Introduction
In previous years, the potency of genetically
modified T cell-mediated immunity against viruses and certain
malignancies has been well established. TCR gene adoptive therapy
is a clinically promising approach for the treatment of malignant
tumors and viral diseases. Using ex vivo gene transfer, T
cells isolated from patients can be genetically engineered to
express a novel TCR, and the engineered T cells are re-infused back
into the patient to specifically recognize a tumor-associated
antigen and thereby selectively lyse tumor cells (1). However toxicity has been observed in
clinical trials using genetically modified TCR therapies (2). An important toxic effect is on-target
off-tumor activity, which occurs if the peptide target sequence of
the TCR is also expressed on other cells (3), which has been reported to occur in
clinical trials (4–6). Another undesirable toxic effect is
off-target reactivity, and one cause for this effect is the
occurrence of cross-reactivity, which is due to the ability of the
TCR to react against the peptides expressed on non-target proteins
(7). This toxic effect may also
result from the mixture of TCRs generated by the introduced TCR α-
and β-chains mispairing with the endogenous TCR β- and α-chains.
The mispaired TCR increases the risk of unknown specificity causing
autoreactivity (8). No formal
observations of toxicities mediated by TCR mispairing have been
observed in clinical trials to date, however, preclinical studies
have demonstrated that mispaired TCRs have the potential to induce
the harmful recognition of self-antigens, resulting in graft, vs.
host disease (9). These findings
indicate the requirement to prevent or reduce TCR mispairing, to
improve T cell avidity and reduce potential off-target toxicity,
including the genetic modification of TCR transgenes (10–13),
disruption of endogenous TCR chains via short hairpin RNA or zinc
finger nucleases (14,15), αβ TCR transfer to γδT cells or
γ9δ2TCR transduction of αβT cells (16,17).
Although it has been reported that the transfer of
γ9δ2TCR into αβT cells can prevent the formation of mixed TCR
dimers and efficiently kill cancer cell lines in vitro
(17), the role of the Vγ9Vδ2 TCR
in antigen recognition remains to be fully elucidated, as does the
biology of γδTCR, compared with αβTCR. Thus, the present study
aimed to examine whether the domains of the γδTCR constant
exchanged in αβTCR can improve the pairing and function of αβTCR.
Three chimeric TCR variants were constructed, and domain-exchange
and three-dimensional (3D) modeling strategies were applied, in
which the αβTCR constant was replaced with partial or complete
constant regions of γδTCR, leaving the variable domains intact.
Subsequently, genetically-encoded reporters coupled with a pair of
fluorescent proteins were constructed to monitor the expression and
pairing between chimeric TCRα chains and TCRβ chains using a
confocal laser scanning microscope (CLSM) in living cells. The data
showed that swapping of the αβTCR constant region of
immunoglobulin-like (Ig) domain for the corresponding γδTCR domain
enhanced expression and reduced mispairing on the cell surface. The
other two chimeric TCRs harboring the connecting peptide,
transmembrane and intracellular (cp+tm+ic) domains or complete
constant (C) domain of γδTCR did not show improved expression,
however, the level of mispairing decreased. Finally the function of
the chimeric TCR variants were examined in peripheral blood
mononuclear cells (PBMCs), which revealed that the introduction of
γδTCR constant region of Ig domains in the αβTCR was able to
mediate the same levels of interferon (IFN)-γ secretion and
cytotoxic activity as the wild-type (wt)TCR when co-cultured with
human leukocyte antigen (HLA)2+ human hepatocellular
cell lines. However, the other two TCRs containing the γδTCR
cp+tm+ic domains or C domains did not trigger the lymphocytes to
produce IFN-γ or activate cytotoxic T cells when co-cultured with
HLA-A2+ or HLA-A2− target cells. Taken
together, these findings demonstrated that exchange of the constant
region of the Ig domain of γδTCR in αβTCR decreased mispairing
without compromising T cell function, however, this was not the
case in those containing the γδTCR cp+tm+ic or C domains.
Materials and methods
Cells
PBMCs of HLA-A2+ were isolated
from the blood of healthy donors (Table I), following the provision of
informed consent, using Ficoll gradient centrifugation at 600 × g
for 20 min at room temperature, followed by washing in PBS,
re-suspension at a concentration of 1×106 cells/ml and
activated by soluble anti-CD3ε mAb (OKT3, 30 ng/ml, R&D
Systems, Inc., Minneapolis, MN, USA) and soluble anti-CD28 mAb (1
ng/ml, R&D Systems, Inc.) and 300 IU/ml recombinant human IL-2
(R&D Systems, Inc.) at 37°C for 48 h. Human PBMCs and
Jurkat/E6-1 cells (cat. no. TIB-152; American Type Culture
Collection, Manassas, VA, USA) were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 100 U/ml penicillin-streptomycin at 37°C and 5%
CO2.
 | Table I.Details of blood donors. |
Table I.
Details of blood donors.
| Number | Gender | Age | Hospital admitted
to | Dates of blood
donation |
|---|
| 1 | Female | 24 | The First Affiliated
Hospital/School of Clinical Medicine of Guangdong Pharmaceutical
University | 2013.9.15, 2013.9.21,
2013.9.30, 2013.10.9, 2013.10.20 |
| 2 | Male | 25 |
| 2013.9.15 |
| 3 | Male | 30 |
| 2013.9.15, 2013.9.21,
2013.9.30, 2013.10.9, 2013.10.20 |
| 4 | Female | 28 |
| 2013.9.15 |
| 5 | Male | 21 |
| 2013.9.15, 2013.9.21,
2013.9.30, 2013.10.9, 2013.10.20 |
The HEK293 human embryonic kidney cell line, and the
HepG2 (HLA-A2+), Huh-1 (HLA-A2+) and BEL-7402
(HLA-A2−) human hepatocellular carcinoma cell lines
(Guangdong Province Key Laboratory for Biotechnology Drug
Candidates, Guangzhou, China) were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 100 U/ml penicillin-streptomycin at
37°C and 5% CO2.
Vector construction
The αβTCR was isolated from tumor-infiltrating
lymphocytes of a patient (HLA-A2+;
α-fetoprotein+) with hepatocellular carcinoma, which was
preserved in our laboratory, as described previously (Table II) (18). The γ9δ2TCR was isolated from
healthy human PBMCs and was composed of TRGV9/J2/C1 and
TRDV2/D3/J1/C1. TCR-V(D)J gene nomenclature was according to
http://www.imgt.org. Unmodified wtTCR was used as a
control TCR.
 | Table II.Patient details of patient with
hepatocellular carcinoma. |
Table II.
