Introduction
Thrombophilia is a blood coagulation disorder
characterized by an increased tendency to form clots, and results
in a predisposition to adverse thrombotic events, including deep
venous thromboembolism (DVT) and pulmonary embolism (PE). The risk
factors for thrombophilia are genetic and environmental (1). Previous research has indicated that
thrombophilia is the result of a deficiency in protein
differentiation or function, including protein C and S, and
mutations in ~8 human genes have been identified as causative
(2–4). However, the genetic basis of
thrombophilia requires further investigation (1). A 52-year-old Han Chinese man was
referred to the Shenzhen People's Hospital (Shenzen, China), due to
recurrent DVT, with the first hospitalization for DVT occurring in
2001. Following treatment, the patient recovered. In 2003, the
patient was readmitted due to PE and an inferior vena cava filter
was placed in order to prevent further PE in Shenzhen People's
Hospital. The patient's illness was diagnosed as thrombophilia
following laboratory measurements and computed tomography (CT)
angiography. The factors that may be involved in his disease were
analyzed. In order to investigate the possible underlying causes,
whole-exome sequencing (WES) technology was used to research the
genetic factors, and a mutation in the F2 gene was
identified. The results of mutation detection in the Tianjin
Institute of Hematology (Tianjin, China) validated this finding. No
Factor V Leiden or prothrombin G20210A mutations were detected.
The F2 gene has been assigned to the
chromosomal region 11p11-q12 and encompasses 20.3 kb (5). This gene is transcribed and
translated to produce coagulation factor II, thrombin (462 amino
acids), which is involved in the final stage of blood coagulation
in humans and controls synthesis of prothrombin (5,6).
F2 mutations lead to various forms of thrombosis and
dysprothrombinemia. However, in Asians, including Chinese
populations, F2 mutations are rare. Little was known regarding the
genetic background of DVT, however, previous research has since
revealed common genetic risk factors in DVT (7–9).
This research did not focus on Chinese populations, and the
underlying molecular mechanisms remain to be elucidated. This
evidence suggests that the study of genetic risk factors in this
Chinese family is vital for the further understanding of
thrombophilia. WES technology was used to examine two affected
individuals from the same family as the proband to study the
genetic risk factor for thrombophilia. The results of the present
study may explain the underlying molecular mechanisms of
thrombophilia in this family.
Patients and methods
Human samples
In order to investigate the association between the
familial disease and its molecular mechanisms, a Han Chinese family
with thrombophilia was recruited. The family comprised of 11 living
members (including the proband and the affected individuals II-3,
III-1) and 100 normal Han Chinese donors were also examined. The
age range (35 to 60) and gender (50 males, 50 females) of normal
donors matched the selection of patients II-2, II-3, III-1 with the
disease and their respective ages were 52, 50 and 32. The diagnosis
of thrombophilia was based on typical clinical and laboratory
measurements and ascertained by CT angiography (10). Peripheral blood was collected in
anticoagulation tubes from all study participants and genomic DNA
was extracted from leukocytes of family members and normal donors
using the phenol-chloroform protocol, following standard procedures
(11). The protocol of the present
study was approved by the Ethics Committee of The Second Clinical
Medical College, Jinan University, Shenzhen People's Hospital
(Shenzen, China) and written informed consent was obtained from all
the participants.
Whole exome sequencing
The DNA of the proband and his son (III-1) was
sequenced using the WES technique. The exons of ~26,000 protein
coding genes were sequenced in matched thrombophilia and DNA of
unaffected individuals. The coding sequences from each sample were
captured using the NimbleGen SeqCap EZ Human Exome Library v2.0 kit
(Roche Diagnostics, Indianapolis, IN, USA), and each captured
library was loaded on to a Solexa Hiseq2000 platform (Illumina,
Inc., San Diego, CA, USA) for sequence analysis. Genomic DNA
samples from the two affected individuals were fragmented with the
use of Covaris S2 system (Covaris Inc., Woburn, Massachusetts,
USA). The adapters from DNA Library Prep Reagent Set (New England
Biolabs Inc., USA) then were ligated to each end of the resulting
fragments. The captured fragments were amplified by
ligation-mediated polymerase chain reaction (LM-PCR) and hybridized
to the Nimblegen SeqCap EZ Library so that enriched, non-hybridized
fragments were washed out (12).
