Introduction
Renal cell carcinoma (RCC), a common malignant tumor
originating from renal tubular epithelial cells, is the most common
type of renal cancer and the third most common type of urological
cancer, accounting for 2–3% of all adult malignancies according to
a survey conducted in 2013 in the USA (1,2).
Worldwide, RCC accounts for ~2% of cancer-associated mortality
(2,3), and clear cell carcinoma is the most
common subtype of RCC, which accounts for ~80% (4). Early diagnosis and treatment for this
type of RCC is difficult as it lacks characteristic symptoms, and
is resistant to radiotherapy and chemotherapy (5). Therefore, it is essential to
investigate the mechanism of RCC to identify a biomarker for early
diagnosis and targeted therapy.
The roles of deregulated microRNAs (miRNAs; miRs) in
tumorigenesis have attracted increasing attention. miRNAs are a
class of short, single-stranded non-coding RNAs with a length of
~22 nucleotides (6,7). miRNAs can exert effects by imperfect
binding with the 3′ untranslated region of mRNA and cause
translational repression or mRNA cleavage (8,9). The
function of miRNAs as oncogenes or tumor suppressors depends on the
target gene they regulate. Previous studies have demonstrated that
miRNAs are associated with various cellular processes, including
proliferation, apoptosis, differentiation and stress response
(6,10). miR-195, located on chromosome
17p13.1, has been shown to be downregulated and function as a tumor
suppressor in different types of tumor, including bladder cancer
(11,12), osteosarcoma (13) and cervical cancer (14). However, previous miRNA microarray
chip analysis of RCC showed that miR-195-3p, the mature sequence of
miR-195 also termed miR-195, was upregulated (15), which revealed that the role of
miR-195-3p in RCC may be different, compared with other tumors.
Therefore the present study performed reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis to detect the expression level of miR-195 in RCC tissues
and cell lines, and the investigated the role of miR-195-3p in RCC
tumorigenesis by performing cell proliferation, mobility and
apoptotic assays.
Materials and methods
Tissue samples
In the present study, 31 paired tissues were
collected from Peking University Shenzhen Hospital (Shenzhen,
China) from December 2012 to December 2014. Each pair of tissues
included RCC tissue and adjacent normal tissue, which was a 2 cm
distance from the visible RCC lesion. The collection and use of
tissue samples were reviewed and approved by the Ethics Committees
of Peking University Shenzhen Hospital, and written informed
consent was obtained from all patients. The tissues were immersed
in RNAlater (Qiagen GmbH, Hilden, Germany) for 30 min on dissection
and then stored at −80°C for further use. These tissues were
reviewed and classified using hematoxylin and eosin staining. The
clinical and pathological characteristics of the patients are
presented in Table I.
 | Table I.Clinicopathological features of
patients with renal cell carcinoma. |
Table I.
Clinicopathological features of
patients with renal cell carcinoma.
| Characteristic | n |
|---|
| Mean age (range),
years | 51 (25–70) |
| Gender |
|
|
Male | 19 |
|
Female | 12 |
| Histological
type |
|
| Clear
cell | 26 |
|
Papillary | 5 |
| Primary tumor
stage |
|
| T1 | 17 |
| T2 | 11 |
|
T3+T4 | 3 |
| Fuhrman grade |
|
| I | 14 |
| II | 12 |
|
III | 3 |
| IV | 2 |
| AJCC stage |
|
| I | 17 |
| II | 10 |
|
III+IV | 4 |
Cell lines
The cell lines used in the present study comprised
293T human embryo kidney cells (the Type Culture Collection of the
Chinese Academy of Medical Sciences, Shanghai, China), and 786-O,
ACHN and 769P RCC cell lines (the American Type Culture Collection,
Manassas, VA, USA). The cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 1% antibiotics (100
µl/ml penicillin and 100 mg/ml streptomycin sulfates) and 1%
glutamine in the humidified incubator containing 5% CO2
at the temperature of 37°C.
RNA extraction and RT-qPCR
analysis
Total RNA was extracted from the tissues and cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and purified using an RNeasy Maxi kit (Qiagen GmbH) according to
the manufacturer's protocol. The concentrations were measured on a
NanoDrop2000/2000c spectrophotometer (Thermo Fisher Scientific,
Inc. Subsequently, reverse transcription was performed using the
miScript Reverse Transcription kit (Qiagen GmbH), according to the
manufacturer's protocol, to obtain cDNA. qPCR was then performed on
the Roche lightcycler 480 Real-Time PCR system with the miScript
SYBR®-green PCR kit (Qiagen GmbH) to detect the
expression level of miR-195-3p. PCR amplification was performed
using 1 µl cDNA in a 20 µl reaction system, containing 10 µl
QuantiTect SYBR Green PCR Master mix, 2 µl miScript Universal
Primer, 1 µl specific microRNA primer and 6 µl RNase-free water. U6
was used as the internal control and the primers used are shown in
Table II. PCR thermocycling
conditions were set as follows: 95°C for 1 min, then 40 cycles of
95°C for 15 sec, 55°C for 30 sec and 72°C for 30 sec. The data were
analyzed using the ΔΔCq method (16).
 | Table II.Sequences of transfectants and
primers used in the present study. |
Table II.
Sequences of transfectants and
primers used in the present study.
| miR-195-3p
mimic | Sense:
5′-CCAAUAUUGGCUGUGCUGCUCC-3′ |
|
| Antisense:
5′-AGCAGCACAGCCAAUAUUGGUU-3′ |
| NC | Sense:
5′-UUCUCCGAACGUGUCACGUTT-3′ |
|
| Antisense:
5′-ACGUGACACGUUCGGAGAATT-3′ |
| miR-195-3p
inhibitor |
5′-GGAGCAGCACAGCCAAUAUUGG-3′ |
| Inhibitor NC |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
| miR-195-3p forward
primer |
5′-CCAATATTGGCTGTGCTGCTCC-3′ |
| miR-195-3p reverse
primer | Universal primer
(miScript SYBR Green PCR kit) |
| U6 forward
primer |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6 reverse
primer |
5′-ACGCTTCACGAATTTGCGT-3′ |
Cell transfection
Transfection of 786-O and ACHN cells with miR-195-3p
mimic, inhibitor, negative control (NC) and inhibitor NC
(GenePharma, Shanghai, China) was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), when cells reached 70–90% confluency. Cells were
transfected for 4–6 h at 37°C. Alterations in the expression levels
of miR-195-3p following transfection were determined by performing
RT-qPCR analysis with the aforementioned thermocycling conditions.
The primer sequences used are shown in Table II.
Cell mobility assay
A cell scratch assay and Transwell assay were
performed to assess the mobility of the 786-O and ACHN cells. In
the cell scratch assay, ~6×105 cells were plated in each well of a
6-well plate. After 24 h, the cells were transfected with 200 pmol
miR-195-3p mimic, inhibitor, NC or inhibitor NC. At 6 h
post-transfection, a vertical horizontal line was scratched in the
cell layer using a sterile 200 µl pipette tip. Images of the
scratches at 0 and 24 h were captured using a digital camera
system. The experiments were performed in triplicate and repeated
at least three times. Transwell invasion and migration assays were
performed to assess the migratory and invasive abilities of the
786-O and ACHN RCC cells. Transwell chamber inserts (BD
Biosciences, Franklin Lakes, NJ, USA) with (to assess invasion) or
without (to assess migration) Matrigel (BD Biosciences) were used
in the assay, according to the manufacturer's protocol. The
transfected cells (1×104) in 200 µl serum-free medium were seeded
in the upper chamber of the insert. In the bottom of the inserts
was medium containing 10% FBS. The cells were allowed to migrate
for 36 h or invade for 48 h in the humidified incubator containing
5% CO2 at the temperature of 37°C. The migratory or
invasive cells on the bottom of the inserts were strained with
crystal violet and counted using a microscope (Leica Microsystems
GmbH, Wetzlar, Germany). The experiments were performed in
triplicate and repeated at least three times.
