
respective controls. miR, microRNA; MAFB, V-maf avian
musculoaponeurotic fibrosarcoma oncogene homolog B; siRNA, small
interfering RNA; NC, negative control. |
To determine whether MAFB functions as an important
mediator of the effects of miR-152 in NPC cells, MAFB siRNA and NC
siRNA were transfected into NPC cells. Western blotting was
performed to explore transfection efficiency post-transfection with
the siRNAs. As shown in Fig. 6B,
MAFB was downregulated in CNE1 and SUNE1 cells following
transfection with MAFB siRNA (P<0.05).
The results of an MTT assay demonstrated that
transfection with MAFB siRNA significantly inhibited cell growth,
as compared with in the NC siRNA group (Fig. 6C; P<0.05). Furthermore,
migration and invasion assays demonstrated that transfection with
MAFB siRNA inhibited the cell migration and invasion abilities of
NPC cells compared with cells transfected with NC siRNA (Fig. 6D and E; P<0.05). These results
suggest that the functions of MAFB siRNA were similar to those
induced by miR-152 mimics in NPC cells, thus indicating that MAFB
is a functional target of miR-152 in vitro.
Discussion
miR-152 belongs to the miR-148/152 family, which
also includes miR-148a, miR-148b and miR-152 (16). miR-152 is located at chromosomal
region 17q21.32 (17). Aberrant
expression of miR-152 has been detected in various types of human
tumor, including non-small cell lung cancer (18), glioblastoma (12), gastric cancer (19), hepatocellular carcinoma (20), prostate cancer (21) and ovarian cancer (22). However, to the best of our
knowledge, there have been no studies regarding the expression
levels of miR-152 in NPC. The present study demonstrated that
miR-152 was significantly downregulated in NPC tissues and cell
lines. This finding indicated that miR-152 may exhibit a
tumor-suppressive capacity in the carcinogenesis and progression of
NPC.
Aberrant expression of miR-152 is thought to
contribute to the malignant phenotype of several types of tumor.
For example, in non-small cell lung cancer, miR-152 was
downregulated in tumor tissues and serum. The low expression levels
of miR-152 were significantly associated with more aggressive
tumors (18,23). Furthermore, the results of a
functional assay indicated that upregulation of miR-152 inhibited
proliferation, colony formation, migration and invasion of
non-small cell lung cancer cells by targeting ADAM metallopeptidase
domain 17 and fibroblast growth factor 2 (16,24).
Zheng et al reported that the expression
levels of miR-152 were decreased in glioma tissues. Ectopic
expression of miR-152 suppressed cell invasion and angiogenesis by
targeting neuropilin 2 and matrix metalloproteinase-3 (25). In gastric cancer, reduced
expression of miR-152 has been observed in tumor tissues and cell
lines, and restoration of miR-152 expression reduced cell growth,
migration and invasion via blockade of cluster of differentiation
151 (19). Furthermore, Dang et
al demonstrated that the expression levels of miR-152 were
markedly downregulated in hepatocellular carcinoma, and were
significantly correlated with advanced clinical stage, larger tumor
size and positive hepatitis B infection. In addition, transfection
with miR-152 mimics suppressed cell proliferation, migration and
invasion, and enhanced caspase activity and apoptosis by directly
targeting the tumor necrosis factor receptor superfamily member 6b
gene (20). These findings
indicated that miR-152 may serve important roles in these types of
cancer, and may function as a potential therapeutic target for the
treatment of specific cancers.
The present study demonstrated that miR-152
significantly inhibited NPC cell growth, migration and invasion.
Since miR-152 contributed to NPC carcinogenesis and development,
the present study aimed to determine the potential molecular
mechanism underlying miR-152-induced inhibition of NPC
proliferation, migration and invasion. Subsequently, an important
molecular association between miR-152 and MAFB was detected in NPC.
Initially, TargetScan and miRanda databases predicted that MAFB
contained a miR-152 seed match at position 1619–1626 of the MAFB
3′-UTR. Secondly, a dual-luciferase reporter assay demonstrated
that miR-152 directly targeted the MAFB 3′-UTR. Thirdly, RT-qPCR
and western blotting revealed that miR-152 negatively regulated
MAFB expression at the mRNA and protein level. Finally, knockdown
of MAFB decreased NPC cell proliferation, migration and invasion,
similar to the effects of miR-152. These results indicated that
miR-152 may target MAFB to inhibit NPC cell growth, migration and
invasion. Identification of miR-152 target genes is essential for
understanding its role in NPC carcinogenesis and progression. In
addition, it is important for developing novel targeted therapies
for the treatment of NPC.
