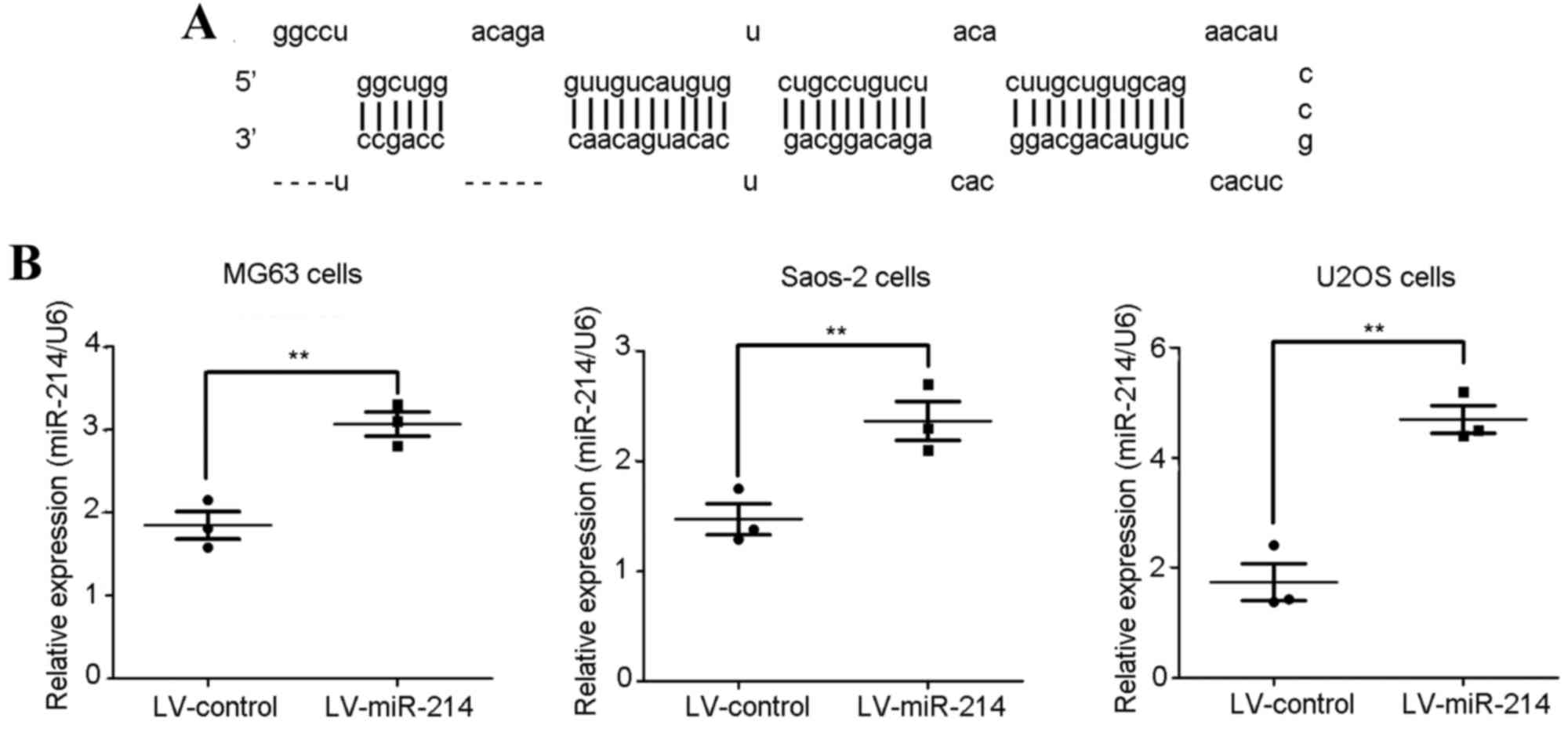
Introduction
Osteosarcoma is the most common primary malignant
bone cancer, which occurs most frequently in adolescents and its
peak incidence is 15–19 years (1,2).
Osteosarcoma usually occurs in the long bones of limbs,
particularly in the distal femur and proximal tibia. Osteosarcoma
is a locally destructive tumor and it usually has a high tendency
for systemic metastasis (3). It is
reported that ~20% of patients have lung metastases at the time of
initial diagnosis and 40% of patients have advanced distant organ
metastases (4,5). Although substantial basic
investigations and clinical trials have been performed to elucidate
the molecular mechanisms underlying the process of osteosarcoma
carcinogenesis, the precise and clear mechanisms remain to be fully
elucidated, which leads to the poor prognosis and therapeutics of
osteosarcoma. The 5-year survival rate of patients with
osteosarcoma remains at 60–70% (6,7).
Therefore, it is important to determine the molecular mechanisms
underlying osteosarcoma carcinogenesis.
MicroRNAs (miRNAs or miRs) constitute a recently
discovered class of small, non-coding RNAs molecules, which are
able to post-transcriptionally regulate gene expression (8,9). In
a variety of organisms, they are widespread and show a high level
of conservation throughout evolution (10,11).
Several studies have reported that miRNAs are usually abnormally
expressed in a number of cancer cells, and they are often important
in the progression of cancer by regulating tumor suppressor genes
and oncogenes (12,13). In previous years, miR-214 has been
located inside the sequence of the long noncoding Dmn3os transcript
and acts as an important regulator in carcinogenesis, although it
may operate in a context-dependent manner and can have
contradictory effects during tumor progression (14). Yang et al (15) reported that miR-214 regulates
gastric cancer cell proliferation, migration and invasion by
targeting phosphatase and tensin homolog. Zhang et al
(16) found that hemolysis-free
plasma miR-214 works as a novel biomarker of gastric cancer and is
correlated with distant metastasis. These studies demonstrated that
miR-214 may be a novel potential therapeutic agent for human
gastric cancer. In addition, decreased expression of miR-214 has
been found to contribute to liver metastasis in patients with
colorectal cancer, which may be involved in determining the
metastatic niche (17). Xu et
al (18) reported that miR-214
is critical in ovarian cancer stem cells via regulation of the
p53-Nanog axis. It can also be used as a therapeutic target for
ovarian cancer. However, Kalniete reported that patients with
triple-negative breast cancer exhibiting a high level of miR-214
showed significantly poorer disease-specific survival rates,
compared with patients with a low level, suggesting that miR-214
may be used as a potential prognostic biomarker for patients with
triple-negative breast cancer.
Until now, the role and cellular effects of miR-214
in osteosarcoma cancer have not been fully clarified. In the
present study, the expression levels of miR-214 were examined in
clinical specimens from patients with osteosarcoma and in
osteosarcoma cell lines. The molecular mechanism underlying the
involvement of miR-214 in osteosarcoma was also examined. The data
obtained in the present study may provide novel clues for improving
the clinical therapy of osteosarcoma.