Cell proliferation assay
MTT and CCK-8 assays were performed to assess cell
proliferation ability. The cells (~3,000) were seeded in each well
of 96-well plate and, 24 h later, were transfected with 5 pmol of
miR-195-3p mimic, inhibitor, NC or inhibitor NC. In the CCK-8
assay, CCK-8 reagent was added into the wells 0, 24, 48 and 72 h
post-transfection. After 1.5 h, the optical density (OD) of each
well was measured using an ELISA microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) at a wavelength of 490 nm.
For the MTT assay at 20 µl MTT (5 mg/m; Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) was added into the wells at 0, 24,
48 and 72 h post-transfection. The medium was then replaced with
150 µl of dimethylsulfoxide (DMSO; Sigma-Aldrich; Merck Millopore)
following incubation at 37°C for 4 h. The OD value of each well was
measured using the ELISA microplate reader (Bio-Rad Laboratories,
Inc.) at a wavelength of 490 nm following agitation for 15 min at
room temperature. The experiments were performed in triplicate and
repeated at least three times.
Cell apoptosis assay
Flow cytometry was performed to assess the apoptotic
rate of the cells following transfection. In each well of a 6-well
plate, ~3×105 cells were seeded and, 24 h later, were transfected
with 200 pmol miR-195-3p mimics, inhibitor, NC or inhibitor NC. At
48 h post-transfection, all cells were harvested and washed twice
with cold PBS. The cells were resuspended in 100 µl 1X binding
buffer, and 5 µl Annexin V-FITC (Invitrogen; Thermo Fisher
Scientific, Inc.) and 3 µl propidium iodide (PI, Invitrogen; Thermo
Fisher Scientific, Inc.) were added into each cell suspension.
After 15 min, 400 µl of binding buffer was added to each tube. The
apoptotic rates were analyzed using flow cytometry (EPICS, Xl-4;
Beckman Coulter, Inc., Brea, CA, USA). The experiments were
performed in triplicate and repeated at least three times.
Statistical analysis
A paired t-test was used to compare the expression
levels of miR-195-3p in the paired tissues. Student's t-test was
used to analyze assays for characterizing the phenotypes of cells.
All statistical analyses were performed using the SPSS 19.0
statistical software package (IBM SPSS, Armonk, NY, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-195-3p is upregulated in RCC
tissues and cell lines
RT-qPCR analysis was performed to detect the
expression levels of miR-195-3p in RCC tissues and cell lines. The
ratios of expression of miR-195-3p in 31 paired RCC tissue samples
are shown in Fig. 1A, which showed
miR-195-3p was upregulated in 21 tissue samples. The mean relative
expression of miR-195-3p in the RCC tissues was 3.88-fold higher,
compared with the expression in adjacent normal tissues, as shown
in Fig. 1B (P<0.05). The
results demonstrated that the expression levels of miR-195-3p in
786-O, 769P and ACHN cells was 33.21-, 3.90- and 3.31-fold higher,
compared with the expression levels in 293T cells, respectively
(Fig. 1C). The results suggested
that miR-195-3p was upregulated in RCC tissues, compared with
adjacent normal tissues, and miR-195-3p may be have an oncogenic
role in RCC.
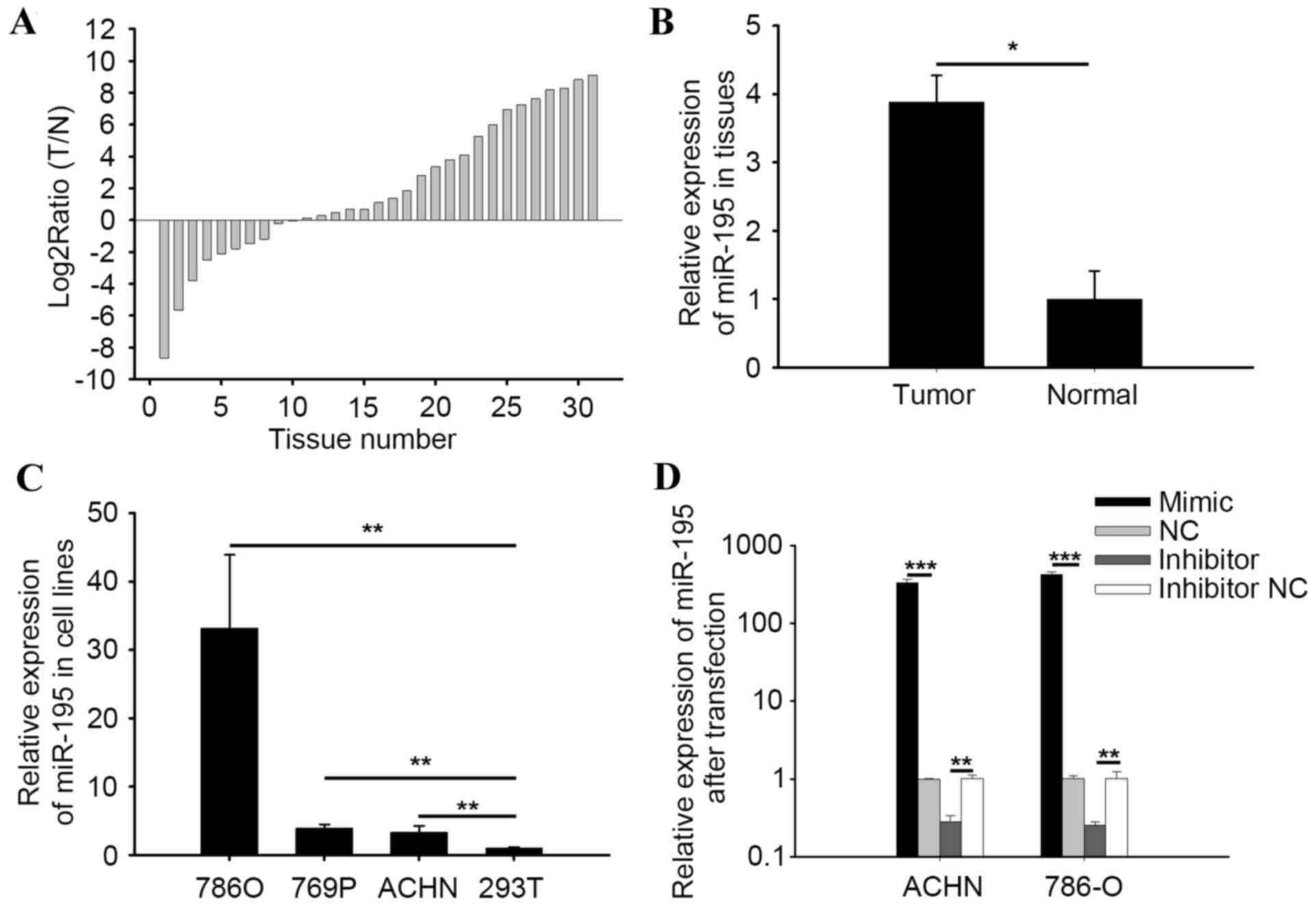 | Figure 1.Expression of miR-195-3p. (A) Log2
ratios (T/N) of miR-195-3p in 31 paired tissue samples. (B)
Relative expression of miR-195-3p in RCC and normal tissues. The
mean relative expression of miR-195-3p in RCC tissues was 3.88-fold
higher than the expression in adjacent normal tissues. (C) Relative
expression of miR-195-3p in 786-O, 769P and ACHN in RCC cell lines,
and the 293T cell line. (D) Relative expression of miR-195-3p in
786-O and ACHN cells following transfection with miR-195-3p mimic,
inhibitor, NC or inhibitor NC. *P<0.05, **P<0.01 and
***P<0.001. RCC, renal cell carcninoma; miR, microRNA; T, RCC
tissue; N, normal tissue; NC, negatice control. |
Validation of cell transfection
efficiency
RT-qPCR analysis was performed to quantify the
transfection efficiency of miR-195-3p mimics or inhibitors,
compared with NC or inhibitor NC. The results indicated that the
expression levels of miR-195-3p in the miR-195-3p mimic group were
424.58-fold higher (786-O) and 328.37-fold higher (ACHN), compared
with the NC group (P<0.001), and expression in the inhibitor
group was 0.28-fold (786-O) and 0.25-fold of the inhibitor NC group
(P<0.01; Fig. 1D).
