Introduction
Ginsenoside Rg3, which has the chemical name of 12β,
20-dihydroxydammar-24-en-3β-yl2-O-β-D-glucopyranosyl-β-
D-glucopyranoside, and ginsenoside Rg3 exist in two forms, the R
type and S type, according to its asymmetric carbon atom (choral
carbon atom) C20. Ginsenoside Rg3 has diverse pharmacological
effects in vitro and in vivo via the activation or
repression of the expression of different genes. Previous studies
have reported that ginsenoside Rg3 inhibits cancer cell
proliferation and induces apoptosis by decreasing the expression of
histone deacetylase 3 (1),
epidermal growth factor receptor (2), fucosyltransferase IV (3) and vascular endothelial growth factor
(VEGF) (4), and upregulates the
protein expression of pro-apoptotic P53 (1), caspase-3, caspase-8 and caspase-9
(5). Ginsenoside Rg3 has been
shown to enhance the radiosensitivity of human esophageal carcinoma
cells by downregulating the expression of VEGF and hypoxia
inducible factor-1α (6), and
ginsenoside Rg3 may have a neuroprotective function in the rat
hippocampus via the inhibition of hippocampus-mediated
N-methyl-D-aspartate receptor activation (7). Additional biological activities
associated with gene regulation have also been demonstrated for
this molecule (2,8).
5-methyl-cytosine (m5Cyt) is the most investigated
epigenetic modification, and alterations in genomic DNA methylation
patterns have been confirmed to be associated with the development
of pathological processes and diseases, including cancer (9,10).
It has also been reported that DNA methylation patterns can be
altered by medication, nutrients and chemicals (11–13).
DNA methyltransferases (DNMTs) are enzymes responsible for
establishing the original methylation patterns of de novo
methylases, DNMT3a and DNMT3b, and for maintaining these throughout
subsequent cellular divisions (maintenance methylase DNMT1).
However, until now, there have been no reports on whether
ginsenoside Rg3 affects DNA methylation or the expression of
methyltransferases.
Global hypomethylation and promoter hypermethylation
have been found in hepatocarcinogenesis (14–16);
therefore, the present study investigated the effects of
ginsenoside Rg3 on methylation in the HepG2 human hepatocarcinoma
cell line. The results showed that ginsenoside Rg3 inhibited HepG2
cell proliferation in a dose-dependent manner, induced a reduction
in global DNA methylation, altered methylated cystosines in the
promoter regions of specific genes, upregulated the expression of
DNMT1, and downregulated the expression of DNMT3a and DNMT 3b. In
addition, the different ginsenoside Rg3 epimers exhibited different
biological activities.
Materials and methods
Reagents
High performance liquid chromatography (HPLC)-grade
standards for 20(S)-ginsenoside Rg3 and 20(R)-ginsenoside Rg3 were
purchased from Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai,
China) and dissolved at a concentration of 50 mg/ml in dimethyl
sulfoxide as a stock solution (stored at −20°C). This solution was
then further diluted in cell culture medium to prepare the working
concentrations (0, 7.81, 15.63, 31.25, 62.5, 125, 250 and 500
µg/ml). 5-methyl-2′-deoxycytidine (5-MedC) was purchased from
United States Biological (Salem, MA, USA), and 2′-deoxyguanosine
(dG), thymidine (T), 2′-deoxycytidine (dC) and 2′-deoxyadenosine
(dA) were obtained from Sigma-Aldrich; Merck Millipore (Darmstadt,
Germany). Sodium acetate, ammonium formate and acetonitrile were
supplied by Beijing Chemical Works (Beijing, China). MNase,
nuclease P1 and alkaline phosphatase were purchased from Takara
Bio, Inc. (Otsu, Japan).
Analysis of cell viability
Cell proliferation was measured using a Cell
Counting Kit-8 (CCK8; Beyotime Institute of Biotechnology, Inc.,
Jiangsu, China). Briefly, cells of the human hepatocarcinoma cell
line HepG2 were cultured in RPMI-1640 supplemented with 10% fetal
bovine serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and 1% antibiotics-antimycotics in a humidified 5%
CO2 atmosphere at 37°C. Exponentially growing cells were
seeded in a 96-well plate at a density of 1.0×104
cells/well. The following day, the cells were treated in triplicate
with the various concentrations of 20(S)-ginsenoside Rg3 or
20(R)-ginsenoside Rg3 for 24 h at 37°C. Following incubation for 24
h, cell viability was assessed using the CCK8 assay, according to
the manufacturer's protocol. Finally, the absorbance value was
recorded at an optical density of 450 nm (OD450) using
an enzyme-linked immunosorbent assay plate reader (Emax Plus;
Molecular Devices, LLC, Sunnyvale, CA, USA). At least three
independent experiments were performed, and the experiments
involved three groups: Untreated HepG2 cells (not treated with
ginsenoside Rg3), HepG2 cells treated with various concentrations
of 20(S)-ginsenoside Rg3 or 20(R)-ginsenoside Rg3 as the
experimental group, and a blank group of cell culture only. The
inhibition rate was calculated using the following formula:
[(untreated group OD450-experimental group
OD450)/(untreated group OD450-blank group
OD450)]x100%.