Materials and methods
Patients
A total of 36 patients (20 male and 16 female) were
recruited and normal human skeletal muscle cells (HskMCs) were
isolated from the skeletal muscle of limbs from adult or child
donors from the Second Affiliated Hospital of Bengbu Medical
College (Bengbu, China). The median age of the patients was 16.87
years (range 6–21 years). The specimens were collected between
April 2013 and March 2015. All patients were diagnosed with
osteosarcoma and underwent surgery. All tissue samples were
analyzed and confirmed by postoperative histopathological
examination of the specimens. The osteosarcoma tissues were
collected and frozen at −80°C until they were analyzed using
western blot analysis. The present study was performed in
compliance with the Declaration of Helsinki and approved by the
ethics committee of the Second Affiliated Hospital of Bengbu
Medical College. All patients were well informed and provided
signed consent prior to the experiments.
Cell lines
The MG63, Saos-2 and U2OS human osteosarcoma cell
lines were obtained from the American Type Culture Collection
(Manassas, VA, USA). The HSkMCs were purchased from Tongpai
Biological Technology Co., Ltd. (Shanghai, China). Cells were
maintained at 37°C in a 5% CO2 atmosphere.
Reagents
Dulbecco's modified Eagle's medium and RPMI-1640
medium were purchased from Hyclone; GE Healthcare Life Sciences
(Logan, UT, USA). Fetal bovine serum (FBS) and
penicillin-streptomycin solution were obtained from Gibco; Thermo
Fisher Scientific, Inc. (Waltham, MA, USA). The transfection
reagents used in the present study was Lipofectamine®
2000 (cat. no. 11668-019), which was purchased from Invitrogen;
Thermo Fisher Scientific, Inc. The total RNA extraction kit (cat.
no. MK700) was obtained from Takara Biotechnology Co., Ltd.
(Dalian, China). The cDNA reverse transcription kit was also
obtained from Takara Biotechnology Co., Ltd. (cat. no. 6110). The
recombinant β-catenin (cat. no. 11279-H20B-50) was purchased from
Sino Biological Inc. (Beijing, China). The specific
antisense-microRNA oligonucleotides (AMOs) were designed and
synthesized by GenePharma Corporation (Shanghai, China).
Cell transfection
Cells (0.5×105 cells per well) were
plated in a 24-well plate and cultured for 24 h. A mixture of
Opti-MEM reduced serum medium (cat. no. 31985-070; Invitrogen;
Thermo Fisher Scientific, Inc.) with 8 µg/ml polybrene was then
prepared. Has-mir-214 lentivirus (cat. no. mir-LV171) and
mir-control lentivirus (cat. no. mir-LV000) were obtained from
Biosettia, Inc. (San Diego, CA, USA). The cells were transfected
using 1 µl lentivirus and were incubated at 37°C overnight with 5%
CO2.
Western blot analysis
The cells stably transfected with the miR-214
lentivirus or control lentivirus were cultured for 6 h. The cells
were then washed with ice-cold PBS and centrifuged at 850 × g for 5
min at room temperature. Cell lysates were then prepared for
western blot analysis, using RIPA lysis buffer (cat. no. P0013C;
Beyotime Institute of Biotechnology, Haimen, China). The membranes
were blocked with 5% FBS in TBST buffer (0.1% Tween-20), and the
amount of protein per lane was 25 µg. The antibodies used were as
follows: Rabbit polyclonal anti-cyclin D1 (cat. no. PA5-32373;
1:1,000) was purchased from Thermo Fisher Scientific, Inc.; rabbit
polyclonal anti-MYC/c-Myc antibody (calculated MW 49 kDa; cat. no.
ALS15021; 1:1,000) and rabbit anti-lymphoid enhancer-binding
factor-1 (LEF-1; 1:1,000; cat. no. AP1048A) polyclonal antibodies
were purchased from Abgent Biotech Co., Ltd. (Suzhou, China);
rabbit polyclonal anti-Wnt1 (C-terminal; cat. no. AP6785b; 1:1,000)
was immunized with a KLH conjugated synthetic peptide 267–296 amino
acids from the C-terminal region of human Wnt1; rabbit polyclonal
anti-Wnt2 (C-terminal region; cat. no. AI10314; 1:1,000); rabbit
polyclonal anti-Wnt4 (center; cat. no. AP6683C; 1:1,000), generated
from rabbits immunized with a KLH conjugated synthetic peptide
211–239 amino acids from the central region of human Wnt 4. The Wnt
1, Wnt 2 and Wnt 4 antibodies were purchased from Abgent Biotech
Co., Ltd. The secondary antibodies were goat anti-rabbit IgG-HRP
(cat. no. sc-2004; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) and goat anti-mouse IgG-HRP (cat. no. sc-2005; 1:1,000;
Santa Cruz Biotechnology, Inc.). Rabbit polyclonal anti-axin (cat.
no. ab1457; 1:1,000; Abcam, Cambridge, UK); rabbit polyclonal
GSK-3β (H-76; cat. no. sc-9166; 1:1,000; Santa Cruz Biotechnology,
Inc.), rabbit polyclonal anti-β-catenin (cat. no. H-102; 1:1,000;
Santa Cruz Biotechnology, Inc.; 200 µg/ml). The primary antibodies
were incubated overnight at 4°C. Anti-β-actin and the
HRP-conjugated goat anti-mouse secondary antibodies were purchased
from Santa Cruz Biotechnology, Inc and incubated for 40 min at room
temperature. The bangs were visualized using a gel imaging system
(BioRad GelDoc XR; Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and the Quantity One 1-D analysis software (version 4.6.9) that
came with the system.
Cell growth curve
The MG63 and U2OS were infected with the hsa-mir-214
lentivirus or control lentivirus for 48 h, and stable cell lines
were screened and selected. Cells in the logarithmic growth phase
were trypsinized and adjusted at a concentration of
4×104 cells/ml. The cells were then plated in a 96
well-plate in three replicates. The cells were incubated at 37°C in
a 5% CO2 in air atmosphere. Trypan blue staining was
used for detect the ratio of living cells. The cells were digested
and counted every 24 h, with four fields of view assessed under a
light microscope. After 5 days, a cell growth curve was prepared.
The culture duration was used as the horizontal axis and the number
of cells was used as the vertical axis.
FACS
The cells (1×106) were collected by
centrifugation at 850 × g for 10 min at 4°C. The supernatant was
discarded and the pellet was resuspended in 1 ml of PBS at room
temperature. Subsequently, the cells were fixed in cold 70% ethanol
for 30 min at 4°C. The cells were washed in PBS and centrifuged at
850 × g for 10 min at 4°C to avoid cell loss. Subsequently, the
cells were treated with ribonuclease by adding 50 µl of a 100 µg/ml
stock of RNase. Finally, 200 µl PI (from a 50 µg/ml stock solution)
was added for assessment using FACS.