miR-195-3p promotes cell
proliferation
MTT and CCK-8 assays were performed to detect cell
proliferation following transfection. The data are shown as the
mean ± standard error of the mean. As shown in Fig. 2A, the results of the CCK-8 assay
suggested that the overexpression of miR-195-3p promoted 786-O cell
proliferation by 5.26, 14.61 (P<0.01) and 8.09% (P<0.001),
and the downregulation of miR-195-3p (Fig. 2B) inhibited 786-O cell
proliferation by 3.79, 10.99 (P<0.01) and 9.07% (P<0.001) at
24, 48 and 72 h post-transfection, respectively. In the ACHN cells,
cell proliferation was promoted by 6.32 (P<0.05), 19.76
(P<0.01) and 11.89 (P<0.001) in the miR-195-3p mimic group.
Cell proliferation in the miR-195-3p inhibitor group was inhibited
by 5.25 (P<0.05), 6.32 (P<0.05) and 9.38% (P<0.01) at 24,
48 and 72 h post-transfection, compared with the NC or inhibitor NC
groups, respectively (Fig. 2C and
D).
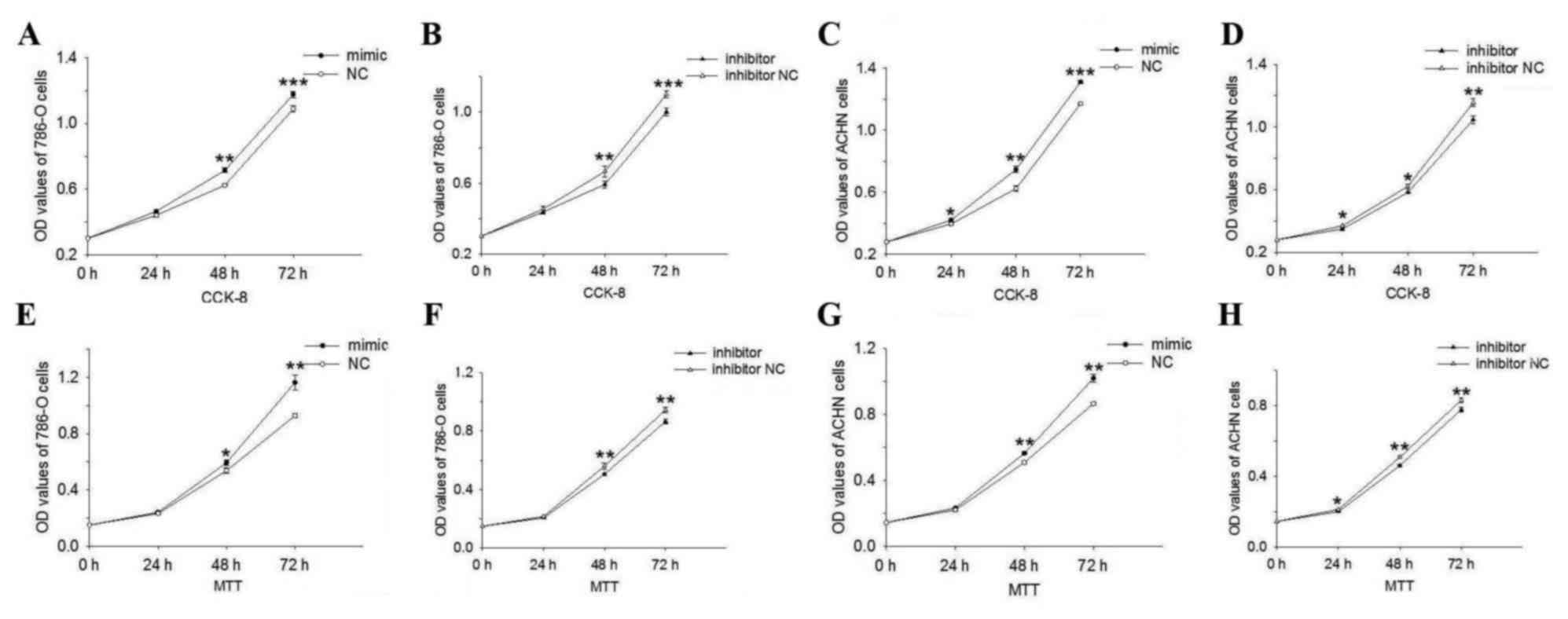 | Figure 2.Cell proliferation assay. CCK-8
assays of the (A and B) 786-O cells and (C and D) ACHN cells showed
that the upregulation of miR-195-3p promoted cell proliferation
compared with NC group and that downregulation of miR-195-3p caused
inhibition compared with inhibitor NC group. Similar results were
obtained in the MTT assay of (E and F) 786-O and (G and H) ACHN
cells. A, C, E and G, *P<0.05, **P<0.01 and ***P<0.001 vs.
NC group; B, D, F and H, *P<0.05, **P<0.01 and ***P<0.001
vs. inhibitor NC group. miR, microRNA; NC, negative control; OD,
optical density. |
The results of the MTT assay showed that the
proliferation of 786-O cells (Fig. 2E
and F) in the inhibitor group was reduced by 4.38, 9.83
(P<0.01) and 8.29% (P<0.01), and that in the mimic group was
promoted by 3.65, 10.37 (P<0.05) and 25.34% (P<0.01),
compared with the inhibitor NC or NC groups at 24, 48 and 72 h
post-transfection. In ACHN cells, the results of the MTT assay
showed that the overexpression of miR-195-3p (Fig. 2G) promoted cell proliferation by
5.42, 10.88 (P<0.01) and 17.78% (P<0.01), whereas
downregulation of miR-195-3p (Fig.