Patient details of patient with
hepatocellular carcinoma.
| Number | Gender | Age | Hospital admitted
to | Dates of blood
donation |
|---|
| 012 | Male | 54 | The First
Affiliated Hospital/School of Clinical Medicine of Guangdong
Pharmaceutical University | 2010.5.9 |
Three TCR variants were constructed using a domain-
exchange strategy in which the IgC, cp+tm+ic and C regions of αβTCR
were exchanged for corresponding regions of γδTCR. These three
chimeras were termed TCR∆IgC; TCR∆cp+tm+ic; and TCR∆C,
respectively. ∆ indicates a lack of αβTCR domain/s and replacement
by corresponding γδTCR domain/s. The exact boundaries of the IgC,
cp+tm+ic and C domains of TCRα, TCRβ, TCRγ and TCRδ are described
in the legend of Fig. 1. To
measure the pairing between the TCR α- and β-chains, the three
modified TCRs were coupled to a pair of fluorescent proteins, ECFP
and EYFP, and adenoviral particles without fluorescent proteins
were constructed and produced, as described previously (18). Primer sequences used for cloning
the TCRα- and β- fusion genes and TCR are provided in Tables III, IV and V. All TCR constructs were sequence
verified.
 | Table III.Primers for amplifying TCRβ∆IgC-EYFP
and TCRα∆IgC-ECFP fusion genes (regular polymerase chain reaction
(PCR) and SOE-PCR). |
Table III.
Primers for amplifying TCRβ∆IgC-EYFP
and TCRα∆IgC-ECFP fusion genes (regular polymerase chain reaction
(PCR) and SOE-PCR).
| Gene name | Primer name | Direction | Primer
sequence |
|---|
| TRBV | P1 | Forward | ATAGCTAGCGCCACCATGGGCTGCAGGCTGCTCTG |
|
| P2 | Reverse |
ACATCTGCATCAAGTTGTTTCTCCAGTACGGTCAGCCT |
| TRGC | P3 | Forward |
AGGCTGACCGTACTGGAGAAACAACTTGATGCAGATGT |
|
| P4 | Reverse |
CGGAGGTGAAGCCACAGTCTGTCTTTATTGGAGGAAAG |
| TRBCm | P5 | Forward |
CTTTCCTCCAATAAAGACAGACTGTGGCTTCACCTCCG |
|
| P6 | Reverse |
CTCGCCCTTGCTCACCATGCCTCTGGAATCCTTTCT |
| EYFP | P7 | Forward |
AGAAAGGATTCCAGAGGCATGGTGAGCAAGGGCGAG |
|
| P8 | Reverse | CGGCGTCGACTTACTTGTACAGCTCGTC |
| TRAV | X1 | Forward | ACGCCACAACCTTGGCCACCATGATATCCTTGAGAGTT |
|
| X2 | Reverse |
GGTTTGGTATGAGGCTGACTATTTGGTTTTACTGTCAGTCTGG |
| TRDC | X3 | Forward |
CCAGACTGACAGTAAAACCAAATAGTCAGCCTCATACCAAACC |
|
| X4 | Reverse |
GCTTGACATCACAGGAACTTTCTGTAGAATCTGTCTTCACTTC |
| TRACm | X5 | Forward |
GAAGTGAAGACAGATTCTACAGAAAGTTCCTGTGATGTCAAGC |
|
| X6 | Reverse |
CTCCTCGCCCTTGCTCACCATGCTGGACCACAGCCGCAGC |
| ECFP | X7 | Forward |
GCTGCGGCTGTGGTCCAGCATGGTGAGCAAGGGCGAGGAG |
|
| X8 | Reverse | AGTGCGGCCGCTTACTTGTACAGCTCGTCCAT |
 | Table IV.Primers for amplifying
TCRβ∆cp+tm+ic-EYFP and TCRα∆cp+tm+ic-ECFP fusion gene (regular
polymerase chain reaction (PCR) and SOE-PCR). |
Table IV.
Primers for amplifying
TCRβ∆cp+tm+ic-EYFP and TCRα∆cp+tm+ic-ECFP fusion gene (regular
polymerase chain reaction (PCR) and SOE-PCR).
| Gene | Primer name | Direction | Primer
sequence |
|---|
| TRBVC | P1 | Forward | ATAGCTAGCGCCACCATGGGCTGCAGGCTGCTCTG |
|
| O2 | Reverse |
GATCCATTGTGATGACATCTGCTCTACCCCAGGCCTCG |
| TRGCm | O3 | Forward |
CGAGGCCTGGGGTAGAGCAGATGTCATCACAATGGATC |
|
| O4 | Reverse |
CCTCGCCCTTGCTCACCATTGATTTCTCTCCATTGCAG |
| EYFP | O5 | Forward |
CTGCAATGGAGAGAAATCAATGGTGAGCAAGGGC |
|
| P8 | Reverse | CGGCGTCGACTTACTTGTACAGCTCGTC |
| TRAVC | X1 | Forward | ACGCCACAACCTTGGCCACCATGATATCCTTGAGAGTT |
|
| H2 | Reverse |
CCTTTGGTTTTACGTGATCTGGGCTGGGGAAGAAGGTG |
| TRDCm | H3 | Forward |
CACCTTCTTCCCCAGCCCAGATCACGTAAAACCAAAGG |
|
| H4 | Reverse |
CCTCGCCCTTGCTCACCATCAAGAAAAATAACTTGGCAGT |
| ECFP | H5 | Forward |
ACTGCCAAGTTATTTTTCTTGATGGTGAGCAAGGGCGAGG |
|
| X8 | Reverse | AGTGCGGCCGCTTACTTGTACAGCTCGTCCAT |
 | Table V.Primers for amplifying TCRβ∆C-EYFP
and TCRα∆C-ECFP fusion genes (regular polymerase chain reaction
(PCR) and SOE-PCR). |
Table V.
Primers for amplifying TCRβ∆C-EYFP
and TCRα∆C-ECFP fusion genes (regular polymerase chain reaction
(PCR) and SOE-PCR).
| Gene | Primer name | Direction | Primer
sequence |
|---|
| TRBV | P1 | Forward | ATAGCTAGCGCCACCATGGGCTGCAGGCTGCTCTG |
|
| P2 | Reverse |
ACATCTGCATCAAGTTGTTTCTCCAGTACGGTCAGCCT |
| TRGC+GCm | P3 | Forward |
AGGCTGACCGTACTGGAGAAACAACTTGATGCAGATGT |
|
| O4 | Reverse |
CCTCGCCCTTGCTCACCATTGATTTCTCTCCATTGCAG |
| EYFP | O5 | Forward |
CTGCAATGGAGAGAAATCAATGGTGAGCAAGGGC |
|
| P8 | Reverse | CGGCGTCGACTTACTTGTACAGCTCGTC |
| TRAV | X1 | Forward | ACGCCACAACCTTGGCCACCATGATATCCTTGAGAGTT |
|
| X2 | Reverse |
GGTTTGGTATGAGGCTGACTATTTGGTTTTACTGTCAGTCTGG |
| TRDC+DCm | X3 | Forward |
CCAGACTGACAGTAAAACCAAATAGTCAGCCTCATACCAAACC |
|
| H4 | Reverse |
CCTCGCCCTTGCTCACCATCAAGAAAAATAACTTGGCAGT |
| ECFP | H5 | Forward |
ACTGCCAAGTTATTTTTCTTGATGGTGAGCAAGGGCGAGG |
|
| X8 | Reverse | AGTGCGGCCGCTTACTTGTACAGCTCGTCCAT |
Cell transfection
The TCR variants fused to the pair of ECFP and EYFP
fluorescent proteins were transduced into Jurkat cells and BEL-7402
cells at a density of 1×106 cells/ml, using
Lipofectamine LTX/PLUS (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
Image acquisition and fluorescence
resonance energy transfer (FRET) analysis
Confocal images of cells were captured using an
Olympus FluoView1000 CLSM with FV10-ASW 1.7 software (Olympus,
Tokyo, Japan), and apparent FRET efficiency was calculated, as
described previously (18).