The Maf oncogene was initially identified in an
acutely oncogenic avian retrovirus AS42 and was isolated from a
spontaneous chicken tumor (26).
The Maf family encompasses three small and four large Maf proteins.
These Maf proteins are divided into two groups: Large MAF proteins,
including c-MAF, MAFB and MAFA/L; and small MAF proteins, including
MAFK, MAFF and MAFG (27–29). MAFB, which is a member of the Maf
protein family, is a transcription factor that shares a conserved
basic region and leucine zipper motif (30). Previous studies have demonstrated
that MAFB serves an important role in early tissue specification
and terminal differentiation (31). The present study demonstrated that
MAFB was upregulated in NPC cell lines compared with in the NP69
normal nasopharyngeal epithelial cell line. The results indicated
that miR-152 may function as a tumor suppressor through the
downregulation of MAFB in NPC. Therefore, the upregulation of MAFB
detected in NPC may be due to the downregulation of miR-152. In
addition, knockdown of MAFB significantly inhibited NPC cell
proliferation, migration and invasion. The present study revealed
that miR-152 negatively regulates MAFB expression to decrease NPC
cell growth, migration and invasion; therefore, it may be
beneficial to investigate a novel targeted therapy against MAFB in
NPC.
In conclusion, the present study demonstrated that
miR-152 was downregulated in NPC tissues and cell lines. In
addition, miR-152 expression and MAFB knockdown inhibited cell
proliferation, migration and invasion, and miR-152 suppressed the
expression of MAFB at the mRNA and protein levels. These results
indicated that miR-152 targets MAFB, which may be associated with
NPC carcinogenesis and progression. Therefore, miR-152 and MAFB may
be investigated as therapeutic targets for the treatment of NPC.
Future work is required to address whether the potential of miR-152
may be fully realized in NPC treatment.
References
|
1
|
Tao Q and Chan AT: Nasopharyngeal
carcinoma: Molecular pathogenesis and therapeutic developments.
Expert Rev Mol Med. 9:1–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu J, Cosmopoulos K, Pegtel M, Hopmans E,
Murray P, Middeldorp J, Shapiro M and Thorley-Lawson DA: A novel
persistence associated EBV miRNA expression profile is disrupted in
neoplasia. PLoS Pathog. 7:e10021932011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan G, Tang X and Tang F: The role of
microRNAs in nasopharyngeal carcinoma. Tumour Biol. 36:69–79. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McDermott AL, Dutt SN and Watkinson JC:
The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol Allied
Sci. 26:82–92. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su SF, Han F, Zhao C, Huang Y, Chen CY,
Xiao WW, Li JX and Lu TX: Treatment outcomes for different
subgroups of nasopharyngeal carcinoma patients treated with
intensity-modulated radiation therapy. Chin J Cancer. 30:565–573.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee AW, Foo W, Law SC, Peters LJ, Poon YF,
Chappell R, Sze WM, Tung SY, Lau WH and Ho JH: Total biological
effect on late reactive tissues following reirradiation for
recurrent nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys.
46:865–872. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hui EP, Leung SF, Au JS, Zee B, Tung S,
Chua D, Sze WM, Law CK, Leung TW and Chan AT: Lung metastasis alone
in nasopharyngeal carcinoma: A relatively favorable prognostic
group. A study by the Hong Kong nasopharyngeal carcinoma study
group. Cancer. 101:300–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wright CM, Dan T, Dicker AP and Simone NL:
microRNAs: The short link between cancer and RT-induced DNA damage
response. Front Oncol. 4:1332014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He J, Tang Y and Tian Y: MicroRNA-214
promotes proliferation and inhibits apoptosis via targeting Bax in
nasopharyngeal carcinoma cells. Mol Med Rep. 12:6286–6292.