3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
assay
An MTT assay was performed to determine cell
viability. Briefly, the stable cell lines were plated into a
96-well plate at a density of 5×103 cells/well. After 6
h, the cells were treated with miR-214-specific AMOs for 24, 48 and
72 h at 37°C. At 4 h prior to assessment, 10 µl of MTT (5 mg/ml)
was added to the medium. The cells were incubated at 37°C in 5%
CO2. The absorbance of each well was read at 490 nm
using a microplate reader.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA samples were extracted with TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) Reverse transcription
was carried out using the SuperScript First-Strand Synthesis System
for RT-PCR (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. SYBR-green was purchased from
Thermo Fisher Scientific, Inc. (cat. no. 4364346). The primers used
for mir-214 were as follows: reverse transcription,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTGCC-3′; PCR
forward, 5′-TGCGGGTGCTCCGCTTCGGCAGC-3′ and reverse
5′-CAGTGCAGGGTCCGAGGT-3′. The thermocycler conditions used were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 5 sec,
annealing at 60°C for 34 sec and detection at 74°C for 3 sec. Three
replicates were performed for each group. The ΔΔCq method was used
for quantification (19).
Statistical analysis
The data were analyzed using SPSS 20.0 software (IBM
SPSS, Armonk, NY, USA). A Student's t-test was performed to analyze
data. P<0.05 was considered to indicate a statistically
different difference. Images of the bands were captured and
analyzed by using Image J software (National Institutes of Health,
Bethesda, MD, USA).
Results
Higher expression levels of miR-214
are observed in patients with osteosarcoma and osteosarcoma cell
lines
In the present study, tissues were collected from a
total of 36 patients at the Second Affiliated Hospital of Bengbu
Medical College, with the median age of 16.87 years (range 6–21
years). In order to determine the role of miR-214 in the
progression of osteosarcoma, the expression levels of miR-214 were
detected in the osteosarcoma specimens and the paired adjacent
normal tissues. The total RNA was extracted from the specimens of
osteosarcoma and paired adjacent normal tissues. As shown in
Fig. 1A, the RNA purity and
integrity were detected using formaldehyde denaturing agarose gel
electrophoresis. Subsequently, the expression levels of miR-214
were detected in the osteosarcoma and paired peritumoral tissues.
The results showed that increased expression levels of miR-214 were
observed in the osteosarcoma tissues (P<0.01), compared with the
peritumoral tissues (Fig. 1B). The
levels of miR-214 were also assessed in various osteosarcoma tumor
cell lines, including the MG63, Saos-2 and U2OS cell lines, and
were compared with the levels in the nontumor HSkMC cell. As shown
in Fig. 1C, the miR-214 levels
were significantly upregulated in the osteosarcoma cancer cells.
The HSkMCs were used as normal controls, which were isolated from
the skeletal muscle of limbs from adult or fetal donors. These data
showed that the expression levels of miR-214 were significantly
increased in the patients with osteosarcoma and the osteosarcoma
cell lines.
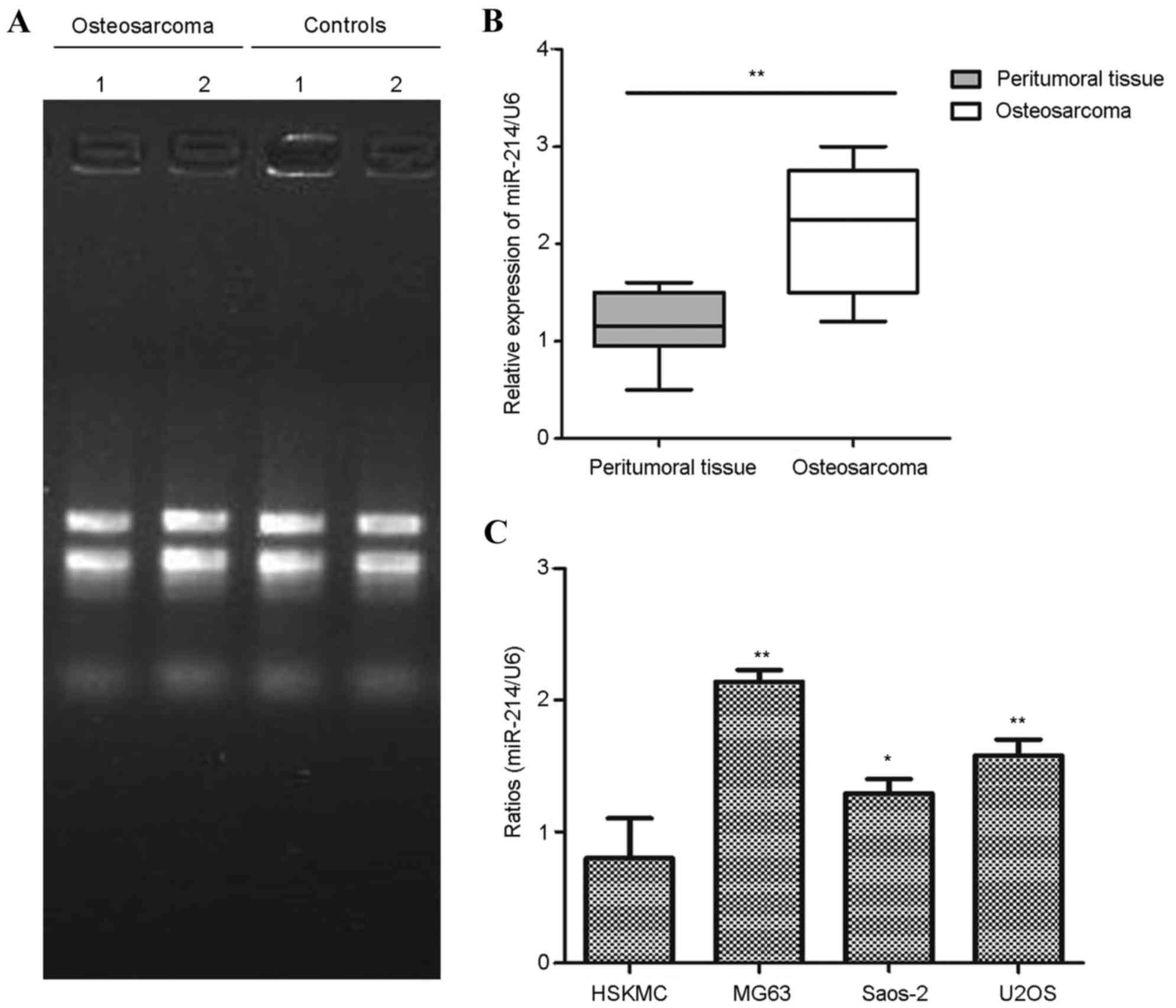 | Figure 1.Expression of miR-214 is higher in
patients with osteosarcoma and osteosarcoma cell lines. (A)
Specimens from osteosarcoma tissues and paired peritumoral tissues
were collected, and two of the independent samples are shown. Total
RNA was extracted from the tissues, and the RNA purity and
integrity were determined using formaldehyde denaturing agarose gel
electrophoresis. Visible bands are shown at 28S and 18S, and a
faint band is shown at 5S. (B) Relative expression of miR-214 was
detected in osteosarcoma tissues and paired peritumoral tissues
using RT-qPCR analysis. **P<0.01, compared with peritumoral
tissues. (C) Analysis of expression levels of miR-214 in
osteosarcoma cell lines (MG63, Saos-2 and U2OS), compared with
normal HSkMCs using RT-qPCR analysis. *P<0.05; **P<0.01,
compared with the HSkMCs. All experiments were performed in
duplicate with three technical replicates. miR, microRNA; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
HSkMCs, human skeletal muscle cells. |
Overexpression of miR-214 promotes
osteosarcoma cell proliferation
In order to assess the role of miR-214 (Fig. 2A) in regulating cell proliferation,
the hsa-mir-214 lentivirus was used to infect the osteosarcoma
cancer cells, and cell proliferation was determined using an MTT
assay. The MG63 cells, Saos-2 cells and U2OS cells were infected
with hsa-mir-214 lentivirus for 48 h, as shown in Fig. 2B, and the levels of miR-214 were
significantly upregulated in the osteosarcoma cancer cells.