Capture efficiency was evaluated by quantitative PCR (qPCR) using
SYBR green setting on the real-time instrument (Roche Light Cycler
480, Indianapolis, IN, USA) (13).
The primers of LM-PCR and qPCR were obtained from Multiplex Oligos
Kit (New England Biolabs Inc., Ipswich, MA, USA) and NEBNext
Library Quant Kit (New England Biolabs Inc.), respectively. The
conditions of LM-PCR and qPCR were provided in the Tables I and II. Each captured library was then loaded
on the Illumina Genome Analyzer (Illumina, Inc.) to ensure that the
2 samples met the desired average sequencing depth of 84x.
Sequencing reads were analyzed by using the Solexa Hiseq2000
platform (Illumina, Inc.).
 | Table I.Thermocycling conditions of ligation
mediated polymerase chain reaction. |
Table I.
Thermocycling conditions of ligation
mediated polymerase chain reaction.
| Cycle step | Temperature | Time | Cycles |
|---|
| Initial
denaturation | 98°C | 30 sec | 1 |
| Denaturation | 98°C | 10 sec |
|
|
Annealing/extension | 65°C | 75 sec | 4 |
| Final extension | 65°C | 5
min | 1 |
| Hold | 4°C | ∞ |
|
 | Table II.Thermocycling conditions of
quantitative polymerase chain reaction. |
Table II.
Thermocycling conditions of
quantitative polymerase chain reaction.
| Cycle step | Temperature | Time | Cycles |
|---|
| Initial
denaturation | 95°C | 1
min | 1 |
| Denaturation | 95°C | 15 sec |
|
| Extension | 63°C | 45 sec | 35 |
Read mapping and variant analysis
WES of the participants was conducted from genomic
DNA isolated from blood and was submitted to the Huada Gene
Research Institute (Shenzen, China) for analysis. Reads were
aligned using the Burrows-Wheeler Aligner to the human reference
genome (build 37) (14).
Alignments in the combined BAM file were tested for single
nucleotide polymorphisms (SNPs) and insertions or deletions
(indels) using the SOAPsnp package and SOAP denovo respectively
(www.soap.genomics.org.cn/soapsnp.html).
Candidate SNPs were filtered and flagged using the
following modifications: i) SNP quality is >20; ii) the
sequencing depth is not <4x; iii) the distance between two SNPs
is >5 bp; and iv) the estimated copy number is <2.
Candidate gene validation
The filtering of candidate mutations requires
further validation. The reliability of WES was verified by Sanger
sequencing to confirm the mutations of candidate genes. The PCR
primers used were, forward, 5′-TGTGAACATCACCCGGTCAG-3′ and reverse,
5′-GACTGACGCCAGCTCTGAAG-3′, and were designed to validate the
candidate mutation in the gene of T165M. The results were
consistent with the candidate mutations identified by WES. The
reliability of WES results were also verified by these methods: i)
Within-family validation, the co-segregated mutations of phenotype
and genotype were observed in the genomes of each family member;
and ii) Validation in 100 normal donors, validation that the
co-segregated mutations of phenotype and genotype were not observed
in normal donors. Furthermore, the cross-species amino acid
comparison of coagulation factor II was analyzed by ClustalW2
(http://www.ebi.ac.uk/Tools/msa/clustalw2/). The amino
acid sequences of F2 were obtained from the NCBI database, and then
entered into ClustalW2. The result of ClustalW2 revealed the most
appropriate match for the selected sequences and aligned them in
such a way that the identities, similarities and differences were
easily identified and understood.