Briefly, the Jurkat cells transduced with chimeric TCR constructs
were immobilized onto a glass-bottomed dish, and the TCR
construct-transduced BEL-7402 cells grown on the glass-bottomed
dish were washed twice with PBS solution. The ECFP channel was
excited with a 458 nm argon laser as a donor and the EYFP channel
was excited with a 515 nm argon laser as an acceptor. Subsequently,
seven images were captured to calculate FRET efficiency. This was
calculated using the sensitized acceptor emission method using the
following equation:
Efficiency=1-IDA/{IDA+pFRETx[(ψdd/ψaa)x(Qd/Qa)]},
where pFRET is the processed FRET obtained by removing the donor
SBT (DSBT) and the acceptor SBT (ASBT) from the contaminated or
uncorrected FRET; IDA is the intensity of the donor in
the presence of the acceptor; ψdd and ψaa are
the collection efficiencies in the donor and acceptor channel; and
Qd and Qa are the quantum yield of the donor
and acceptor, respectively. Statistical analysis of the mean FRET
efficiency were calculated from multiple (n=4) cell images in each
group and five randomly selected regions of interest (ROI) in each
cell image.
TCR adenovirus construction and
transduction of T cells
For production of the adenovirus, HEK-293 cells were
transfected with the respective TCR-encoding shuttle plasmid and
second-generation Ad5F35 adenoviral packaging plasmid
(pBHGIoxdelE13Cre; Biovector Science Lab, Inc., Beijing, China).
The HEK-293 cells were seeded in 6-well plates at a density of
1×106 per well. After 24 h, the cells were
co-transfected with an equimolar ratio of the two plasmids (2.5 µg
total DNA per well) and 6 µl/well Lipofactamine 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The adenovirus supernatants were
harvested ~12 days following transfection, cellular debris was
removed by centrifugation at 14,000 × g for 10 min at room
temperature. Adenovirus particle titers were determined using the
TCID50 method and the supernatants were directly used for
transduction of the Jurkat T cells and PBMCs, respectively, as
previously described (18).
Flow cytometry and analysis
The surface expression of the transgenic TCRs on the
cells were assessed by fluorescein isothiocyanate (FITC)-conjugated
anti-TCRVα12.1 mAb (cat. no. TCR2764; dilution, 1:500; Invitrogen;
Thermo Fisher Scientific, Inc.) and PE-conjugated anti-TCRVβ7.1 mAb
(cat. no. IM2287; dilution, 1:500; Beckman Coulter, Inc., Brea, CA,
USA). The transduced Jurkat T cells and PBMCs (5×105)
were stained with the mAbs on ice for 30 min. Following washing
with PBS, the cells were fixed in 2% PFA prior to measurements on
an Epics-XL flow cytometer (Beckman Coulter, Inc.). Non-transduced
Jurkat T cells and PBMCs were used as controls.
Cytokine release assays
The unmodified PBMCs and TCR-modified PBMCs were
assessed for reactivity in IFN-γ release assays using commercially
available ELISA kits (Boster Systems, Inc., Pleasanton, CA, USA).
The HepG2 (HLA-A2+), Huh-1 (HLA-A2+) and
BEL-7402 (HLA-A2−) target human hepatocellular carcinoma
cell lines were cultured in medium at 37°C, followed washing with
PBS prior to the initiation of co-cultures. For these assays,
3×105 responder cells (PBMCs) and 1×104
stimulator cells were incubated in a 0.2 ml RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
and 100 U/ml penicillin/streptomycin in individual wells of 96-well
plates and were co-cultured for 24 h at 37°C and 5% CO2.
The secretion of IFN-γ was measured in the culture supernatants
diluted to be in the linear range of the assay.
CTL assay
The abilities of the transduced PBMCs to lyse the
HLA-A2+/HLA-A2− human hepatocellular
carcinoma targets were measured using a calcein AM (CAM) release
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), as
described previously (19).
Briefly, 1×106 tumor cells were labeled with 2 µM CAM,
which was diluted from a 1 mM stock solution of CAM in dimethyl
sulfoxide (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for
30 min at 37°C. The labeled target cells were washed three times
with PBS and re-suspended at a concentration of 1×105
cells/ml in complete medium. The labeled target cells
(1×104 in a volume of 100 µl) were plated in 96-well
V-bottomed plates with effector cells in 200 µl of complete medium
at effector to target (E:T) ratios between 1:1 and 30:1. Following
incubation for 4 h at 37°C with 5% CO2, the supernatants
was harvested and the quantities of calcein released were measured
using a Varioskan Flash multimode reader (Thermo Fisher Scientific,
Inc.). Spontaneous release was determined by incubating the target
cells in medium alone, and maximum release was determined by
suspending the cells with 1% Triton X-100. Each data point was an
average of four wells. The percentage of PBMC-specific lysis was
calculated as follows: Specific lysis (%)=(experimental
release-spontaneous release)/(maximum release-spontaneous release)
× 100.
Splicing by overlap extension
(SOE)-PCR
This method was used to generate the fusion genes of
three chimeric TCR variants. Variable fragments for generating
TCRα∆IgC-ECFP variants (including TCRAV, TRDC, TRACm and ECFP) and
TCRβ∆IgC-EYFP variants (including TRBV, TRGC, TRBCm and EYFP) were
amplified using a set of forward primers and reverse primers
(Table III). The first step of
SOE-PCR reactions were performed with 100 ng of the template
without primers, 10X Buffer, 2 mM dNTPs, 25 mM MgSO4,
0.5 U KOD-Plus-Neo Polymerase (Toyobo Co., Ltd., Osaka, Japan) in
25 µl reaction volume. The PCR cycling conditions were as follows:
Denaturation at 94°C for 2 min, followed by 5 cycles at 94°C for 30
sec, at 57°C for 30 sec, and at 68°C for 90 sec, and completed with
a final extension at 68°C for 7 min. The PCR generated overlapping
gene segments that are then used as template DNA for the second
step of SOE-PCR to create a full-length product. Therefore, another
25 µl reaction mixture (contained 10X Buffer, 2 mM dNTPs, 25 mM
MgSO4, 0.5 KOD-Plus-Neo Polymerase and 1 µM forward
primer X1/P1 and reverse primer X8/P8; Invitrogen; Thermo Fisher
Scientific, Inc.) were added to the first reaction mixture for the
second step of SOE-PCR. The reaction conditions were initial
denaturation at 94°C for 2 min, followed by 30 cycles at 94°C for
30 sec, at 62°C for 30 sec, and at 68°C for 90 sec and a final
extension at 68°C for 7 min. The other two TCR (TCR∆cp+tm+ic and
TCR∆C) fusion genes were generate using the same methods, as the
primer sequences for the amplification of variable regions and
fusion chains are given in Tables
IV and V.