The MTT assay was used to detect the cell
proliferation of osteosarcoma cells. The MG-63 cells and Saos-2
cells were infected with the hsa-mir-214 lentivirus and cultured
for 1, 2, 3, 4 and 5 days, respectively. As shown in Fig. 3A, the cells infected with the
has-miR-214 lentivirus grew at a significantly higher rate,
compared with those infected with the negative control lentivirus
(*P<0.05; **P<0.01).
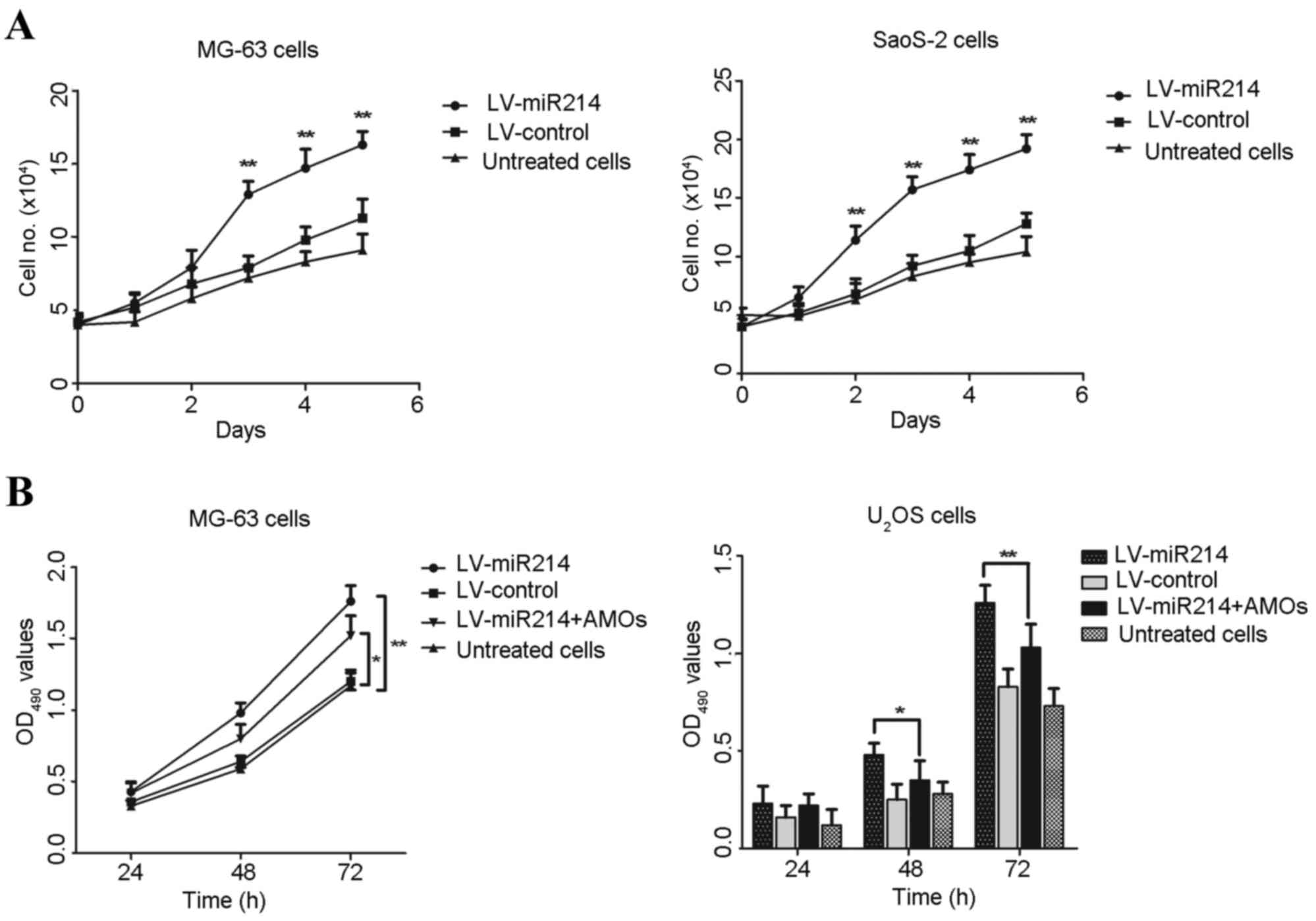 | Figure 3.Overexpression of miR-214 inhibits
osteosarcoma cell proliferation. Cell lines were infected with
miR-214 lentivirus or control lentivirus, and stable cell lines in
the logarithmic growth phase were collected. The concentration of
the cell suspension was adjusted into 4×104/ml. (A)
Stable cells were plated into 96-well plate and cultured for 5
days, with cells digested and counted every 24 h. Each group
contained three replicates. (B) Stable cell lines were treated with
specific AMOs for 24, 48 and 72 h, respectively. The proliferation
of MG63 cells and U2OS cells were determined using a
3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
assay. *P<0.05 and **P<0.01, compared with the AMO-treated
group. miR, microRNA; AMOs, antisense-microRNA oligonucleotides;
OD, optical density. |
Subsequently, specific AMOs were used to reverse the
effect of the hsa-mir-214 lentivirus. The miR-214
lentivirus-infected cells grew at a faster rate, compared with the
control lentivirus-infected cells. However, following treatment of
the miR-214-infected osteosarcoma cells with miR-214-specific AMOs
for 48 and 72 h, respectively, cell proliferation was significantly
decreased, compared with that in the miR-214-infected osteosarcoma
cells (Fig. 3B). These data showed
that higher levels of miR214 promoted the proliferation of
osteosarcoma cancer cells, and miR-214 specific-AMOs reversed its
effect on the osteosarcoma cells.