2H) inhibited 786-O cell proliferation by 5.69 (P<0.05),
9.21 (P<0.01) and 6.41% (P<0.01) at 24, 48 and 72 h
post-transfection, respectively. The results of the proliferation
assays showed that miR-195-3p promoted RCC cell proliferation.
miR-195-3p increases cell
mobility
Cell scratch, Transwell migration and invasion
assays were performed to investigate the effect of miR-195-3p on
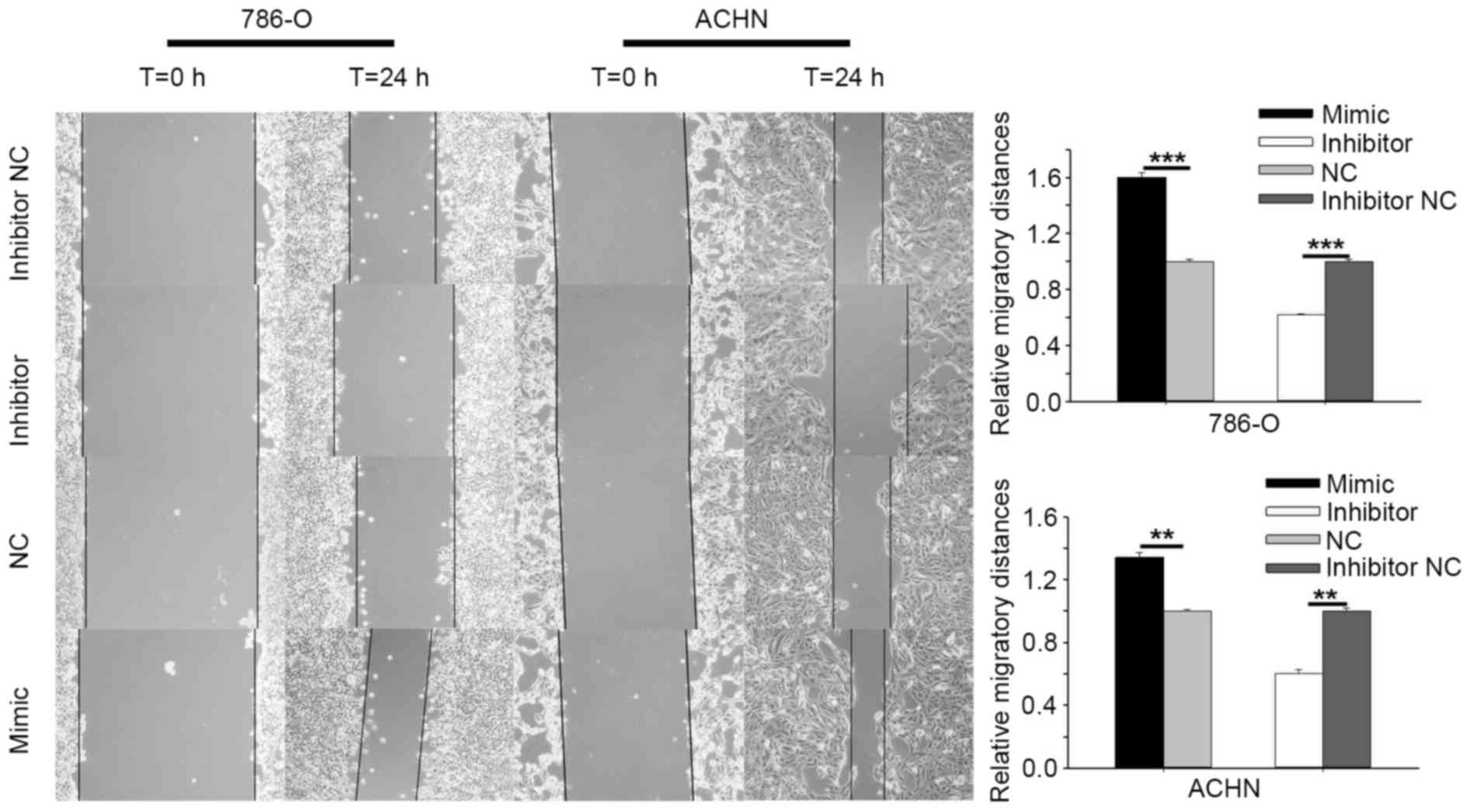
RCC (786-O and ACHN) cell mobility. The results of cell scratch
assay are shown in Fig. 3.
Overexpression of miR-195-3p by transfection with the miR-195-3p
mimic promoted the 786-O cell migratory distance by 60.31%
(P<0.001) and the ACHN distance by 34.23% (P<0.01) at 24 h
post-transfection, compared with the NC cells. Downregulation of
miR-195-3p by transfection with miR-195-3p inhibitor reduced cell
migratory distance by 38.13% (P<0.001) in the 786-O cells and
40.01% (P<0.01) in the ACHN cells at 24 h post-transfection,
compared with the cells transfected with inhibitor NC.
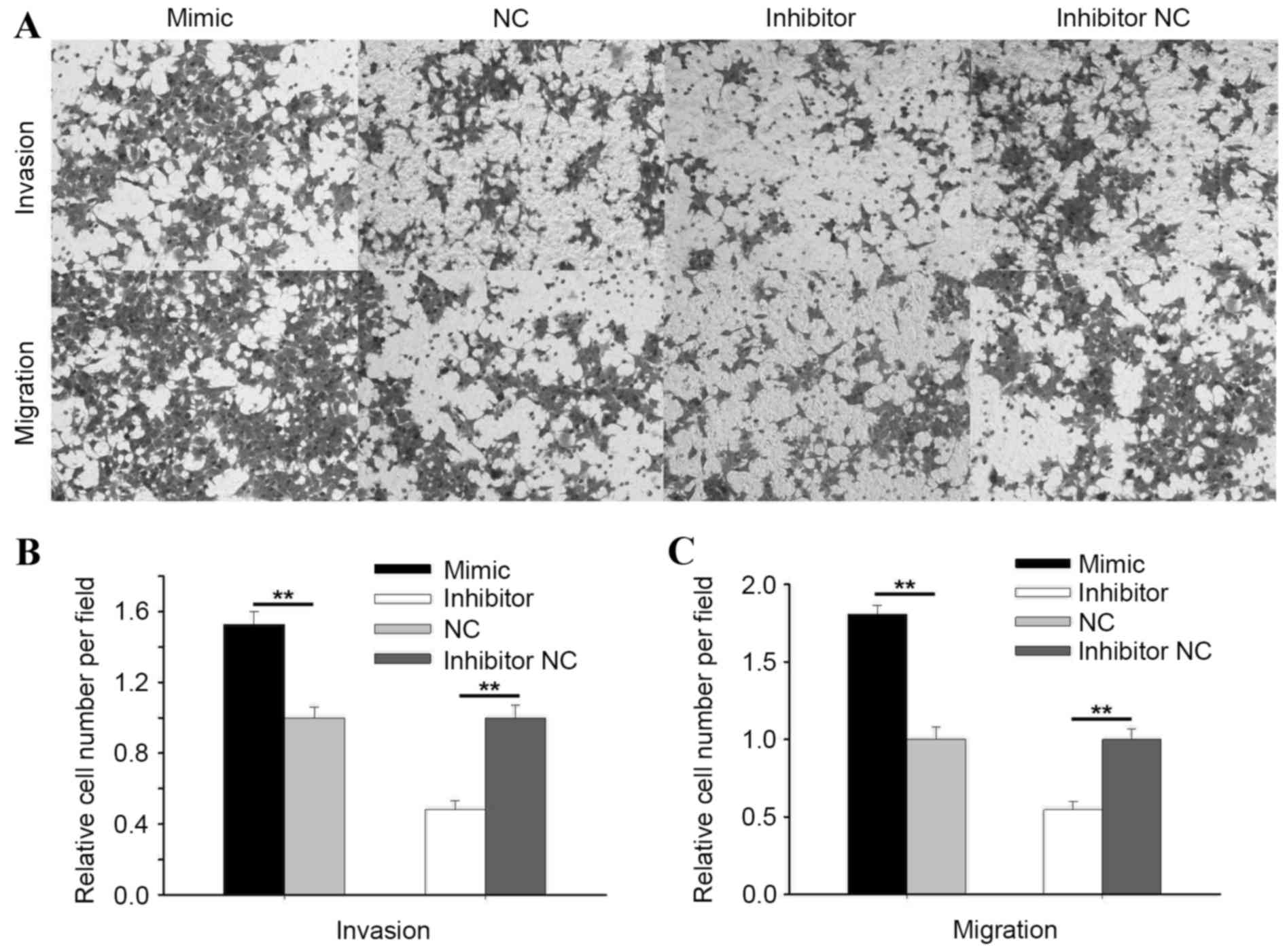
As shown in Fig. 4,
the results of the Transwell invasion assay showed that the
invasive ability of 786-O cells was promoted by 52.83% (P<0.01)
by upregulating miR-195-3p, and was suppressed by 51.69%
(PP0.01) by downregulating miR-195-3p (Fig. 4B). The migratory ability of 786-O
cells was promoted by 80.79% by upregulating miR-195-3p, and
suppressed by 45.44% (P<0.01) by downregulating miR-195-3p
(Fig. 4C).