Results
Clinical characteristics of the
studied family with thrombophilia
The genetic factors that increase the risk of a
thrombotic event are well known, and include deficiencies of
protein C and protein S. Factor V Leiden and prothrombin G20210A
mutations, elevated levels of factors VIII, IX or XI, homocysteine,
and fibrinogen may also be risk factors for a thrombotic event.
Thrombophilia was confirmed by examining laboratory measurements
and CT angiography in Shenzhen People's Hospital. The levels of
factors VIII, IX and XI were measured by using enzyme-linked
immunosorbent assays, a one-stage coagulation assay and a
monoclonal antifactor XI capture antibody, respectively. Protein C
and protein S were measured by a polyclonal enzyme-linked
immunosorbent assay and the coagulation study was measured by the
STAGO automatic coagulometer. Blood coagulation factor and protein
C and S levels were normal. However, fibrinogen levels were
abnormal in the proband and other affected family members. Their
activated partial thromboplastin time and plasma prothrombin time
were observed to be prolonged. Plasma immunoreactive fibrinogen
levels were also lower (Table
III).
 | Table III.The results of a coagulation study in
a Han Chinese family with an increased risk of thrombophilia. |
Table III.
The results of a coagulation study in
a Han Chinese family with an increased risk of thrombophilia.
| Subject | APTT (sec) (ref:
28–43.5) | PT (sec) (ref:
11–15.1) | PT INR (ref:
0.75–1.25) | PT% (ref:
70–120) | TT (sec) (ref:
14.0–21.0) | TT-R ATIO (ref:
0.67–1) | D-DIC (µg/ml) (ref:
0–0.5) | ATIII% (ref:
85–135) | Fg (g/l) (ref:
2–4) |
|---|
| I-2 | 43.80 | 13.30 | 1.28 | 69 | 17.80 | 0.85 | 2.85 | 87 | 1.52 |
| II-2 | 44.30 | 12.50 | 1.30 | 68 | 17.40 | 0.83 | 3.94 | 104 | 1.64 |
| II-3 | 43.90 | 14.9 | 1.29 | 77 | 14.10 | 0.75 | 2.78 | 86 | 2.66 |
| II-6 | 32.70 | 12.80 | 1.06 | 105 | 16.30 | 0.69 | 0.11 | 96 | 2.56 |
| II-7 | 20.50 | 11.70 | 1.13 | 112 | 15.40 | 0.71 | 0.06 | 92 | 0.33 |
| III-1 | 44.50 | 13.00 | 1.31 | 70 | 18.30 | 0.82 | 2.55 | 112 | 1.77 |
| III-2 | 31.50 | 12.30 | 1.24 | 107 | 15.20 | 0.75 | 0.01 | 115 | 3.12 |
| III-4 | 32.40 | 12.70 | 1.33 | 114 | 14.90 | 0.82 | 0.01 | 121 | 3.34 |
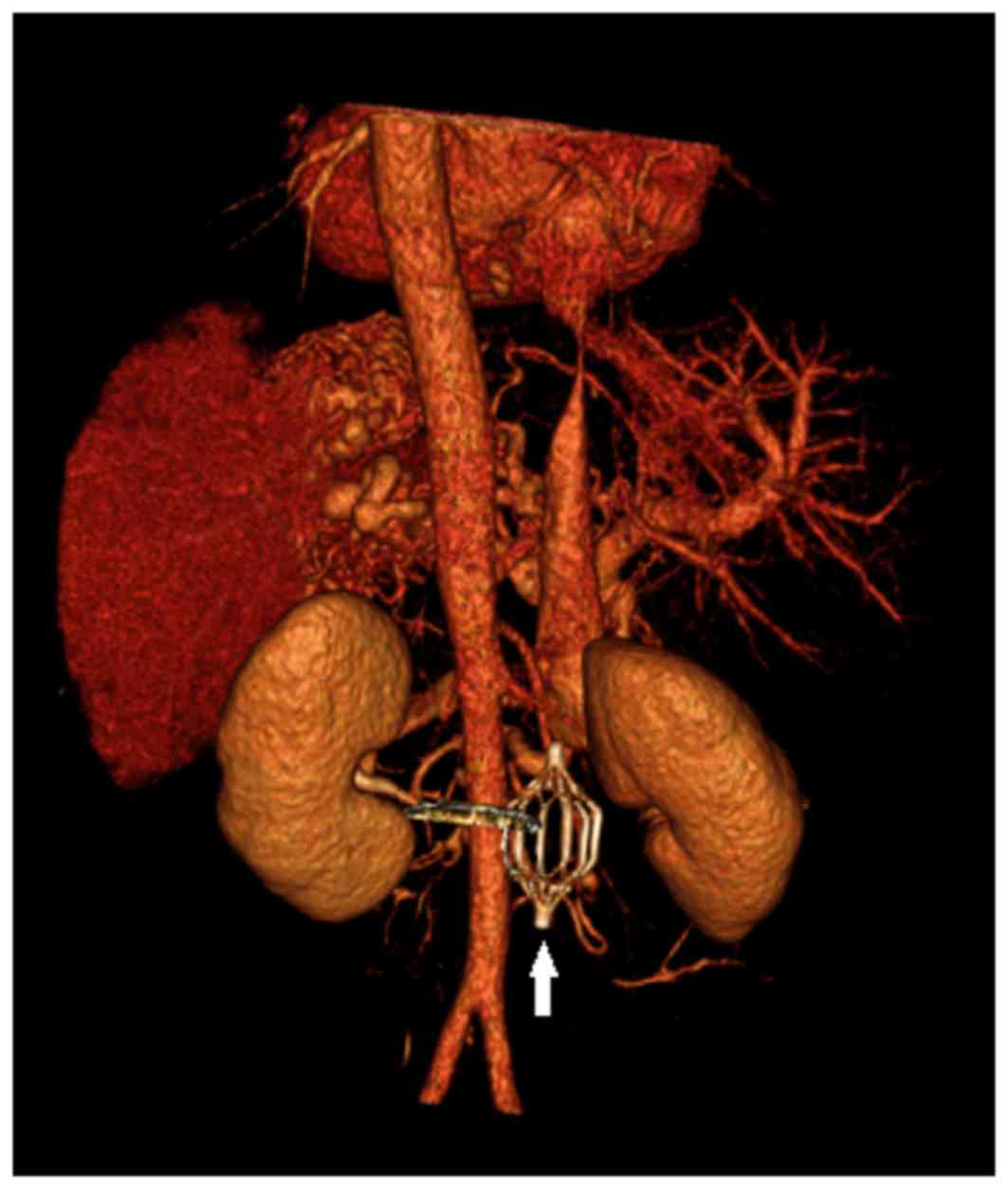
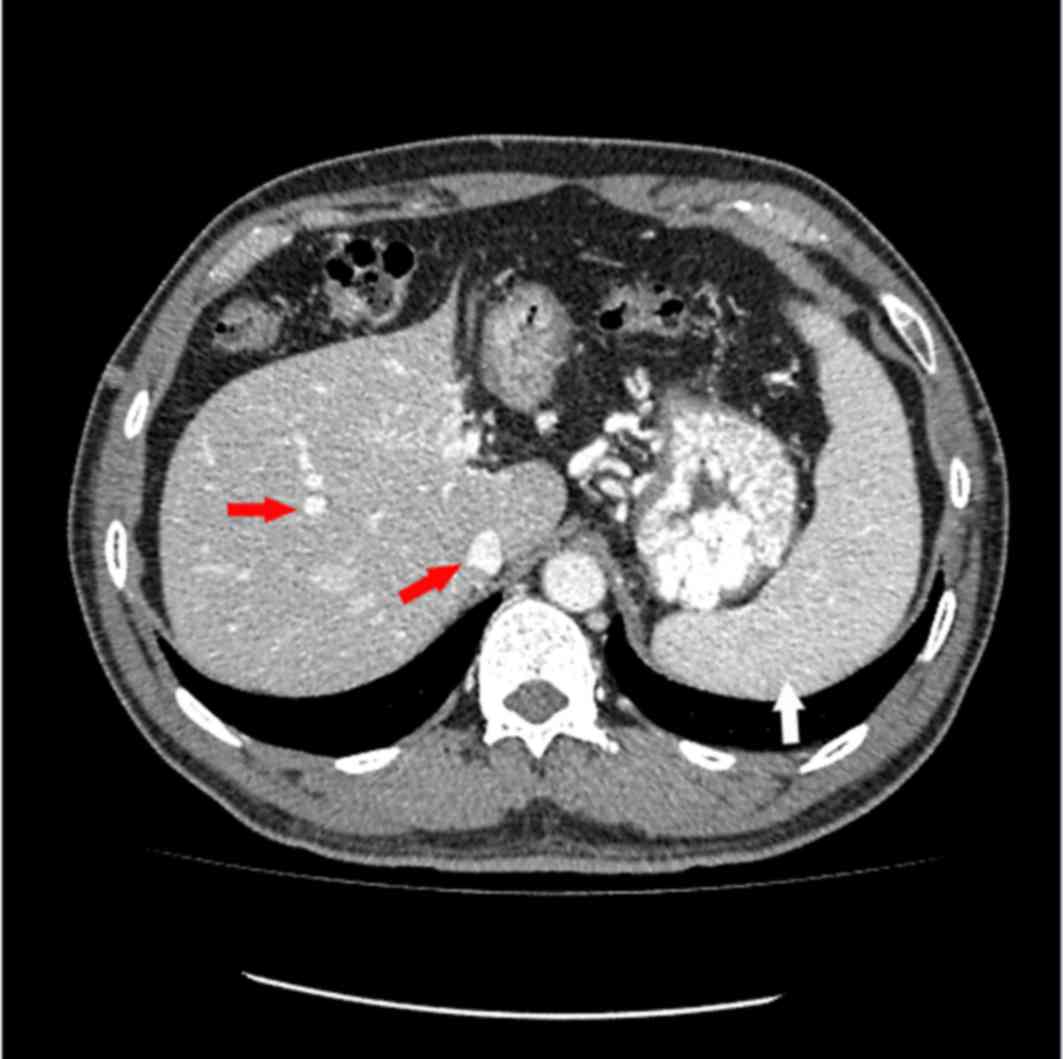
The proband underwent combined CT angiography and CT
venography of the stomach and splenomegaly and cirrhosis was
observed (Figs. 1 and 2). CT is a form of imaging that is used
as part of the daily clinical routine to detect DVT and PE.
Whole exome sequencing identified F2
as a candidate gene
The exomes of the proband and his son (II-2 and
III-1) were captured and sequenced. Following the sequencing and
filtering as described previously, candidate variants were
identified for the two affected individuals. Overall, high coverage