miR-214-specific AMOs regulate cell
cycle progression and induce cell cycle arrest at the G0/G1
phase
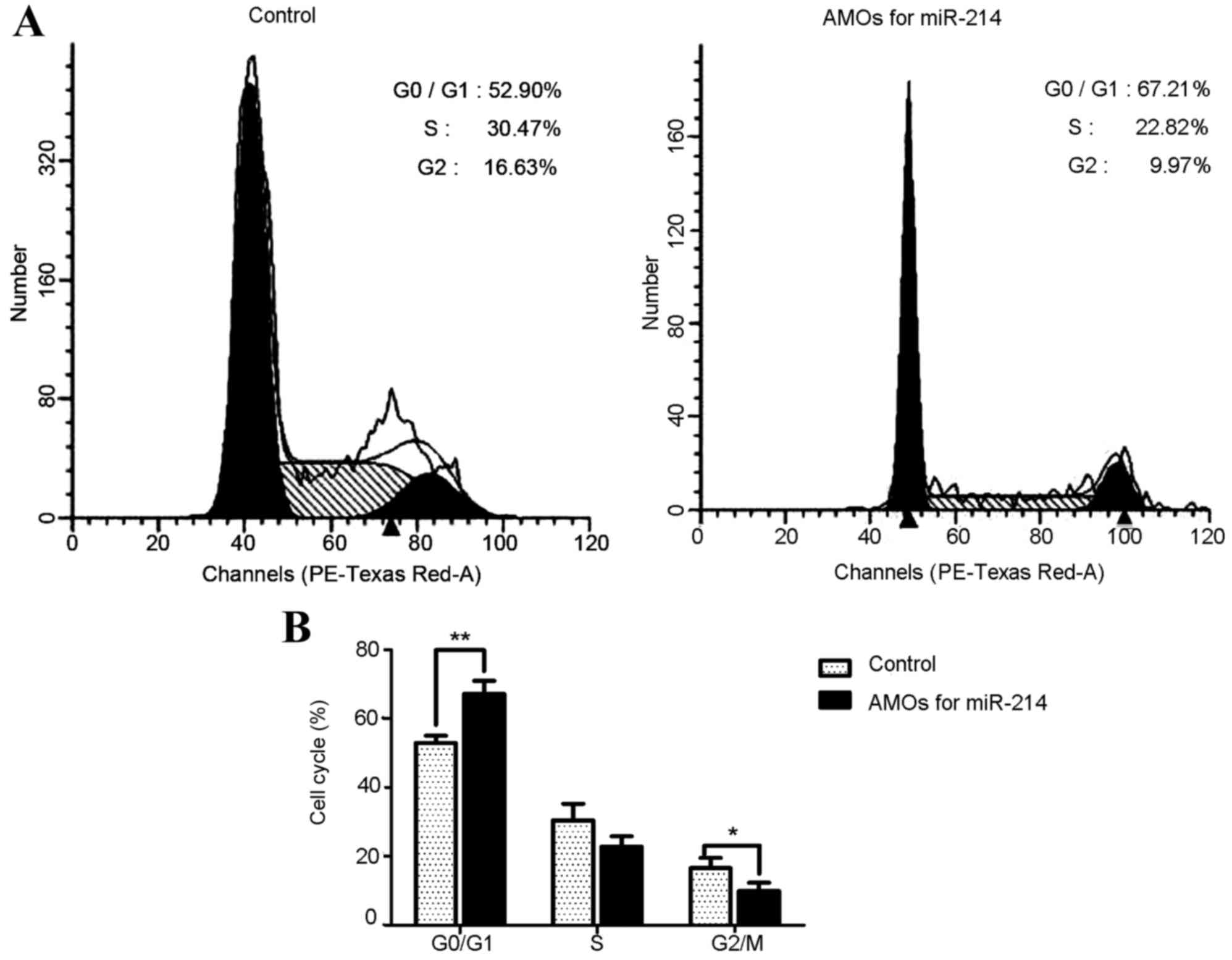
To further examine the effect of miR-214 on cell
cycle progression, a FACS assay was used to detect cell
distribution in the miR-214 lentivirus-infected cancer cells and
control lentivirus-infected cells. As shown in Fig. 4A and B, MG63 cells infected with
miR-214 lentivirus were arrested in the G0/G1 phase; the percentage
of cells in the G0/G1 phase was significantly increased from 52.90
to 67.21% (P<0.01), compared with the cells infected with
control lentivirus. These data suggested that miR-214 was important
in regulating the G1/S transition.
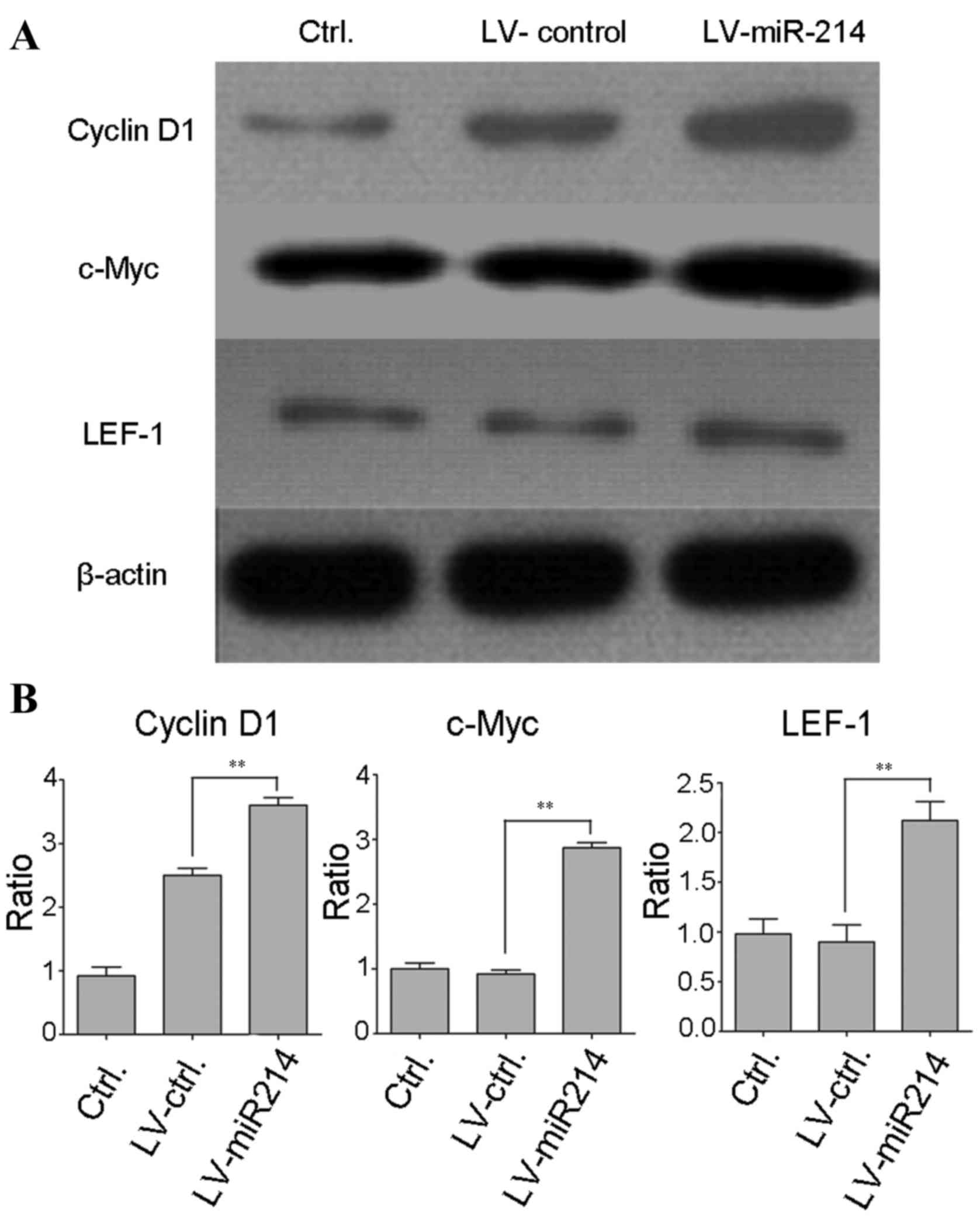
Overexpression of miR-214 upregulates
the Wnt/β-catenin signaling pathway
Studies have shown that aberrant activation of the
Wnt/β-catenin pathway is common in the progression of osteosarcoma.