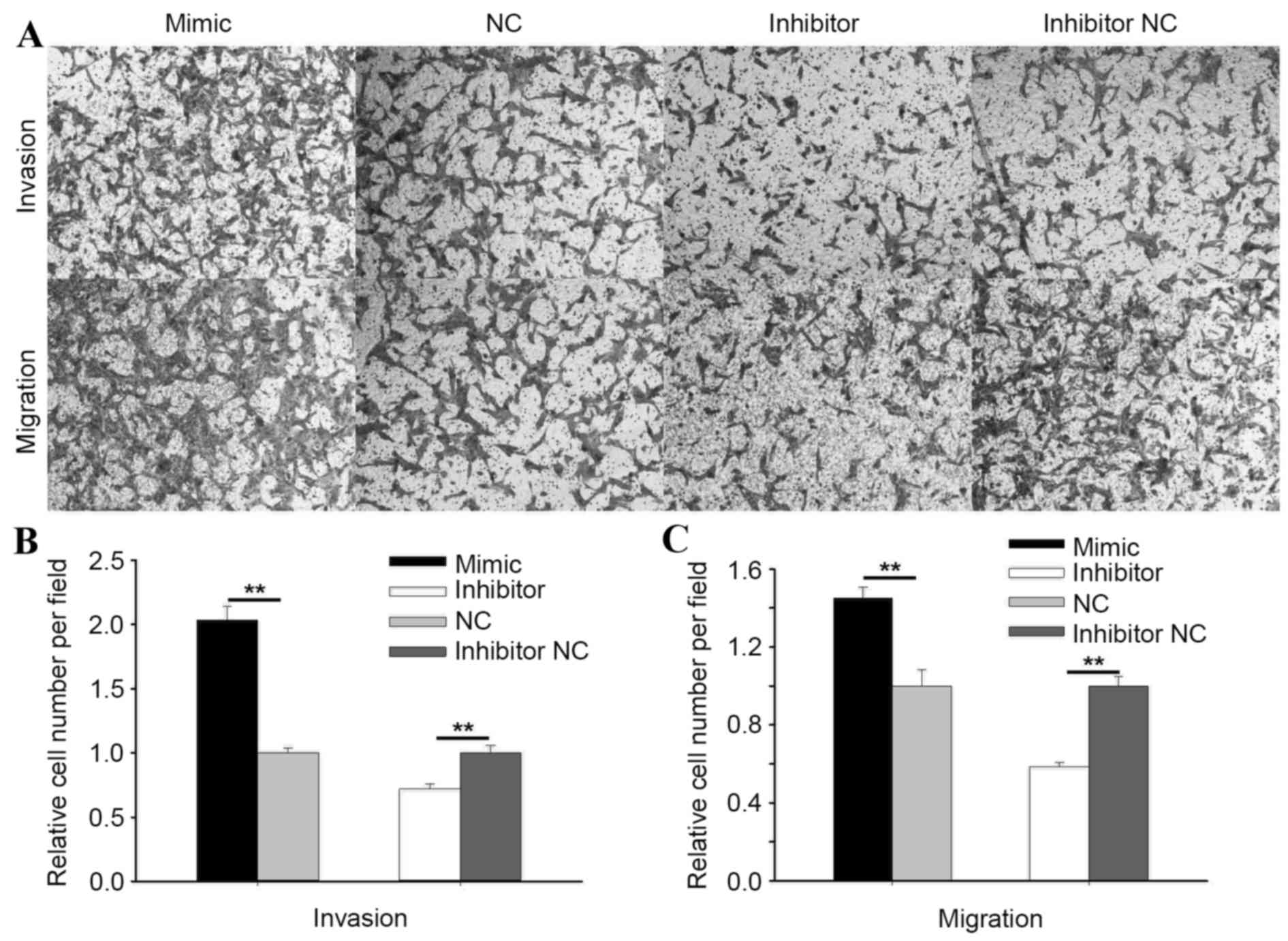
In ACHN cells, the Transwell invasion assay showed
that the invasive ability of cells transfected with the miR-195-3p
mimic was increased by 103.53% (P<0.01) and reduced by 27.85%
(P<0.01) in cells transfected with the miR-195-3p inhibitor,
compared with the NC or inhibitor NC group, respectively (Fig. 5B). As shown in Fig. 5C, the migratory ability of cells
transfected with the miR-195-3p mimic was increased by 44.91%
(P<0.01) and reduced by 41.35% (P<0.01) in cells transfected
with miR-195-3p inhibitor, compared with cells transfected with NC
or inhibitor NC. The results of the Transwell and wound scratch
assays indicated that miR-195-3p promoted the mobility of RCC
cells.