of the exome was attained, 93 and 99 million sequencing reads were
produced for two affected individuals, comprising 7.3 and 7.4
billion bases, respectively. In total, ~90.7% of these were aligned
to the human reference genome and 60.5% of these fell onto targeted
and enriched exons. The average sequence read depth was 84x in
targeted exons. A total of 101,037 and 97,227 genetic variations,
including nonsynonymous mutations, splice-acceptor and splice-donor
site variations, and indels were identified in the two affected
individuals in the family, respectively. Furthermore, these
variants of II-2 and III-1 were further evaluated by National
Center for Biotechnology Information (NCBI) dbSNP build 132
(http://www.ncbi.nlm.nih.gov/projects/SNP/), SIFT
(http://blocks.fhcrc.org/sift/SIFT.html.) and PolyPhen2
(http://genetics.bwh.harvard.edu/pph2/). SIFT and
PolyPhen2 programs were used for protein analysis. The applied
process of SIFT and PolyPhen2 were compliant with the previously
described protocols (15,16). Following this filtration, only one
variant (NM_000506:c.C494T:p.T165M;rs5896) was located in the
candidate region. The variant is a homozygote missense mutation
that has not previously been observed in Han Chinese population by
other researchers, but this variant was reported as a genetic risk
factor associated with thrombophilia in the Xinjiang Kazakh
population (17).