In order to determine whether miR-214 affects the activation of the
Wnt/β-catenin signaling pathway, western blot analysis was
performed to detect the protein expression of downstream target
genes of the Wnt/β-catenin pathway. As shown in Fig. 5A and B, in the miR-214
lentivirus-infected cells, the levels of cyclin D1, c-myc and LEF-1
were significantly increased, compared with those in the control
lentivirus-infected cancer cells (LEF, P<0.05; cyclin D1,
P<0.01; c-myc, P<0.01). These results revealed that miR-214
may be involved in regulating the Wnt/β-catenin pathway.
LV-miR-214 infection upregulates the
protein level of β-catenin
It is known that the expression of miR-214 regulates
the Wnt/β-catenin signaling pathway. The present study aimed to
further examine the further mechanism underlying the effect of
miR-214, western blot analysis was performed to detect the levels
of relative proteins in the Wnt/β-catenin signaling pathway. As
shown in Fig. 6A, no significant
variations were found in the levels of Wnt1, Wnt2, Wnt4, Axin and
Gsk-3β in the miR-214 lentivirus-infected U2OS cells, compared with
those in the control lentivirus-infected U2OS cancer cells.
Variation in the levels of β-catenin in the MG63 cells and Saos-2
cells were also examined. Consistent with the data in Fig. 6A, the levels of β-catenin were
increased in the LV-miR-214-infected MG63 cells and Saos-2 cells
(Fig. 6B). However, the mRNA
levels of β-catenin were not altered in the LV-miR-214-infected
MG63 or Saos-2 cells (Fig. 6C),
and there were no statistical differences between the LV-control
and LV-miR-214-infected cells. These data suggested that β-catenin
may be a target of miR-214 in osteosarcoma cells.
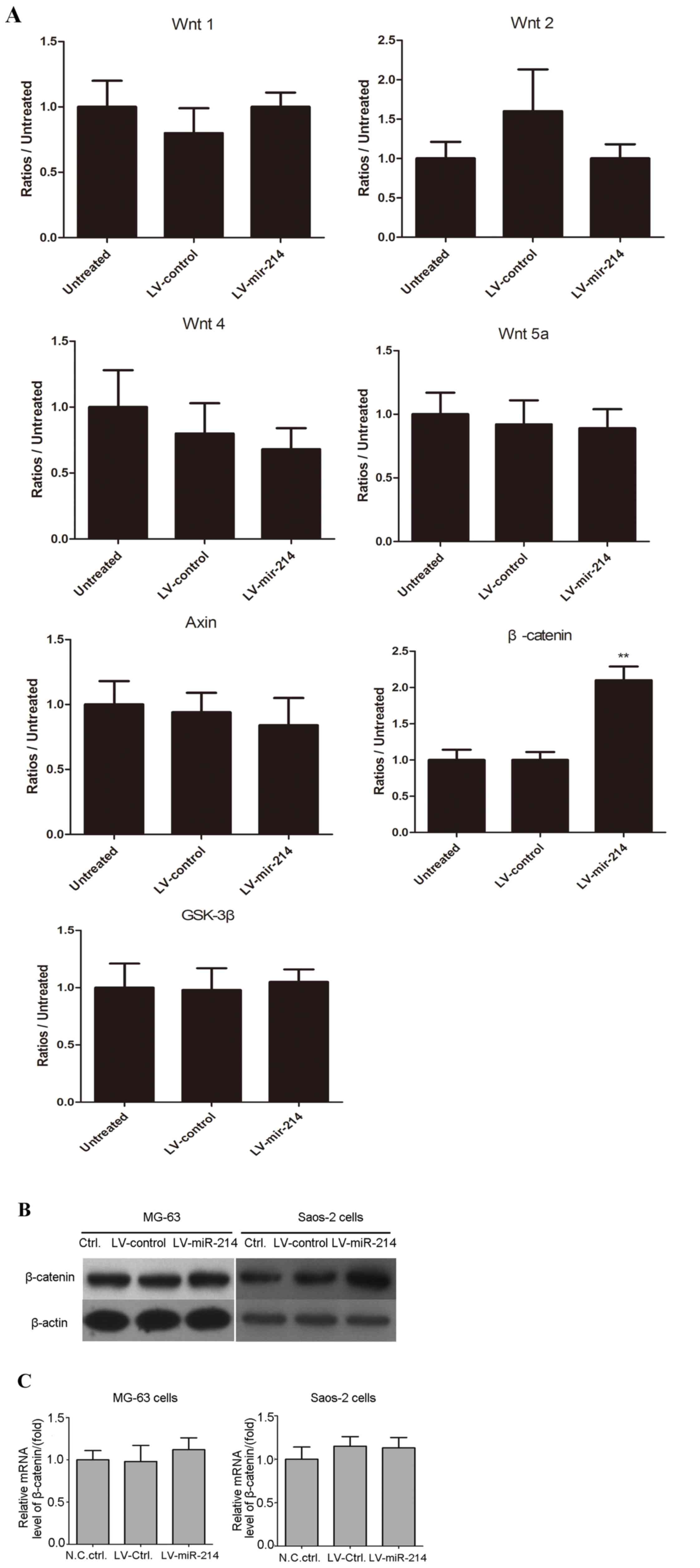 | Figure 6.LV-miR-214 infection downregulates the
protein level of β-catenin. (A) Stable cells (LV-miR-214-U2OS and
LV-control-U2OS; 3×105 cells/per well) were plated into
a 24-well plate, cultured for 24 h and cell lysate was prepared
using 1X SDS lysis buffer. The levels of Wnt1, Wnt2, Wnt4, Wnt5a,
Axin, β-catenin and Gsk-3β were detected using western blot
analysis. **P<0.01, compared with untreated cells. (B) Levels of
β-catenin were detected in MG-63 cells and Saos-2 cells. β-actin
was used as internal reference. (C) Histogram of relative mRNA
levels of β-catenin in MG-132 and Saos-2 cells infected with the
miR-214 lentivirus and control lentivirus. Expression of β-catenin
was detected using reverse transcription-quantitative polymerase
chain reaction analysis. β-actin was used as internal reference. No
statistical differences were found between groups. miR, microRNA;
Gsk-3β, glycogen synthase kinase β; Ctrl, control; N.C, negative
control. |
Exogenous addition of β-catenin
effectively reverses the efficiency of miR-214-specific AMOs
In order to assess the specificity of β-catenin in
the LV-miR-214-infected MG63 cells, a rescue experiment was
performed by adding β-catenin protein to serum-starved MG63 cells.