Statistical analysis
Differences among the TCRs in various assays were
examined using Student's t-test (unpaired; two-tailed) and two-way
analysis of variance with a Bonferroni's multiple comparisons test
using GraphpPad Prism 6 software (GraphPad Software Inc., La Jolla,
CA, USA). P<0.05 was considered to indicate a statistically
significant difference. Data are expressed as the mean ± standard
deviation.
Results
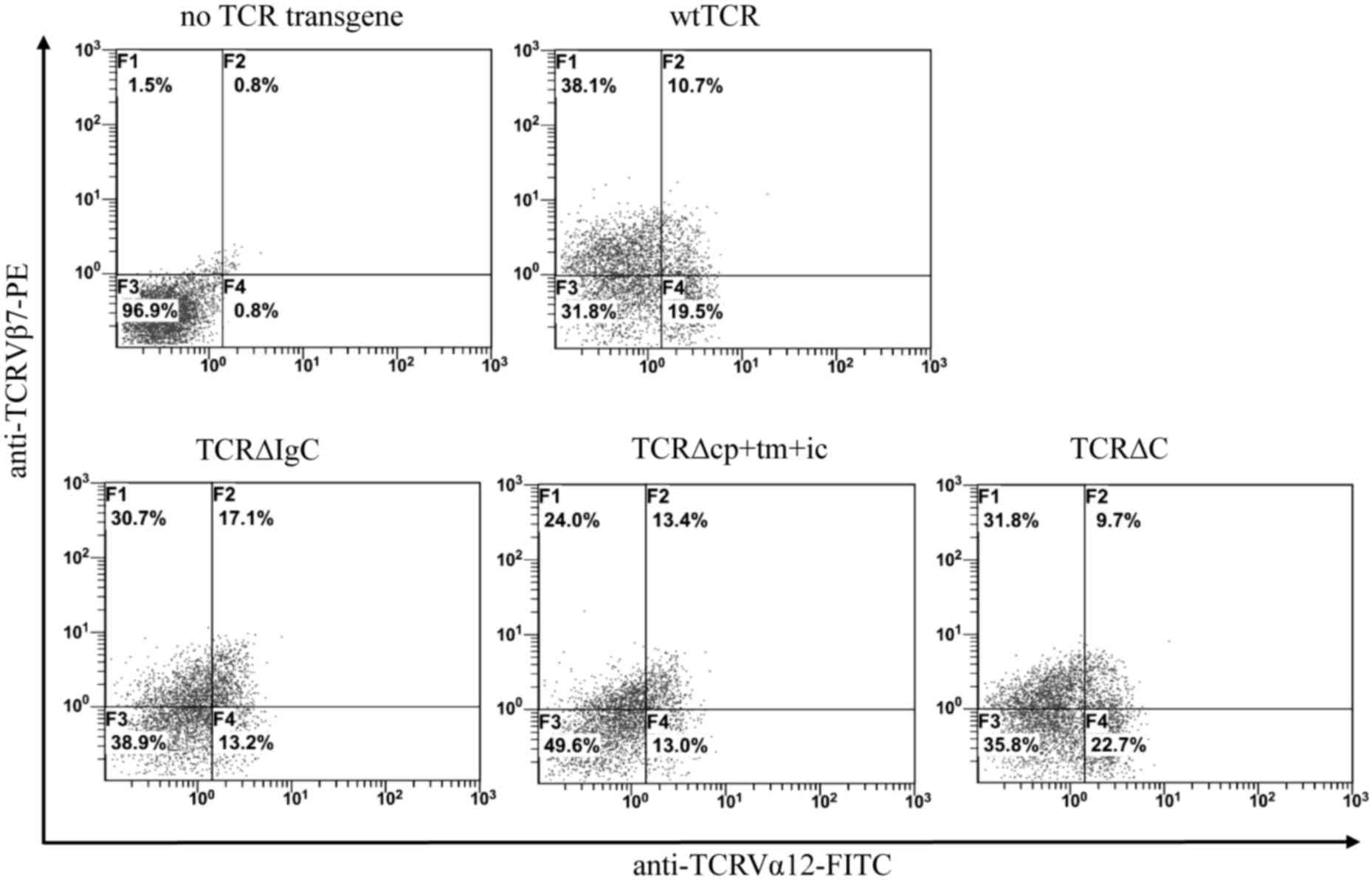
Expression of wtTCR and chimeric TCR
constructs
In the present study, three chimeric TCR variants
were generated, in which the IgC, cp+tm+ic and C regions of αβTCR
were replaced with corresponding regions of γδTCR (Fig. 1). The wtTCR and chimeric TCR genes
were cloned separately into the pDC315 shuttle plasmid to produce
the Ad5F35 adenovirus, and transduction of Jurkat T cells was
performed with subsequent fluorescence-activated cell sorting
analysis. Double immunofluorescent staining with anti-TCRVα12
mAbFITC and anti-Vβ7mAbPE showed that Jurkat
cells transduced with TCR∆IgC exhibited higher surface coexpression
(17.1%), compared with wtTCR (10.7%), and TCR∆cp+tm+ic exhibited a
marginally higher surface expression (13.4%), compared with wtTCR.
By contrast, TCR∆C exhibited a lower level of surface expression,
compared with wtTCR (Fig. 2). To
further examine the expression of the chimeras in living cells, the
C terminus of chimeric TCRα chains and TCRβ chains were fused to a
pair of cyan and yellow fluorescent proteins, ECFP and EYFP
respectively (Fig. 3A and B). In
Jurkat cells, the fluorescence observation using a CLSM showed that
TCR∆IgC was expressed more markedly on the cell surface, which was
in accordance with the flow cytometry results. By contrast, the
other two modified TCRs, TCR∆cp+tm+ic and TCR∆C, exhibited no
apparent difference in expression levels, compared with wtTCR.
These data suggested that the modified αβTCRs harboring the γδTCR
constant region were expressed on the Jurkat cell surface and that
the FRET reporters were able to used for monitoring the expression
and interaction of TCR variants.