2015.PubMed/NCBI
|
|
10
|
Chen JJ, Liu SX, Chen MZ and Zhao ZY:
Has-miR-125a and 125b are induced by treatment with cisplatin in
nasopharyngeal carcinoma and inhibit apoptosis in a p53-dependent
manner by targeting p53 mRNA. Mol Med Rep. 12:3569–3574.
2015.PubMed/NCBI
|
|
11
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z,
Li Z and Xue Y: MiR-152 functions as a tumor suppressor in
glioblastoma stem cells by targeting Kruppel-like factor 4. Cancer
Lett. 355:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Allaya N, Khabir A, Sallemi-Boudawara T,
Sellami N, Daoud J, Ghorbel A, Frikha M, Gargouri A,
Mokdad-Gargouri R and Ayadi W: Over-expression of miR-10b in NPC
patients: Correlation with LMP1 and Twist1. Tumour Biol.
36:3807–3814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su Y, Wang Y, Zhou H, Lei L and Xu L:
MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS
Lett. 588:1983–1988. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsuruta T, Kozaki K, Uesugi A, Furuta M,
Hirasawa A, Imoto I, Susumu N, Aoki D and Inazawa J: miR-152 is a
tumor suppressor microRNA that is silenced by DNA hypermethylation
in endometrial cancer. Cancer Res. 71:6450–6462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Chen YY, Li SQ, Huang C and Qin YZ:
Expression of miR-148/152 family as potential biomarkers in
non-small-cell lung cancer. Med Sci Monit. 21:1155–1161. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhai R, Kan X, Wang B, Du H, Long Y, Wu H,
Tao K, Wang G, Bao L, Li F and Zhang W: miR-152 suppresses gastric
cancer cell proliferation and motility by targeting CD151. Tumour
Biol. 35:11367–11373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dang YW, Zeng J, He RQ, Rong MH, Luo DZ
and Chen G: Effects of miR-152 on cell growth inhibition, motility
suppression and apoptosis induction in hepatocellular carcinoma
cells. Asian Pac J Cancer Prev. 15:4969–4976. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao
L, Cai H, Li P, Cao Q, Ju X, et al: miR-152 controls migration and
invasive potential by targeting TGFα in prostate cancer cell lines.
Prostate. 73:1082–1089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou X, Zhao F, Wang ZN, Song YX, Chang H,
Chiang Y and Xu HM: Altered expression of miR-152 and miR-148a in
ovarian cancer is related to cell proliferation. Oncol Rep.
27:447–454. 2012.PubMed/NCBI
|
|
23
|
Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu
RX, Liu SH, Yi QT, Li J and Song CH: Serum miR-152, miR-148a,
miR-148b, and miR-21 as novel biomarkers in non-small cell lung
cancer screening. Tumour Biol. 36:3035–3042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng Z, Ma R, Tan W and Zhang L: MiR-152
suppresses the proliferation and invasion of NSCLC cells by
inhibiting FGF2. Exp Mol Med. 46:e1122014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng X, Chopp M, Lu Y, Buller B and Jiang
F: MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis
via NRP-2 and MMP-3. Cancer Lett. 329:146–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nishizawa M, Kataoka K and Vogt PK: MafA
has strong cell transforming ability but is a weak transactivator.
Oncogene. 22:7882–7890. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blank V and Andrews NC: The Maf
transcription factors: Regulators of differentiation. Trends
Biochem Sci. 22:437–441. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ring BZ, Cordes SP, Overbeek PA and Barsh
GS: Regulation of mouse lens fiber cell development and
differentiation by the Maf gene. Development. 127:307–317.
2000.PubMed/NCBI
|
|
29
|
Motohashi H, Shavit JA, Igarashi K,
Yamamoto M and Engel JD: The world according to Maf. Nucleic Acids
Res. 25:2953–2959. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zanocco-Marani T, Vignudelli T, Parenti S,
Gemelli C, Condorelli F, Martello A, Selmi T, Grande A and Ferrari
S: TFE3 transcription factor regulates the expression of MAFB
during macrophage differentiation. Exp Cell Res. 315:1798–1808.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eychene A, Rocques N and Pouponnot C: A
new MAFia in cancer. Nat Rev Cancer. 8:683–693. 2008. View Article : Google Scholar : PubMed/NCBI
|