Sanger sequencing
To validate the findings of WES, Sanger sequencing
was used to analyze the candidate mutation (T165 M) in F2 in
the family. As a result, the Sanger sequencing further confirmed
the candidate variant (T165M) did not result in the possibility of
a false positive. To further study the characterization of the
variant, Sanger sequencing was used to analyze the mutation in
F2 in other family members and 100 normal donors. As a
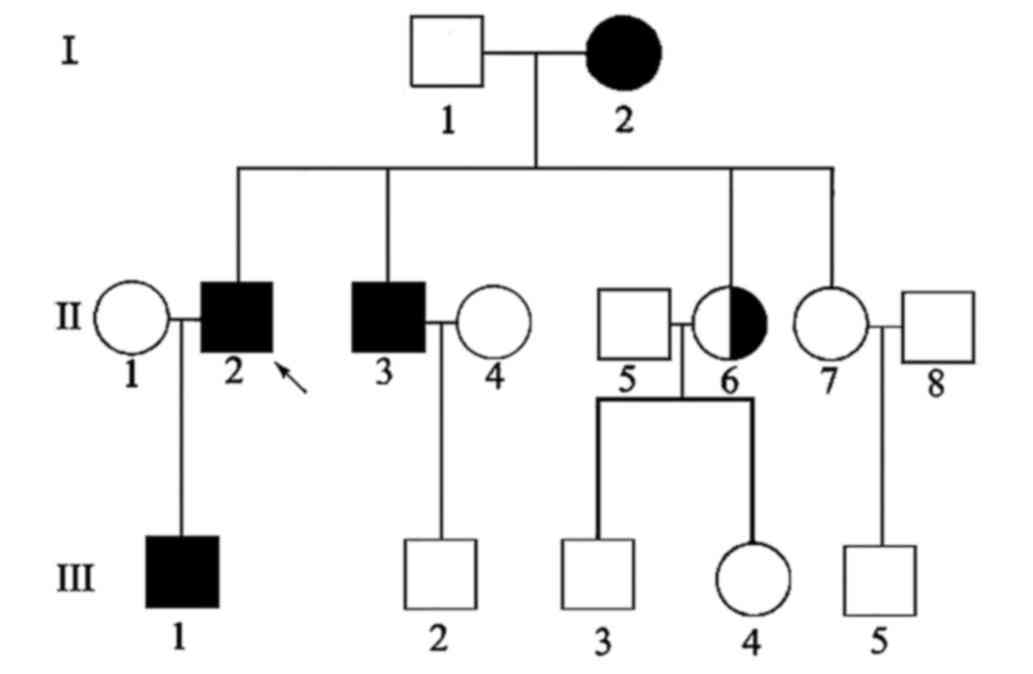
consequence, one unaffected family member (II-6) was identified as
heterozygous for the mutation and two affected family members (I-1
and II-3) had the same mutation. None of the 100 population-matched
controls had this mutation. The genogram is presented in Fig. 3.
Position and cross-species
conservation of T165M
The structure of F2 is reported to be a human
prothrombin (5). The affected
amino acid (T165M) was identified in exon 6 and resulted in a
threonine to methionine substitution at amino acid 165. Previous
research has revealed that exon 6 and its flanking regions are
highly polymorphic (18). However,
T165M was located within a highly conserved region of F2 by
means of ClustalW2 which aligned the affected amino acid sequences
and the flanking regions (residues 145–185) from human, mouse,
orangutan, rabbit, rat, bovine, sheep, horse, pig,
tropical-clawed-frog and chimpanzee genomes.