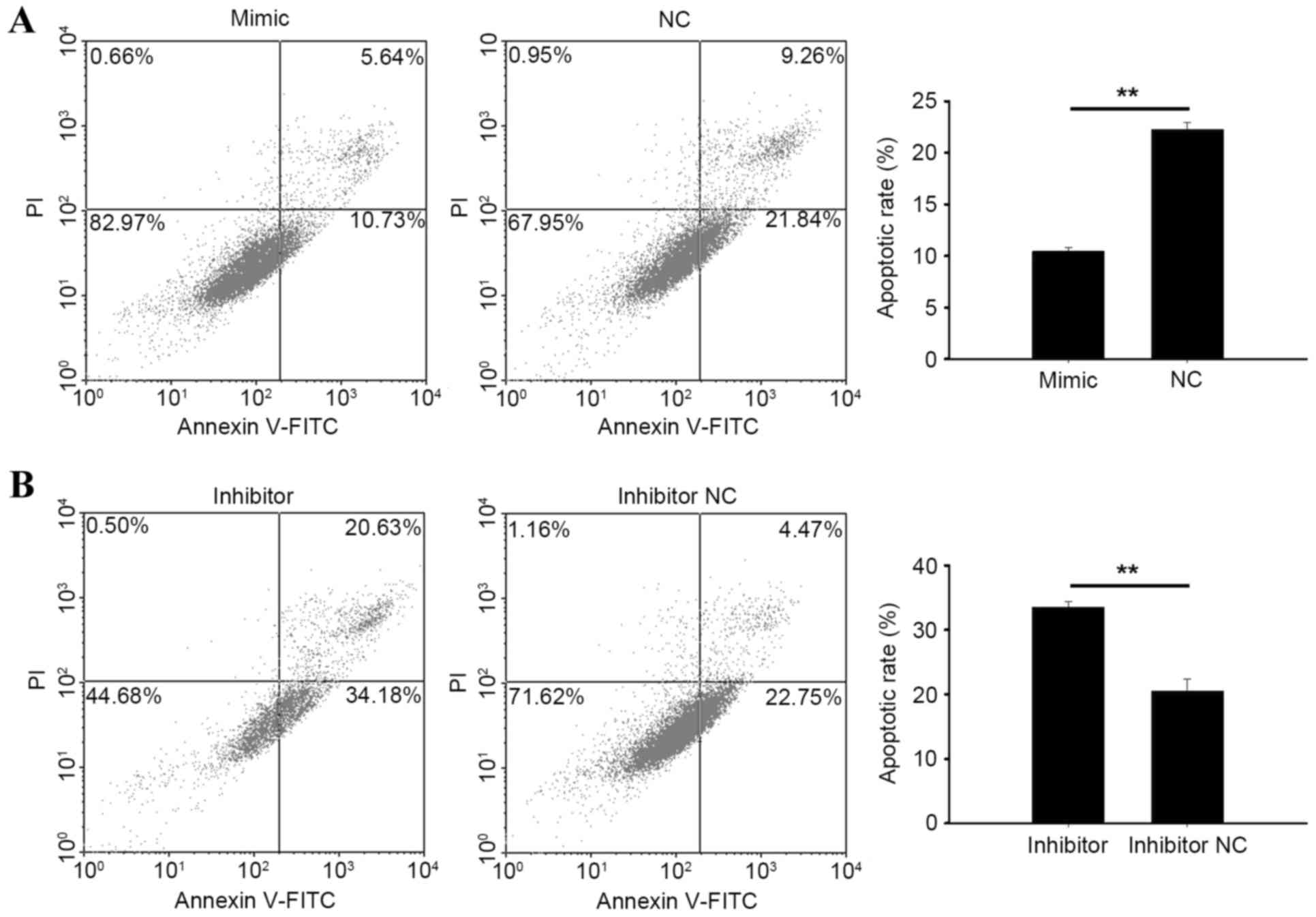
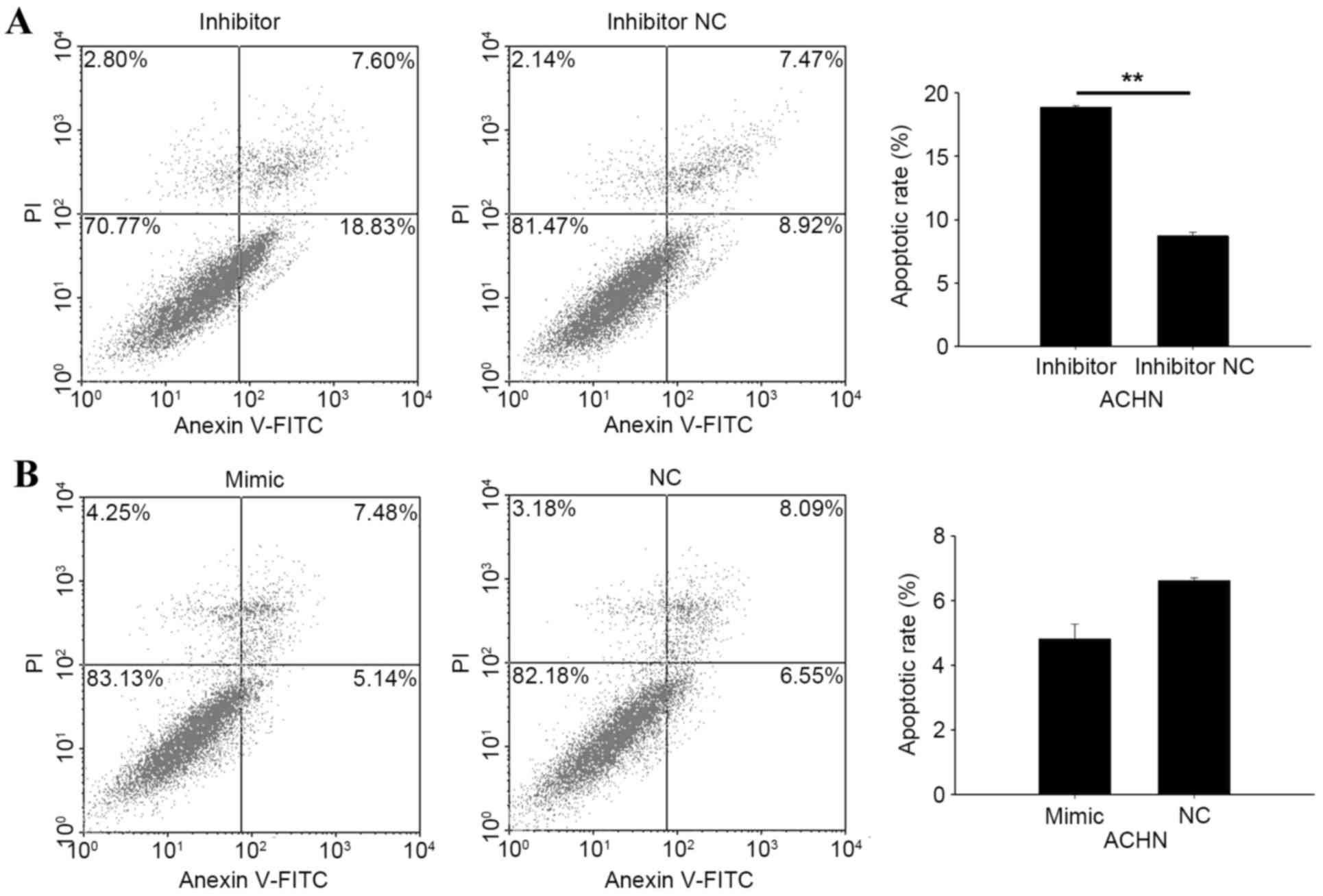
Knockdown of miR-195-3p induces cell
apoptosis
Flow cytometry was performed to qualify the
apoptotic rate of RCC cells following transfection. At 48 h
post-transfection with the miR-195-3p mimic, miR-195-3p inhibitor,
NC or inhibitor NC, cells were collected for measurement. As shown
in Fig. 6A and B, the apoptotic
rate of 786-O cells transfected with the miR-195-3p mimic was
10.45%, and was 22.27% in cells transfected with NC (P<0.01).
The apoptotic rates of 786-O cells transfected with the miR-195-3p
inhibitor or inhibitor NC were 33.58 and 20.50%, respectively
(P<0.01). In ACHN cells transfected with the miR-195-3p
inhibitor, the apoptotic rate was 18.91%, and was 8.73% in cells
transfected with the inhibitor NC (P<0.01). However, no
significant differences were found between cells transfected with
the miR-19-3p 5 mimic and NC, with apoptotic rates of 4.83% in the
mimic group and 6.62% in the NC group (Fig. 7A and B). These results revealed
that the knockdown of miR-19-3p 5 induced RCC cell apoptosis.
Discussion
Tumorigenesis is involved with the activation of a
series of oncogenes and inactivation of various tumor suppressors.
The genes identified to be associated with RCC, comprising Von
Hippel-Lindau, MET, folliculin, TSC1, TSC2, FH and SDH, are all
possibly regulated by miRNAs, therefore, miRNAs are potential
biomarkers for RCC for use as targeted therapy.