As shown in Fig. 7, the
LV-miR-214-infected MG63 cells were treated with 100 ng/ml
β-catenin for 24, 48 and 72 h, respectively. The results showed
that treatment with exogenously added β-catenin significantly
reversed the efficiency of miR-214-specific AMOs. This reverse
experiment can be used to assess the specificity of β-catenin in
miR-214 lentivirus-infected cells.
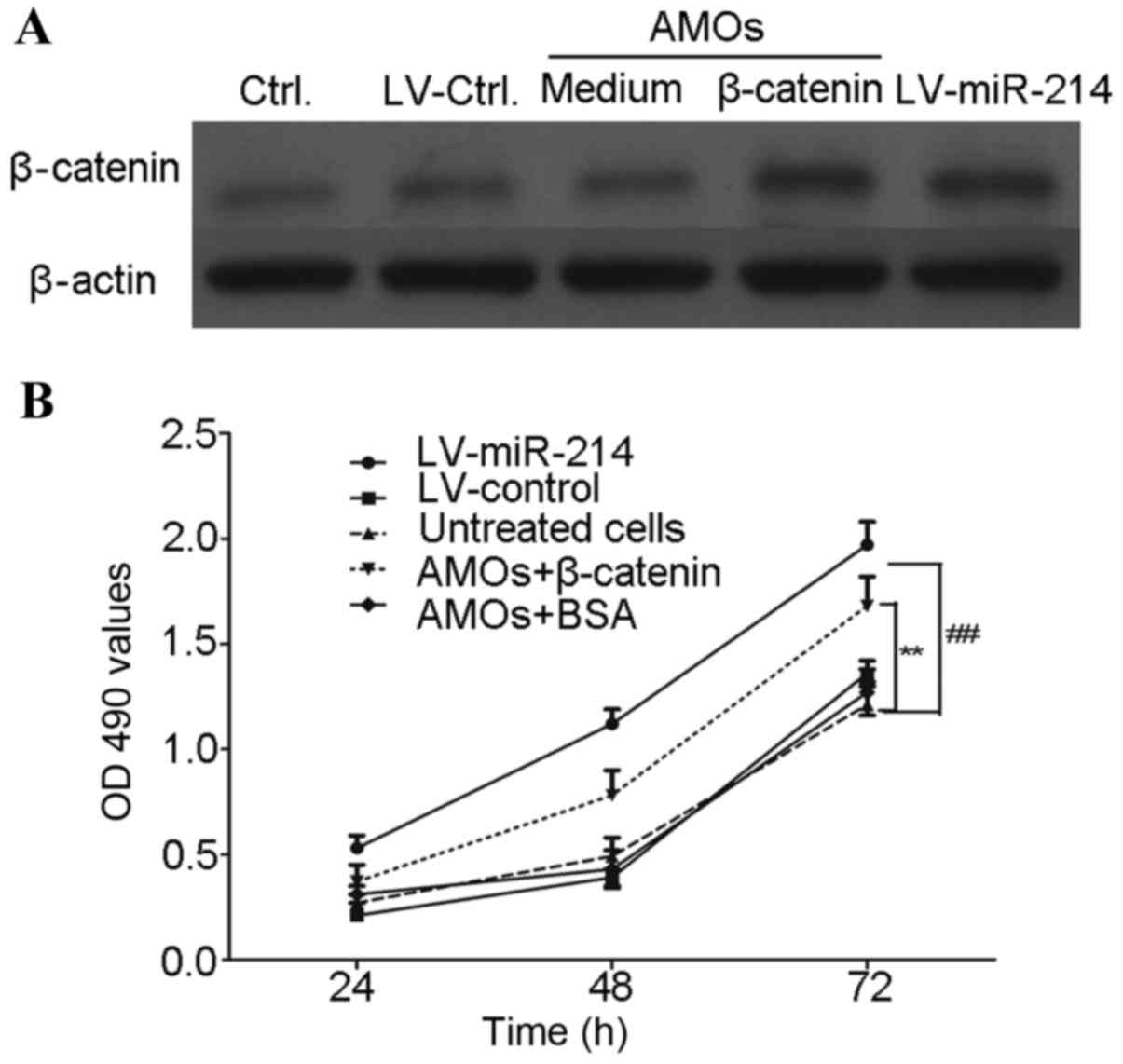 | Figure 7.Exogenous addition of β-catenin
effectively reverses the efficiency of the miR-214 lentivirus. The
stable cells (LV-miR-214-MG63 and LV-control-MG63; 3×105
cells/per well) were plated into a 24-well plate for 6 h, (A)
LV-miR-214-MG63 cells were treated with 100 ng/ml of β-catenin for
48 h and the expression of β-cateinin was determined using western
blot analysis. (B) Cells were treated with β-catenin for 24, 48 and
72, respectively. The proliferation of MG63 cells was determined
using a 3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium
bromide assay. **P<0.01, compared with LV-miR-214+β-catenin
group; ##P<0.01, compared with LV-control group. miR,
microRNA; AMOs, antisense-microRNA oligonucleotides; Ctrl, control;
OD, optical density. |
Discussion
Osteosarcoma is the most common type of bone cancer,
and is most often found in teenagers (20). The incidence rate of osteosarcoma
in teenagers is high, suggesting that rapid bone growth induces
osteosarcoma (3). Investigations
on the molecular mechanisms of osteosarcoma have gradually
increased in number, however, a comprehensive understanding of the
mechanisms underlying osteosarcoma remains to be fully elucidated.
Following substantial progression in understanding of the roles of
miRNAs, their role in cancer has received significant attention
(21–23). In the present study, the role of
miR-214 in the progression of human osteosarcoma was established
and the specific molecular mechanisms of miRNA-214 in human
osteosarcoma were clarified.
The present study detected the expression of
miRNA-214 in human osteosarcoma specimens and paired peritumoral
tissues. The results demonstrated that increased levels of miR-214
were observed in the osteosarcoma tissues. This was in contrast to
data by Wen et al (24) on
cervical cancer. However, Yang et al (15) found that miR-214 was overexpressed
in gastric cancer tissues and cell lines, which was consistent with
the results of the present study. Thus, the emerging data
demonstrated that miR-214 has a controversial role in different
types of tumor, which may be due to organ-specific actions and the
possibility that miR-214 has different target genes to exert
opposing functions.