Discussion
In the present study, CT angiography and clinical
trials were used to diagnose thrombophilia within the family. CT
angiography has emerged as a useful diagnostic imaging method for
the appraisal of DVT and PE and is more effective than other
imaging modalities, particularly for the assessment of mediastinal
and parenchymal structures. It was possible to visualize that the
location of the inferior vena cava filter and splenomegaly and
cirrhosis in the proband. The laboratory measurements may indicate
the underlying factor for the greater risk for thrombophilia in
this family. Notably, factor V Leiden and prothrombin G20210A
mutations were not detected in the proband. The underlying cause
may be the resultant abnormalities of blood coagulation components,
which may affect the thrombin-mediated mechanism and the process of
coagulation and anticoagulation.
The present study investigated the association
between the possible F2 variants and the underlying
molecular mechanisms of thrombophilia using the WES technique, and
revealed that the T165M mutation results in a predisposition to
thrombophilia. The present study demonstrated that the heterozygous
genotype for this variant does not affect the risk of thrombophilia
in this Han Chinese family. However, an increased risk of
thrombophilia was observed in the homozygotic genotype for this
variant. The majority of the thrombophilia-associated gene variants
were filtered by a method on the basis of a number of attributes in
the present study. Initially, the thrombophilia-associated gene
variants were identified in the two affected individuals.
Subsequently, the remaining variants were filtered by comparing to
the NCBI dbSNP build 132, excluding variants predicted as
irrelevant by SIFT and PolyPhen2. This strategy resulted in the
identification of the candidate variant in F2. This variant
in F2 may result in a form of thrombosis. This site was
located in the exon 6 region of F2. The location and
missense of the F2 variant suggest that it may exert an
effect on the prothrombin function and destroy the vascular
integrity during development and postnatal life (from the NCBI
Reference Sequence Database). Previous research also observed the
T165M variant located in F2 gene in the Xinjiang Kazakhs
population, where it was associated with thrombophilia (17).
In 1996 a mutation in the prothrombin gene, the
F2 G20210A mutation, was identified, and this mutation was
determined to be a risk factor for thrombophilia (19). In the present study, the T165M
variant was observed in the F2 gene, which is the first
result to demonstrate this site associated with thrombophilia in
F2 gene in the Han Chinese family. The T165M variant was
observed in the Han Chinese family studied, and its presence in
other affected members besides the proband indicated that this
variant may be a risk factor for thrombophilia, and thus
thrombosis. In addition, the T165M variant was suggested to be
associated with an adverse effect on the prothrombin function and
an increased risk of thrombophilia in this Han Chinese family, and
has not been identified in previous studies.
The results of the present study may also have other
important implications. The T165M variant in the
thrombophilia-associated gene F2 was identified as a genetic
risk factor for thrombophilia in a Han Chinese family. Therefore,
sequencing of the F2 gene using the WES technique may be
useful to identify novel genetic defects for thrombophilia, and the
present study demonstrates the power of WES as a high-throughput
platform for the screening and identification of variants in a time
and cost efficient manner. Cross-species amino acid comparison also
revealed that the T165M mutation was located in a highly conserved
region of F2. As the T165M site was conserved over vast
evolutionary distances and this site has been maintained by
evolution despite speciation, it is therefore likely to be involved
in the normal function of F2.
In conclusion, the results of the present study
assisted in the elucidation of the genetic background of
thrombophilia. The F2 variant, T165M, was associated with
impaired prothrombin function and a genetic risk of thrombophilia
in a Han Chinese family. Further studies involving large sample
groups remain to be conducted regarding this variant and any
possible correlations it may exhibit.
Acknowledgements
The present study was supported by the Guangxi
Natural Science Foundation (grant no. 2015GXNSFBA139176).
References
|
1
|
Raffini L and Thornburg C: Testing
children for inherited thrombophilia: More questions than answers.
Br J Haematol. 147:277–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kvasnička T, Hájková J, Bobčíková P,
Cverhová V, Malíková I, Ulrych J, Bříza J, Dušková D, Poletínová S,
Kieferová V and Kvasnička J: The frequencies of six important
thrombophilic mutations in a population of the Czech Republic.