Measurement of global DNA methylation
using HPLC
HPLC analysis was performed on a liquid
chromatograph coupled with a Waters 2489 UV/visible detector
(Waters Corporation, Milford, MA, USA). Separation was achieved on
a Symmetry® C18 (5 µm; 4.6×250) column (Waters
Corporation). The mobile phase consisted of solvent A (50 mM
ammonium formate; pH 5.4) and solvent B (HPLC-grade acetonitrile).
The following gradient program used was: Solvent B 2% at 0 min,
solvent B 3% at 18 min, solvent B 27% at 25 min, solvent B 35% at
40 min and solvent B 2% at 45 min. The total duration of analysis
was 50 min. The flow rate was set at 0.2 ml/min and the column oven
temperature was maintained at 25°C. UV detection was performed at
277 nm.
The concentrations of MedC and dC were calculated
from linear regression curves, and the percentage of methylation
was then calculated using the following equation: 5-MedC
(%)=[5-MedC/(dC+MedC)]x100.
Preparation of standards
The 5-MedC, dC, dA, dT and dG stock standard
solutions were prepared by diluting purchased powder in appropriate
volumes of HPLC grade water. All stock solutions were then mixed at
an appropriate ratio to ensure all components were present at the
same concentrations: 0.009765, 0.039625, 0.156250, 0.625000, 2.5
and 10 mM for 5-MedCand dC. All standard solutions were stored at
−20°C and used within 1 week.
DNA extraction and hydrolysis
Exponentially growing HepG2 cells were seeded at a
density 5.0×105 into 75 cm2 culture flasks
and, following a 24 h period, the cells were either left untreated
or were treated with 20(S)-ginsenoside Rg3 or 20(R)-ginsenoside
Rg3, in accordance with the cell viability assessment. The cells
were then collected, and DNA was purified using a Qiagen
DNAeasy® blood and tissue kit (Qiagen GmbH, Hilden,
Germany).
Cryonase™ cold-active nuclease (Takara Bio, Inc.),
micrococcal nuclease (Takara Bio, Inc.) and digestion buffer
comprising 20 mM Tris-HCl, 5 mM NaCl and 2.5 mM CaCl2
(pH 8.0) were added to the purified DNA and incubated at 37°C
overnight. Alkaline phosphatase (calf intestine; 1 unit/pmol;
Takara Bio, Inc.) was added to the samples, and incubation was
continued for an additional 4 h. The hydrolyzed DNA was then
centrifuged at 18,000 × g for 20 min at 4°C and adjusted to
an appropriate concentration (~4 mM) in HPLC grade water for
analysis using HPLC.
MspI/HpaII polymerase chain reaction
(PCR) assay
The MspI/HpaII PCR assay was performed
as follows: Purified genomic DNA (2 µg) from the control and
ginsenoside Rg3-treated samples were digested with 20 units of
methylation-sensitive enzymes (MspI or HpaII) in a
20-µl reaction volume using the buffers supplied by the
manufacturer (Takara Bio, Inc.). The reaction mixtures were
incubated overnight at 37°C. The digested DNA samples were purified
with phenol:chloroform:isoamyl alcohol (25:24:1) and chloroform
extraction, followed by ethanol precipitation.
PCR was performed with the undigested and digested
DNA from the control and ginsenoside Rg3-treated samples. A total
of 40 primers were designed based on the sequences of the promoter
regions of Homo sapiens B cell lymphoma 2 (Bcl2; NIH
accession no. EU119400; www.ncbi.nlm.nih.gov/nuccore/EU119400), Homo
sapiens VEGF gene (NIH accession no. AF095785; www.ncbi.nlm.nih.gov/nuccore/AF095785) and Homo
sapiens tumor protein P53 (TP53; NIH accession no. NG_017013;
www.ncbi.nlm.nih.gov/nuccore/NG_017013). Each set of
primers spanned one or more CCGG sites of the target gene. The
primer names, primer sequences and positions of the CCGG sequence
for each gene are listed in Table
I. Each reaction mixture contained 1 µl DNA (~10 ng), 10 µl
Premix Taq™ DNA Polymerase Hot-Start (Takara Bio, Inc.), 1 µl DNA
primer (0.5 µM) and 7 µl ddH2O. The reaction was
denatured at 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec, 55°C for 45 sec and 72°C for 1 min, with a final incubation
step at 72°C for 5 min. Following agarose gel electrophoretic
separation and staining with ethidium bromide, the amplified
products were visualized under UV irradiation. Images were captured
for further analysis.
 | Table I.Primers. |
Table I.