In the present study RT-qPCR analysis revealed that
miR-195 was upregulated in RCC, whereas previous studies have shown
that miR-195 is downregulated in the majority of types of cancer,
including colorectal cancer (17),
glioblastoma (18), bladder cancer
(11,12), osteosarcoma (13), cervical cancer (14), gastric cancer (19), hepatocellular carcinoma (20), esophageal squamous cell carcinoma
(10), breast cancer (21), non-small cell lung cancer (22) and prostate cancer (23). Therefore, the present study, to the
best of our knowledge was the first to report that miR-195 was
upregulated in RCC. Subsequently, the function of miR-195 in RCC
was examined, and the results revealed that the overexpression of
miR-195 promoted RCC cell proliferation, migration and invasion,
and reduced apoptosis, whereas the downregulation of miR-195
suppressed cell proliferation, migration and invasion induced
apoptosis. With the exception of ACHN cells, the overexpression of
miR-195 marginally reduced the apoptotic rate of the ACHN cells
with a characteristic low apoptotic rate.
Previous studies of miR-195 have focused on
urological cancer, with the exception of renal tumors. Guo et
al (24) found that miR-195
suppressed prostate cancer cell proliferation and metastasis by
targeting BCOX1. Another study of miR-195 in prostate cancer
revealed that miR-195 suppresses prostate cancer cell migration and
invasion through its direct target gene, Fra-1 (25). miR-195 was also found to inhibit
prostate cancer cell metastasis and EMT by targeting FGF2 (26). Therefore, in prostate cancer,
miR-195 functions as a tumor suppressor partially by targeting
BCOX1, Fra-1 and fibroblast growth factor 2 FGF2. In bladder cancer
it has been demonstrated that miR-195 induces G1-phase arrest by
targeting CDK4 (27), and inhibits
bladder cancer cell proliferation, at least partially, through the
inhibition of Cdc42/STAT3 signaling (12). miR-195 has been indicated to be
associated with the glycometabolism in bladder cancer by
suppressing glucose uptake through regulating the expression of
GLUT3 (11). In other tumors of
the urological system miR-195 is predominantly a tumor suppressor
and can affect cellular migration, invasion, metastasis, EMT and
glycometabolism.
Various studies of miR-195 have revealed that
miR-195 actes as a tumor suppressor in hepatocellular carcinoma
(HCC) and colorectal cancer (CRC). miR-195 has been reported to
regulate HCC cell apoptosis, proliferation, invasion and migration
(9,19,28–30).
It has also been reported that miR-195 is involved as a tumor
suppressor by targeting LAST2 (27), SRC-3 (28), CBX4 (30), tumor necrosis factor-α/nuclear
factor-κB (31) and PCMT1
(32). Wang et al (20) found that miR-195 suppresses HCC
angiogenesis and metastasis by inhibiting VEGF, VAV2 and CDC42
(20). Another study showed that
the miR-497-195 cluster can regulate HCC cell proliferation and
cell cycle by targeting CCNE1, CDC25A, CCND3, CDK4 and BTRC
(33). All studies on miR-195 in
HCC have indicated that miR-195 functions as a tumor suppressor. In
CRC, miR-195 has been described as a tumor suppressor by regulating
cell proliferation, migration, invasion and apoptosis (17,34,35).
A study investigating miR-195 as a biomarker in CRC demonstrated
that the downregulation of miR-195 was associated with poor
prognosis and lymph node metastasis (6). miR-195 has also been described as a
biomarker in cervical cancer, osteosarcoma, adrenocortical cancer
and breast cancer (21,36–40).
Zhao et al (38) found that
miR-195 has a higher sensitivity for breast cancer detection, and
the expression level of miR-195 has been found to significantly
predict the survival rates of patients with HER2-positive breast
cancer (36). Zhang et al
(37) reported that use of a serum
miRNA panel, comprising miR-16–2*, miR-195, miR-2861 and miR-497,
was able to distinguish cervical cancer from cervical
intraepithelial neoplasia and healthy controls with high accuracy.
Down-regulated miR-195 can predict a poor prognosis in patients
with osteosarcoma or adrenocortical cancer (39,40).
Therefore, miR-195 is a potential biomarker for multiple types of
cancers, and can be used for diagnosis, targeted therapy or
predicting prognosis.
miR-195 has been reported to have the ability to
regulate the sensitivity of cancer cells to chemotherapeutic drugs.
Yang et al (41) found that
miR-195 sensitizes HCC cells to 5-FU by targeting BCL-w. In colon
cancer, miR-195 has been shown to sensitize cells to doxorubicin by
targeting BCL2L2 (42). In breast
cancer, the overexpression of miR-195 sensitizes cells to
adriamycin by inhibiting Raf-1, and enhances the radiosensitivity
of cells by inhibiting BCL2 (43,44).
Thus, miR-195 is a novel anticarcinogen in certain types of cancer
that are particularly resistant to certain chemotherapeutics.
miR-195 has been associated with diseases other than
cancer. In Alzheimer's disease miR-195 can negatively regulate
BACE1, which offers potential therapy for Alzheimer's disease.
In conclusion, the present study is the first, to
the best of our knowledge, to describe miR-195-3p as an oncogene in
RCC by regulating RCC cell proliferation, mobility and apoptosis.
Further investigations aim to focus on the pathway of miR-195-3p in
RCC and the possibility of using as a biomarker for RCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81101922), the Science and
Technology Development Fund Project of Shenzhen (grant nos.
JCYJ20130402114702124 and JCYJ20150403091443329) and the Fund of
Guangdong Key Medical Subject.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun TY, Xie HJ, Li Z and Kong LF:
Expression of FOXC2 in renal cell carcinoma and its relationship to
clinical pathological features. Int J Clin Exp Med. 8:13388–13392.
2015.PubMed/NCBI
|
|
3
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tostain J, Li G, Gentil-Perret A and
Gigante M: Carbonic anhydrase 9 in clear cell renal cell carcinoma:
A marker for diagnosis, prognosis and treatment. Eur J Cancer.
46:3141–3148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iwamoto H, Kanda Y, Sejima T, Osaki M,
Okada F and Takenaka A: Serum miR-210 as a potential biomarker of
early clear cell renal cell carcinoma. Int J Oncol. 44:53–58.
2014.PubMed/NCBI
|
|
6
|
Wang X, Wang J, Ma H, Zhang J and Zhou X:
Downregulation of miR-195 correlates with lymph node metastasis and
poor prognosis in colorectal cancer. Med Oncol. 29:919–927. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ujifuku K, Mitsutake N, Takakura S,
Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K,
Nagata I and Yamashita S: miR-195, miR-455-3p and miR-10a(*) are
implicated in acquired temozolomide resistance in glioblastoma
multiforme cells. Cancer Lett. 296:241–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of MiR-195 and
MiR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu MG, Li S, Yu TT, Qian LJ, Cao RS, Zhu
H, Xiao B, Jiao CH, Tang NN, Ma JJ, et al: Differential expression
of miR-195 in esophageal squamous cell carcinoma and miR-195
expression inhibits tumor cell proliferation and invasion by
targeting of Cdc42. FEBS Lett. 587:3471–3479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fei X, Qi M, Wu B, Song Y, Wang Y and Li
T: MicroRNA-195-5p suppresses glucose uptake and proliferation of
human bladder cancer T24 cells by regulating GLUT3 expression. FEBS
Lett. 586:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao C, Qi L, Chen M, Liu L, Yan W, Tong S
and Zu X: microRNA-195 inhibits cell proliferation in bladder
cancer via inhibition of cell division control protein 42
homolog/signal transducer and activator of transcription-3
signaling. Exp Ther Med. 10:1103–1108. 2015.PubMed/NCBI
|
|
13
|
Mao JH, Zhou RP, Peng AF, Liu ZL, Huang
SH, Long XH and Shu Y: microRNA-195 suppresses osteosarcoma cell
invasion and migration in vitro by targeting FASN. Oncol Lett.