Physiol Res. 63:245–253. 2014.PubMed/NCBI
|
|
3
|
Coriu L, Ungureanu R, Talmaci R, Uscatescu
V, Cirstoiu M, Coriu D and Copaciu E: Hereditary Thrombophilia and
thrombotic events in pregnancy: Single-center experience. J Med
Life. 7:567–571. 2014.PubMed/NCBI
|
|
4
|
Spiezia L, Campello E, Bon M, Tison T,
Milan M, Simioni P and Prandoni P: ABO blood groups and the risk of
venous thrombosis in patients with inherited thrombophilia. Blood
Transfus. 11:250–253. 2013.PubMed/NCBI
|
|
5
|
Royle NJ, Irwin DM, Koschinsky ML,
MacGillivray RT and Hamerton JL: Human genes encoding prothrombin
and ceruloplasmin map to 11p11-q12 and 3q21-24, respectively. Somat
Cell Mol Genet. 13:285–292. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walldorf U, Hovemann B and Bautz EK: F1
and F2: Two similar genes regulated differently during development
of Drosophila melanogaster. Proc Natl Acad Sci USA. 82:5795–5799.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Allawi NA, Badi AI, Goran MA, Nerweyi
FF, Ballo HM and Al-Mzury NT: The contributions of thrombophilic
mutations to genetic susceptibility to deep venous thrombosis in
iraqi patients. Genet Test Mol Biomarkers. 19:500–504. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simsek E, Yesilyurt A, Pinarli F, Eyerci N
and Ulus AT: Combined genetic mutations have remarkable effect on
deep venous thrombosis and/or pulmonary embolism occurence. Gene.
536:171–176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cosmi B, Legnani C, Pengo V, Ghirarduzzi
A, Testa S, Poli D, Prisco D, Tripodi A and Palareti G: PROLONG
Investigators (on behalf of FCSA Italian Federation of
Anticoagulation Clinics): The influence of factor V Leiden and
G20210A prothrombin mutation on the presence of residual vein
obstruction after idiopathic deep-vein thrombosis of the lower
limbs. Thromb Haemost. 109:510–516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Christiansen SC, Cannegieter SC, Koster T,
Vandenbroucke JP and Rosendaal FR: Thrombophilia, clinical factors,
and recurrent venous thrombotic events. JAMA. 293:2352–2361. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Dai Y, Liu Y and Ren J:
Mandibulofacial dysostosis, microtia, and limb anomalies in a
newborn: A new form of acrofacial dysostosis syndrome? Clin Genet.
78:570–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen R, Im H and Snyder M: Whole-exome
enrichment with the roche nimbleGen SeqCap EZ exome library SR
platform. Cold Spring Harb Protoc. 2015:634–641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clark MJ, Chen R, Lam HY, Karczewski KJ,
Chen R, Euskirchen G, Butte AJ and Snyder M: Performance comparison
of exome DNA sequencing technologies. Nat Biotechnol. 29:908–914.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ng PC and Henikoff S: SIFT: Predicting
amino acid changes that affect protein function. Nucleic Acids Res.
31:3812–3814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
polyPhen-2. Curr Protoc Hum Genet Chapter. 7:Unit7.202013.
|
|
17
|
Ge XH, Zhu F, Wang BL, Wang CM, Zhu B,
Guan S, Ci HB, Sai LM, Jiang XK and Ren H: Association between
prothrombin gene polymorphisms and hereditary thrombophilia in
Xinjiang Kazakhs population. Blood Coagul Fibrinolysis. 25:114–118.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iwahana H, Yoshimoto K and Itakura M:
Highly polymorphic region of the human prothrombin (F2) gene. Hum
Genet. 89:123–124. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poort SR, Rosendaal FR, Reitsma PH and
Bertina RM: A common genetic variation in the 3′-untranslated
region of the prothrombin gene is associated with elevated plasma
prothrombin levels and an increase in venous thrombosis. Blood.
88:3698–3703. 1996.PubMed/NCBI
|