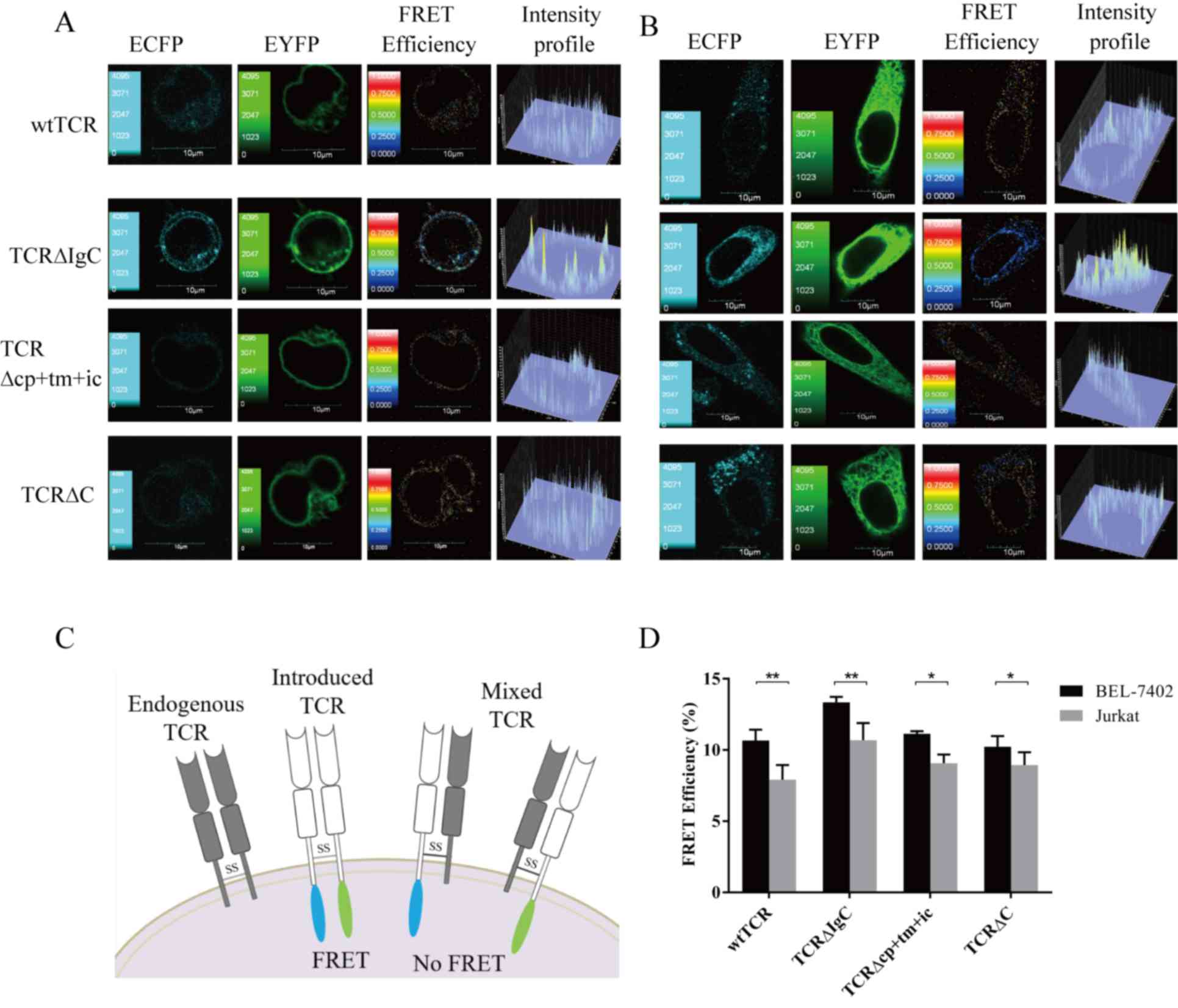 | Figure 3.FRET efficiencies between chimeric
TCRα and TCRβ chains. (A) Jurkat and (B) BEL-7402 cells transiently
expressed wtTCR, TCR∆IgC, TCR∆cp+tm+ic and TCR∆C. The confocal
images in the ECFP and EYFP channels were performed with a confocal
laser-scanning microscope and FV10-ASW 1.7 software. The FRET
efficiency and intensity profile were calculated using the
sensitized acceptor emission method. (C) Detection of FRET between
TCRα-ECFP and TCRβ-EYFP when pairing. If mispairing occurred, no
FRET was detected. (D) Four independent cell images in each group
and five randomly selected regions of interest in each cell image
were selected for statistical analysis of the FRET efficiency. Data
are expressed as the mean ± standard deviation. *P<0.05,
**P<0.01. Data represent one of four independent experiments
with similar results. Scale bar=10 µm. The continuous color scale
(black-white) represents FRET efficiency (0–1). TCR, T cell
receptor; wt, wild-type; Ig, immunoglobulin-like; cp+tm+ic,
connecting peptide, transmembrane and intracellular; FRET,
fluorescence resonance energy transfer. |
Analysis of TCR mispairing
The Jurkat cell line (cloneE6-1) was used as a
recipient T cell model for TCR gene transfer, to determine whether
the chimeric TCRα- and β-chains assembled preferentially when there
was a pair of endogenous TCRs. It was hypothesized that the
introduced TCR α- and β-chains comprising heterodimers on the cell
surface results in FRET efficiency between the donor (ECFP) and
acceptor (EYFP) fluorescent proteins. When the introduced TCR α-
and β- chains and the endogenous TCR β- and α-chains mispair, FRET
is not detected between the mispairing TCRs (Fig. 3C). As shown in Fig. 3A, seven images were used to remove
the DSBT and ASBT from the contaminated FRET to obtain the FRET
efficiency images. The corrected FRET efficiency images showed that
TCR∆IgC exhibited a higher FRET efficiency, compared with wtTCR.
The images of TCR∆cp+tm+ic and TCR∆C exhibited no differences in
FRET efficiencies, compared with wtTCR. A total of four independent
cell images and five ROIs in each cell image were selected for FRET
efficiency analysis, respectively, in each group. The statistical
results showed that the average FRET efficiency between the TCR∆IgC
α- and β-chains (10.69±0.76%) was significantly higher, compared
with the average FRET efficiency between the wtTCR α- and β-chains
(7.92±1.32%; P<0.01). No significant differences were found
between the FRET efficiencies of the other two chimeric TCRs
(9.07±0.61 and 8.95±0.89%), compared with wtTCR (P>0.05;
Fig. 3D).