4:1125–1129. 2012.PubMed/NCBI
|
|
14
|
Li Z, Wang H, Wang Z and Cai H: MiR-195
inhibits the proliferation of human cervical cancer cells by
directly targeting cyclin D1. Tumour Biol. 37:6457–6463. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu L, Chen L, Xu Y, Li R and Du X:
microRNA-195 promotes apoptosis and suppresses tumorigenicity of
human colorectal cancer cells. Biochem Biophys Res Commun.
400:236–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ,
Yao Q, Zhou H and Qu LH: MicroRNA-195 plays a tumor-suppressor role
in human glioblastoma cells by targeting signaling pathways
involved in cellular proliferation and invasion. Neuro Oncol.
14:278–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang R, Zhao N, Li S, Fang JH, Chen MX,
Yang J, Jia WH, Yuan Y and Zhuang SM: MicroRNA-195 suppresses
angiogenesis and metastasis of hepatocellular carcinoma by
inhibiting the expression of VEGF, VAV2 and CDC42. Hepatology.
58:642–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo Q, Wei C, Li X, Li J, Chen L, Huang Y,
Song H, Li D and Fang L: MicroRNA-195-5p is a potential diagnostic
and therapeutic target for breast cancer. Oncol Rep. 31:1096–1102.
2014.PubMed/NCBI
|
|
22
|
Yongchun Z, Linwei T, Xicai W, Lianhua Y,
Guangqiang Z, Ming Y, Guanjian L, Yujie L and Yunchao H:
MicroRNA-195 inhibits non-small cell lung cancer cell
proliferation, migration and invasion by targeting MYB. Cancer
Lett. 347:65–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai C, Chen QB, Han ZD, Zhang YQ, He HC,
Chen JH, Chen YR, Yang SB, Wu YD, Zeng YR, et al: miR-195 Inhibits
tumor progression by targeting RPS6KB1 in human prostate cancer.
Clin Cancer Res. 21:4922–4934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo J, Wang M and Liu X: MicroRNA-195
suppresses tumor cell proliferation and metastasis by directly
targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res.
34:912015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y,
Liu Y, Li S, Liang Z, Xu X, et al: MicroRNA-195-5p, a new regulator
of Fra-1, suppresses the migration and invasion of prostate cancer
cells. J Transl Med. 13:2892015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu C, Guan H, Wang Y, Chen M, Xu B, Zhang
L, Lu K, Tao T, Zhang X and Huang Y: miR-195 inhibits EMT by
targeting FGF2 in prostate cancer cells. PLoS One. 10:e01440732015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin Y, Wu J, Chen H, Mao Y, Liu Y, Mao Q,
Yang K, Zheng X and Xie L: Cyclin-dependent kinase 4 is a novel
target in micoRNA-195-mediated cell cycle arrest in bladder cancer
cells. FEBS Lett. 586:442–447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang X, Yu J, Yin J, Xiang Q, Tang H and
Lei X: MiR-195 regulates cell apoptosis of human hepatocellular
carcinoma cells by targeting LATS2. Pharmazie. 67:645–651.
2012.PubMed/NCBI
|
|
29
|
Jiang HL, Yu H, Ma X, Xu D, Lin GF, Ma DY
and Jin JZ: MicroRNA-195 regulates steroid receptor coactivator-3
protein expression in hepatocellular carcinoma cells. Tumour Biol.
35:6955–6960. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng C, Li J, Wang Q, Liu W, Zhou J, Liu
R, Zeng Q, Peng X, Huang C, Cao P, et al: MicroRNA-195 functions as
a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma.
Oncol Rep. 33:1115–1122. 2015.PubMed/NCBI
|
|
31
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al: Genome-wide screening
reveals that miR-195 targets the TNF-α/NF-κB pathway by
down-regulating IkappaB kinase alpha and TAB3 in hepatocellular
carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Amer M, Elhefnawi M, El-Ahwany E, Awad AF,
Gawad NA, Zada S and Tawab FM: Hsa-miR-195 targets PCMT1 in
hepatocellular carcinoma that increases tumor life span. Tumour
Biol. 35:11301–11309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Furuta M, Kozaki K, Tanimoto K, Tanaka S,
Arii S, Shimamura T, Niida A, Miyano S and Inazawa J: The
tumor-suppressive miR-497-195 cluster targets multiple cell-cycle
regulators in hepatocellular carcinoma. PLoS One. 8:e601552013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang B, Tan Z and Song Y: Study on the
molecular regulatory mechanism of MicroRNA-195 in the invasion and
metastasis of colorectal carcinoma. Int J Clin Exp Med.
8:3793–3800. 2015.PubMed/NCBI
|
|
35
|
Wang L, Qian L, Li X and Yan J:
MicroRNA-195 inhibits colorectal cancer cell proliferation,
colony-formation and invasion through targeting CARMA3. Mol Med
Rep. 10:473–478. 2014.PubMed/NCBI
|
|
36
|
Tashkandi H, Shah N, Patel Y and Chen H:
Identification of new miRNA biomarkers associated with
HER2-positive breast cancers. Oncoscience. 2:924–929.
2015.PubMed/NCBI
|
|
37
|
Zhang Y, Zhang D, Wang F, Xu D, Guo Y and
Cui W: Serum miRNAs panel (miR-16-2*, miR-195, miR-2861, miR-497)
as novel non-invasive biomarkers for detection of cervical cancer.
Sci Rep. 5:179422015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao FL, Dou YC, Wang XF, Han DC, Lv ZG,
Ge SL and Zhang YK: Serum microRNA-195 is down-regulated in breast
cancer: A potential marker for the diagnosis of breast cancer. Mol
Biol Rep. 41:5913–5922. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cai H, Zhao H, Tang J and Wu H: Serum
miR-195 is a diagnostic and prognostic marker for osteosarcoma. J
Surg Res. 194:505–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soon PS, Tacon LJ, Gill AJ, Bambach CP,
Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson
BG and Sidhu SB: miR-195 and miR-483-5p identified as predictors of
poor prognosis in adrenocortical cancer. Clin Cancer Res.
15:7684–7692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang X, Yin J, Yu J, Xiang Q, Liu Y, Tang
S, Liao D, Zhu B, Zu X, Tang H and Lei X: miRNA-195 sensitizes
human hepatocellular carcinoma cells to 5-FU by targeting BCL-w.
Oncol Rep. 27:250–257. 2012.PubMed/NCBI
|
|
42
|
Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W,
Jiang X, Zhang C and Qu J: MicroRNA-195 chemosensitizes colon
cancer cells to the chemotherapeutic drug doxorubicin by targeting
the first binding site of BCL2L2 mRNA. J Cell Physiol. 230:535–545.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian
J and Weng Y: Upregulation of miR-195 increases the sensitivity of
breast cancer cells to Adriamycin treatment through inhibition of
Raf-1. Oncol Rep. 30:877–889. 2013.PubMed/NCBI
|
|
44
|
Zhu J, Ye Q, Chang L, Xiong W, He Q and Li
W: Upregulation of miR-195 enhances the radiosensitivity of breast
cancer cells through the inhibition of BCL-2. Int J Clin Exp Med.
8:9142–9148. 2015.PubMed/NCBI
|