It has been reported that abnormal expression of the
Wnt/β-catenin signaling pathway has been detected in osteosarcoma
tissues, which significantly contributes to the promotion of cell
survival and proliferation in multiple types of malignancy
(25–27). Thus, the Wnt/β-catenin signaling
pathway has received increased attention in targeted therapy for
osteosarcoma. The present study aimed to detect the association
between the expression of miR-214 and the activity of the
Wnt/β-catenin signaling pathway. The results of the present study
showed no significant variation in the levels of Wnt1, Wnt2, Wnt4,
Axin or Gsk-3β in the miR-214 lentivirus-infected cells, compared
with those in the control lentivirus-infected U2OS cancer cells,
however, the levels of β-catenin in the MG63 cells and Saos-2 cells
were upregulated. As expected, miR-214 may be involved in
regulating and activating the Wnt/β-catenin pathway by directly
regulating β-catenin.
The results of the FACS assay showed that
miR-214-specific AMOs inhibited the cell cycle and induced cell
cycle arrest at the G0/G1 phase, which also demonstrated that
miR-214 regulated cell cycle progression and promoted the
progression of cells through the check point of the G0/G1 phase. In
addition, a reverse experiment was designed in which the MG63 cells
were treated with exogenously added β-catenin, which significantly
reversed the efficiency of the miR-214-specific AMOs. It was shown
that this reversal experiment was able to assess the specificity of
β-catenin in miR-214-specific AMOs treated cells. Taken together,
the results of the present study showed that miR-214 in
osteosarcoma cells had an important oncogenic role to regulate
multiple targets in the Wnt/β-catenin pathway. Therefore, miR-214
offers potential as a novel target for the clinical therapy of
human osteosarcoma.
Acknowledgements
The present study was supported by the Anhui
Province Natural Science Foundation (grant no. 1408085MH207).
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khandekar S, Dive A, Munde P and Fande PZ:
Chondroblastic osteosarcoma of the left zygomatic bone: Rare case
report and review of the literature. J Oral Maxillofac Pathol.
18:281–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berner K, Johannesen TB, Berner A,
Haugland HK, Bjerkehagen B, Bøhler PJ and Bruland ØS: Time-trends
on incidence and survival in a nationwide and unselected cohort of
patients with skeletal osteosarcoma. Acta Oncol. 54:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berner K, Bjerkehagen B, Bruland ØS and
Berner A: Extraskeletal osteosarcoma in Norway, between 1975 and
2009, and a brief review of the literature. Anticancer Res.
35:2129–2140. 2015.PubMed/NCBI
|
|
5
|
Haddox CL, Han G, Anijar L, Binitie O,
Letson GD, Bui MM and Reed DR: Osteosarcoma in pediatric patients
and young adults: A single institution retrospective review of
presentation, therapy, and outcome. Sarcoma. 2014:4025092014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu XW, Zi Y, Xiang LB and Han TY:
Periosteal osteosarcoma: A review of clinical evidence. Int J Clin
Exp Med. 8:37–44. 2015.PubMed/NCBI
|
|
7
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: MicroRNAs and Potential Targets in Osteosarcoma: Review.
Front Pediatr. 3:692015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun X, Luo S, He Y, Shao Y, Liu C, Chen Q,
Cui S and Liu H: Screening of the miRNAs related to breast cancer
and identification of its target genes. Eur J Gynaecol Oncol.
35:696–700. 2014.PubMed/NCBI
|
|
10
|
Tiberio P, Callari M, Angeloni V, Daidone
MG and Appierto V: Challenges in using circulating miRNAs as cancer
biomarkers. Biomed Res Int. 2015:7314792015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nagaraj AB, Joseph P and DiFeo A: miRNAs
as prognostic and therapeutic tools in epithelial ovarian cancer.
Biomark Med. 9:241–257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang L, Shrestha S, LaChaud G, Scott MA
and James AW: Review of microRNA in osteosarcoma and
chondrosarcoma. Med Oncol. 32:6132015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ouyang L, Liu P, Yang S, Ye S, Xu W and
Liu X: A three-plasma miRNA signature serves as novel biomarkers
for osteosarcoma. Med Oncol. 30:3402013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Penna E, Orso F and Taverna D: miR-214 as
a key hub that controls cancer networks: Small player, multiple
functions. J Invest Dermatol. 135:960–969. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang KC, Xi HQ, Cui JX, Shen WS, Li JY,
Wei B and Chen L: Hemolysis-free plasma miR-214 as novel biomarker
of gastric cancer and is correlated with distant metastasis. Am J
Cancer Res. 5:821–829. 2015.PubMed/NCBI
|
|
17
|
Cristóbal I, Caramés C, Madoz-Gúrpide J,
Rojo F, Aguilera O and García-Foncillas J: Downregulation of
miR-214 is specific of liver metastasis in colorectal cancer and
could play a role determining the metastatic niche. Int J
Colorectal Dis. 29:8852014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J,
Kruk PA, Wenham RM, Nicosia SV, Lancaster JM, Sellers TA and Cheng
JQ: MicroRNA miR-214 regulates ovarian cancer cell stemness by
targeting p53/Nanog. J Biol Chem. 287:34970–34978. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gougelet A, Pissaloux D, Besse A, Perez J,
Duc A, Dutour A, Blay JY and Alberti L: Micro-RNA profiles in
osteosarcoma as a predictive tool for ifosfamide response. Int J
Cancer. 129:680–690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian K, Wang L, Di R, Xu J, Li G and Li Z:
Effect and mechanism of miRNA to osteosarcoma cell. Pak J Pharm
Sci. 27 Suppl 5:1657–1660. 2014.PubMed/NCBI
|
|
23
|
Zhao F, Lv J, Gan H, Li Y, Wang R, Zhang
H, Wu Q and Chen Y: MiRNA profile of osteosarcoma with CD117 and
stro-1 expression: miR-1247 functions as an onco-miRNA by targeting
MAP3K9. Int J Clin Exp Pathol. 8:1451–1458. 2015.PubMed/NCBI
|
|
24
|
Wen Z, Lei Z, Jin-An M, Xue-Zhen L,
Xing-Nan Z and Xiu-Wen D: The inhibitory role of miR-214 in
cervical cancer cells through directly targeting mitochondrial
transcription factor A (TFAM). Eur J Gynaecol Oncol. 35:676–682.
2014.PubMed/NCBI
|
|
25
|
Blagodatski A, Poteryaev D and Katanaev
VL: Targeting the Wnt pathways for therapies. Mol Cell Ther.
2:282014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagaraj AB, Joseph P, Kovalenko O, Singh
S, Armstrong A, Redline R, Resnick K, Zanotti K, Waggoner S and
DiFeo A: Critical role of Wnt/β-catenin signaling in driving
epithelial ovarian cancer platinum resistance. Oncotarget.
6:23720–23734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Merhi A, De Mees C, Abdo R, Victoria
Alberola J and Marini AM: Wnt/β-Catenin signaling regulates the
expression of the ammonium permease gene rhbg in human cancer
cells. PLoS One. 10:e01286832015. View Article : Google Scholar : PubMed/NCBI
|