To further investigate the extent of mispairing of
the modified TCR with the endogenous TCR, BEL-7402 cells deficient
in TCR and CD3 molecules were selected as the next recipient cell
model. It was hypothesized that as there was no endogenous TCR in
the BEL-7402 cells, the pairing of the introduced TCR α- and
β-chains will not be interfered with, allowing the extent of
mispairing to be measured by comparing the FRET efficiencies of
BEL-7402 and Jurkat cells. The images showed that the TCR∆IgC
exhibited a higher FRET efficiency, compared with the wtTCR in the
BEL-7402 cells (Fig. 3B). The
statistical results showed that the average FRET efficiency of
wtTCR in the Jurkat cells (7.92±1.32%) was lower, compared with
that in the BEL-7402 cells (10.59±1.02%; P<0.05; Fig. 3D), suggesting that the wtTCR was
mispaired with the endogenous TCR in Jurkat cells, and the level of
mispairing with endogenous TCR was 25%. Statistical analysis also
showed that the FRET efficiencies of TCR∆IgC were decreased in the
Jurkat cells (10.69±0.76%), compared with the BEL-7402 cells
(13.34±0.40%), indicating a 20% mismatch rate in the Jurkat cells
(Fig. 3D). TCR∆cp+tm+ic and TCR∆C
also showed mispairing with the endogenous TCR (19 and 14%,
respectively). Together, these data showed that replacement with
various constant domains of γδTCR in αβTCR reduced mispairing to
the same extent, but were unable to prevent mispairing.
Analysis of TCR-transduced primary T
cells
The Ad5F35 adenovirus encoding the wtTCR and the
chimeric TCRs were used to transfer into PBMCs, and PBMCs
expressing the wtTCR and chimeric TCRs were co-cultured with the
HepG2 (HLA-A2+, Huh-1 (HLA-A2+) and BEL-7402
(HLA-A2−) human hepatocellular carcinoma cell lines. The
secretion of IFN-γ into the medium was measured using an ELISA
procedure. As shown in Fig. 4A,
when co-cultured with HLA-A2+ cell lines, the PBMCs
transduced with TCR∆IgC secreted the same quantity of IFN-γ as the
PBMCs transduced with wtTCR. Unexpectedly, the PBMCs transduced
with TCR∆cp+tm+ic and TCR∆C secreted lower levels of IFN-γ,
compared with wtTCR, with levels close to those of the PBMCs
containing no TCR transgene. This indicated that TCR∆cp+tm+ic and
TCR∆C did not trigger cytokine secretion. The levels of IFN-γ in
the PBMCs transduced with the three chimeric TCRs or wtTCR were not
above background levels following incubation with the
HLA-A2− cell line.
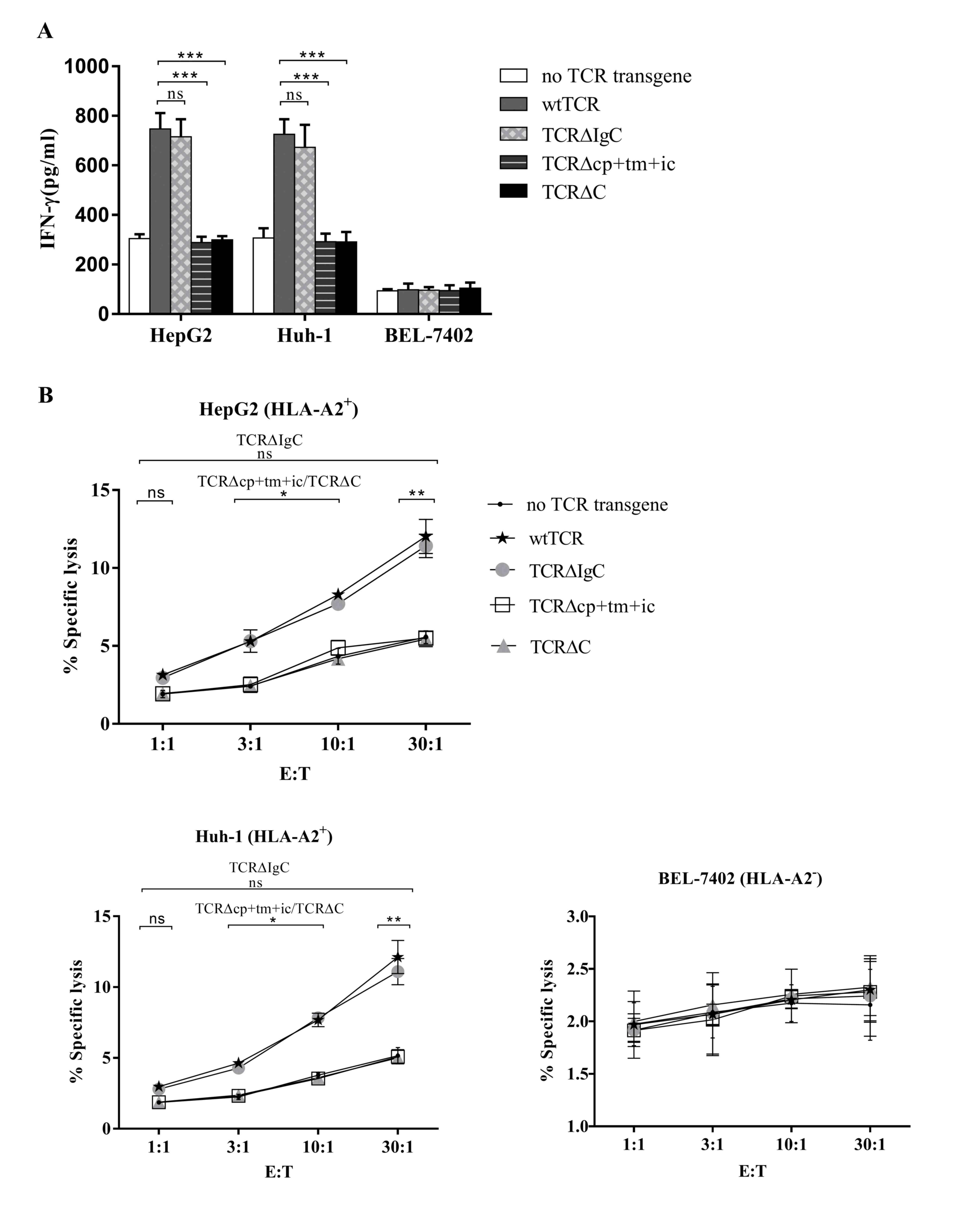 | Figure 4.Function of chimeric TCRs in PBMCs.
(A) Human PBMCs expressing either wtTCR or modified TCR variants
were co-cultured with different tumor cell lines for 24 h, and
concentrations of IFN-γ secreted into the co-culture supernatant
were measured using ELISA. (B) Specific cytotoxicity of tumor cell
lines. The human PBMCs expressing the wtTCR, TCR∆IgC, TCR∆cp+tm+ic
or TCR∆C transgenes, or without a TCR transgene were co-cultured
with CAM-labeled tumor cell lines at the indicated E:T ratios for 4
h, following which specific lysis was calculated. Results represent
the average of three independent experiments, performed with three
donors. *P<0.05, **P<0.01, ***P<0.001). TCR, T cell
receptor; PBMCs, peripheral blood mononuclear cells; wt, wild-type;
Ig, immunoglobulin-like; cp+tm+ic, connecting peptide,
transmembrane and intracellular; C, complete constant; CAM, calcein
AM; IFN-γ, interferon-γ; HLA, human leukocyte antigen; E:T,
effector to target cell; ns, non-significant. |
The cell-mediated cytotoxicity of human PBMCs
expressing either wtTCR or the modified TCR variants was also
compared in a 4-h CAM release assay. The Ad5F35 adenoviruses
encoding the wtTCR and chimeric TCRs were transferred into PBMCs.
PBMCs expressing the wtTCR and chimeric TCRs were co-cultured with
CAM-labeled human hepatocellular carcinoma cell lines. It was
observed that wtTCR and TCR∆IgC were able to mediate specific lysis
of the HLA-A2+ hepatocellular carcinoma cell lines, as
shown in Fig. 4B. The lymphocytes
expressing TCR∆IgC showed equivalent lysis in the
HLA-A2+ HepG2 and Huh-1 target cell lines, compared with
the wtTCR at an E:T ratio of 30:1 (11.42±0.75 and 12.03±1.10%,
respectively for HepG2 target cells; 11.10±0.92 and 12.13±1.17%,
respectively for Huh-1 target cells). These results indicated that
the lymphocytes expressing TCR∆IgC exhibited similar cytotoxic
activity to wtTCR. However, the lymphocytes expressing TCR∆cp+tm+ic
and TCR∆C exhibited lower lytic activity, compared with the wtTCR,
however, lysis was equivalent to the control PBMCs. In the
HLA-A2− target cell line, no significant lysis was
observed by any of the TCR transgene PBMCs.
Taken together, TCR∆IgC substitution of the γδTCR
IgC domain was functionally equivalent to the wtTCR, whereas
TCR∆cp+tm+ic and TCR∆C affected the recognition and cytotoxic
abilities.
Discussion
The αβ and γδTCRs are two types of antigen receptor
expressed on distinct T cell populations, γδTCR is homologous to
αβTCR in the variable and constant regions, and the αβ and γδ TCRs
are heterodimers linked by disulfide bonds (20). The γδT cells carrying Vγ9Vδ2 TCRs
are primarily found in peripheral blood, where they constitute a
minor fraction of total T cells and respond to non-peptidic
intermediates of the mevalonate pathway, termed phosphoantigens
(21). It has been reported that
Vγ9Vδ2TCR can be efficiently expressed in αβT cells without
mispairing with αβTCR, and mediates the tumor-specific
proliferation of αβT cells (17).
In the present study, three chimeric TCR variants were generated by
swapping the partial or complete constant regions of αβTCR with
those of γδTCR (Fig. 1). These
constructs were assessed for surface expression, mispairing with
endogenous TCR chains, and TCR transgene-mediated functions in
Jurkat T cells and primary human T cells. The subsequent
observations revealed for these chimeric TCR variants that the
introduction of the γδTCR IgC domain in the αβTCR improved surface
expression, reduced mispairing and did not compromise the function
of the unmodified wtTCR. The other two TCRs containing cp+tm+ic or
C domain of γδTCR showed decreased mispairing with endogenous TCR,
but impaired function in T cells.
FACS analysis in Jurkat cells showed that TCR∆IgC
exhibited improved surface expression, and the CLSM images also
showed TCR∆IgC exhibited on the cell surface of the Jurkat cells
and the non-T cells (BEL-7402). The fluorescent images of the
reporter were subjected to FRET analysis, which showed that TCR∆IgC
exhibited a higher FRET efficiency, compared with the wtTCR in
BEL-7402 cells and Jurkat cells. Detailed statistical analysis of
FRET efficiencies between BEL-7402 cells and Jurkat cells showed
that TCR∆IgC reduced mispairing, but failed to prevent mispairing
with endogenous TCR. The function of TCR∆IgC in cytotoxic
lymphocytes showed equivalent IFN-γ secretion and cytotoxic
activity as in wtTCR when targeting HLA-A2+ HepG2 and
Huh-1 cell lines, however not the HLA-A2− cell line.
These results suggested that the γδTCR IgC domain substituted for
αβTCR preserved the recognition and lytic abilities of wtTCR, and
even classic HLA restriction. The TCR αβ heterodimer has three
conserved basic residues (R, K and K) in the transmembrane regions.
These residues are considered to drive the associations between TCR
and CD3 components by forming pair-wise ionic interactions,
however, the association between this charged residue and the αβTCR
heterodimer remains to be elucidated (22). The present study hypothesized that
TCR∆IgC prevents mispairing with endogenous TCR to a certain
extent, perhaps due to harboring the γδTCR IgC domain, and the
preserved ability of the original TCR may be due to the αβTCR
transmembrane region interacting with the CD3 complex.
By contrast, the TCR∆cp+tm+ic and TCR∆C did not
increase the surface expression significantly, compared with the
wtTCR when monitoring these TCR variants in living cells. No
significant differences in FRET efficiencies were found for the
TCR∆cp+tm+ic and TCR∆C, compared with the wtTCR in BEL-7402 cells
and Jurkat cells. However, the statistical analysis of FRET
efficiencies between BEL-7402 cells and Jurkat cells showed these
two modified TCRs decreased mispairing. At present, the molecular
mechanisms determining the efficiency of TCR pairing remain to be
elucidated. Studies have shown that the variable region sequences
are important in determining the efficiency of the expression of
TCRs (23). In the present study,
which examined TCR constant region modifications (TCR∆cp+tm+ic and
TCR∆C), minimal effect was found in the their expression
efficiencies, compared with wtTCR. It was hypothesized that the
variable region sequences drive efficient αβ pairing, which can
proceed despite modifications in the constant region. Unexpectedly,
when their function was assessed in cytotoxic lymphocytes, the
present study observed that TCR∆cp+tm+ic and TCR∆C were unable to
trigger the secretion of IFN-γ, and failed to mediated cytotoxicity
in either the HLA-A2+ nor HLA-A2−
hepatocellular carcinoma cell lines. The TCRβ chain contains a
conserved transmembrane glutamic acid, which is not found in the γ
chain, and this residue may be a key determinant in the
differential CD3 composition of the αβ and γδ complexes (24). The chimeric TCRs in the present
study contained γ instead of β residues in the transmembrane
domain, which may have affected the composition of the CD3
subunits. Therefore, the present study hypothesized that the
differences in CD3 subunit composition between the αβ- and
γδTCR/CD3 complexes may have resulted in the chimeric TCRs
containing a substituted cp+tm+ic domain of γδTCR failing to
transduce a signal through the TCR complex. This may explain why
TCR∆cp+tm+ic and TCR∆C lost the functions of recognition and lysis
in primary T cells.
In conclusion, the present study showed that the
modified aβTCR, substituted for by the IgC domain of γδTCR,
improved expression and pairing on the cell surface, and did not
compromise the function of the already present wtTCR.
Acknowledgements
This study was supported by the Medical Science and
Technology Research Foundation of Guangdong Province (grant no.
A2016041) and the National Natural Science Foundation of China
(grant nos. 31300737 and 81303292) and the Natural Science
Foundation of Guangdong Province (grant no. 2015A030310310).
References
|
1
|
Sharpe M and Mount N: Genetically modified
T cells in cancer therapy: Opportunities and challenges. Dis Model
Mech. 8:337–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Casucci M, Hawkins RE, Dotti G and
Bondanza A: Overcoming the toxicity hurdles of genetically targeted
T cells. Cancer Immunol Immunother. 64:123–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cameron BJ, Gerry AB, Dukes J, Harper JV,
Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, et al:
Identification of a Titin-derived HLA-A1-presented peptide as a
cross-reactive target for engineered MAGE A3-directed T cells. Sci
Transl Med. 5:197ra1032013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson LA, Morgan RA, Dudley ME, Cassard
L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich
JR, et al: Gene therapy with human and mouse T-cell receptors
mediates cancer regression and targets normal tissues expressing
cognate antigen. Blood. 114:535–546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parkhurst MR, Yang JC, Langan RC, Dudley
ME, Nathan DA, Feldman SA, Davis JL, Morgan RA, Merino MJ, Sherry
RM, et al: T cells targeting carcinoembryonic antigen can mediate
regression of metastatic colorectal cancer but induce severe
transient colitis. Mol Ther. 19:620–626. 2011. View Article : Google Scholar
|
|
6
|
Morgan RA, Chinnasamy N, Abate-Daga D,
Gros A, Robbins PF, Zheng Z, Dudley ME, Feldman SA, Yang JC, Sherry
RM, et al: Cancer regression and neurological toxicity following
anti-MAGE-A3 TCR gene therapy. J Immunother. 36:133–151. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Linette GP, Stadtmauer EA, Maus MV,
Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM,
Cimino PJ, et al: Cardiovascular toxicity and titin
cross-reactivity of affinity-enhanced T cells in myeloma and
melanoma. Blood. 122:863–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Loenen MM, De Boer R, Amir AL,
Hagedoorn RS, Volbeda GL, Willemze R, van Rood JJ, Falkenburg JH
and Heemskerk MH: Mixed T cell receptor dimers harbor potentially
harmful neoreactivity. Proc Natl Acad Sci USA. 107:10972–10977.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bendle GM, Linnemann C, Hooijkaas AI, Bies
L, De Witte MA, Jorritsma A, Kaiser AD, Pouw N, Debets R, Kieback
E, et al: Lethal graft-versus-host disease in mouse models of T
cell receptor gene therapy. Nat Med. 16:565–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA
and Morgan RA: Enhanced antitumor activity of murine-human hybrid
T-cell receptor (TCR) in human lymphocytes is associated with
improved pairing and TCR/CD3 stability. Cancer Res. 66:8878–8886.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cohen CJ, Li YF, El-Gamil M, Robbins PF,
Rosenberg SA and Morgan RA: Enhanced antitumor activity of T cells
engineered to express T-cell receptors with a second disulfide
bond. Cancer Res. 67:3898–3903. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Voss RH, Willemsen RA, Kuball J, Grabowski
M, Engel R, Intan RS, Guillaume P, Romero P, Huber C and Theobald
M: Molecular design of the Calphabeta interface favors specific
pairing of introduced TCRalphabeta in human T cells. J Immunol.
180:391–401. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aggen DH, Chervin AS, Schmitt TM, Engels
B, Stone JD, Richman SA, Piepenbrink KH, Baker BM, Greenberg PD,
Schreiber H and Kranz DM: Single-chain VαVβ T-cell receptors
function without mispairing with endogenous TCR chains. Gene Ther.
19:365–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ochi T, Fujiwara H, Okamoto S, An J, Nagai
K, Shirakata T, Mineno J, Kuzushima K, Shiku H and Yasukawa M:
Novel adoptive T-cell immunotherapy using a WT1-specific TCR vector
encoding silencers for endogenous TCRs shows marked antileukemia
reactivity and safety. Blood. 118:1495–1503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Provasi E, Genovese P, Lombardo A, Magnani
Z, Liu PQ, Reik A, Chu V, Paschon DE, Zhang L, Kuball J, et al:
Editing T cell specificity towards leukemia by zinc finger
nucleases and lentiviral gene transfer. Nat Med. 18:807–815. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van der Veken LT, Coccoris M, Swart E,
Falkenburg JH, Schumacher TN and Heemskerk MH: Alpha beta T cell
receptor transfer to gamma delta T cells generates functional
effector cells without mixed TCR dimers in vivo. J Immunol.
182:164–170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marcu-Malina V, Heijhuurs S, van Buuren M,
Hartkamp L, Strand S, Sebestyen Z, Scholten K, Martens A and Kuball
J: Redirecting αβ T cells against cancer cells by transfer of a
broadly tumor-reactive γδT-cell receptor. Blood. 118:50–59. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tao C, Shao H, Yuan Y, Wang H, Zhang W,
Zheng W, Ma W and Huang S: Imaging of T-cell receptor fused to CD3ζ
reveals enhanced expression and improved pairing in living cells.
Int J Mol Med. 34:849–855. 2014.PubMed/NCBI
|
|
19
|
Jang YY, Cho D, Kim SK, Shin DJ, Park MH,
Lee JJ, Shin MG, Shin JH, Suh SP and Ryang DW: An improved flow
cytometry-based natural killer cytotoxicity assay involving calcein
AM staining of effector cells. Ann Clin Lab Sci. 42:42–49.
2012.PubMed/NCBI
|
|
20
|
De Libero G, Lau SY and Mori L:
Phosphoantigen presentation to TCR γδ Cells, a conundrum getting
less gray zones. Front immunol. 5:6792014.PubMed/NCBI
|
|
21
|
Scheper W, Sebestyen Z and Kuball J:
Cancer Immunotherapy using γδT cells: Dealing with diversity. Front
Immunol. 5:6012014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuhns MS, Davis MM and Garcia KC:
Deconstructing the form and function of the TCR/CD3 complex.
Immunity. 24:133–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heemskerk MH, Hagedoorn RS, van der Hoorn
MA, van der Veken LT, Hoogeboom M, Kester MG, Willemze R and
Falkenburg JH: Efficiency of T-cell receptor expression in
dual-specific T cells is controlled by the intrinsic qualities of
the TCR chains within the TCR-CD3 complex. Blood. 109:235–243.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Teixeiro E, Daniels MA, Hausmann B, Schrum
AG, Naeher D, Luescher I, Thome M, Bragado R and Palmer E: T cell
division and death are segregated by mutation of TCRbeta chain
constant domains. Immunity. 21:515–526. 2004. View Article : Google Scholar : PubMed/NCBI
|