Primers.
| Primer | Primer sequence
(3′-5′) | Position (bp) | Gene | NCBI accession
no. |
|---|
| XZ-82 |
cagagtcacctgtcttcacag | 1617–1637 | BCL2 | EU119400 |
| XZ-83 |
tctagccgtgtatgagagtgtg | 1750–1772 | BCL2 | EU119400 |
| XZ-84 |
acacactctcatacacggctag | 1750–1751 | BCL2 | EU119400 |
| XZ-85 |
cgccatgaaaacaagggctg | 1915–1934 | BCL2 | EU119400 |
| XZ-86 |
cagcccttgttttcatggcg | 1915–1934 | BCL2 | EU119400 |
| XZ-87 |
gccttctgctcaggcctg | 2060–2077 | BCL2 | EU119400 |
| XZ-88 |
caggcctgagcagaaggc | 2060–2077 | BCL2 | EU119400 |
| XZ-89 |
gcccgctccgctgcgc | 2154–2169 | BCL2 | EU119400 |
| XZ-90 |
gcgcagcggagcgggc | 2154–2169 | BCL2 | EU119400 |
| XZ-91 |
gttaaaggcgccgcggcag | 2250–2268 | BCL2 | EU119400 |
| XZ-94 |
gaaccgtgtgacgttacgcac | 2344–2364 | BCL2 | EU119400 |
| XZ-95 |
caccttcgctggcagcg | 2528–2544 | BCL2 | EU119400 |
| XZ-96 |
caggaggaggagaaagggtg | 2647–2666 | BCL2 | EU119400 |
| XZ-97 |
ggatgactgctacgaagttctc | 2724–2745 | BCL2 | EU119400 |
| XZ-98 |
gcttctagcgctcggcac | 2828–2845 | BCL2 | EU119400 |
| XZ-99 |
gacggaggcaggaatcctc | 2926–2944 | BCL2 | EU119400 |
| XZ-100 |
gaggattcctgcctccgtc | 2926–2944 | BCL2 | EU119400 |
| XZ-101 |
gcacaggcatgaatctctatccac | 3006–3028 | BCL2 | EU119400 |
| XZ-102 |
gtggatagagattcatgcctgtg | 3006–3028 | BCL2 | EU119400 |
| XZ-103 |
gcggcggcagatgaattac | 3109–3127 | BCL2 | EU119400 |
| XZ-104 |
tctcgagctcttgagatctc | 3144–3163 | BCL2 | EU119400 |
| XZ-105 |
gattcccagacttctgcttcac | 3194–3215 | BCL2 | EU119400 |
| XZ-106 |
gccagactccacagtgcatac | 1370–1390 | VEGF | AF095785 |
| XZ-107 |
ctgagaacgggaagctgtgtg | 1442–1462 | VEGF | AF095785 |
| XZ-108 |
ccattccctctttagccagag | 1743–1763 | VEGF | AF095785 |
| XZ-109 |
cattcacccagcttccctgtg | 1806–1826 | VEGF | AF095785 |
| XZ-110 |
cactccaggattccaacagatc | 1930–1951 | VEGF | AF095785 |
| XZ-111 |
gagccgttccctctttgctag | 2017–2037 | VEGF | AF095785 |
| XZ-112 |
acgtaacctcactttcctgctc | 2053–2074 | VEGF | AF095785 |
| XZ-113 |
ccaccaaggttcacagcctg | 2142–2161 | VEGF | AF095785 |
| XZ-114 |
caggcttcactgggcgtc | 2199–2216 | VEGF | AF095785 |
| XZ-115 |
agcctcagcccctccacac | 2240–2258 | VEGF | AF095785 |
| XZ-116 |
tgtggaggggctgaggctc | 2241–2259 | VEGF | AF095785 |
| XZ-117 |
gatcctccccgctaccag | 2347–2364 | VEGF | AF095785 |
| XZ-118 |
ctggtagcggggaggatc | 2347–2364 | VEGF | AF095785 |
| XZ-119 |
gaatatcaaattccagcaccgag | 2483–2505 | VEGF | AF095785 |
| XZ-120 |
cggtgctggaatttgatattcattg | 2485–2509 | VEGF | AF095785 |
| XZ-121 |
aagccgtcggcccgattc | 2606–2623 | VEGF | AF095785 |
| XZ-162 |
ctccatttcctttgcttcctc | 4908–4928 | P53 | NG_017013 |
| XZ-163 |
ctggcacaaagctggacagtc | 4964–4984 | P53 | NG_017013 |
| XZ-164 |
cttctcaaaagtctagagccac | 5058–5079 | P53 | NG_017013 |
| XZ-165 |
gcgtgtcaccgtcgtggaaag | 5141–5161 | P53 | NG_017013 |
| XZ-166 |
tggagctttggggaaccttgag | 5422–5443 | P53 | NG_017013 |
| XZ-167 |
gatgtgcaaagaagtacgctttag | 5449–5472 | P53 | NG_017013 |
| XZ-168 |
ctaaagcgtacttctttgcacatc | 5449–5472 | P53 | NG_017013 |
| XZ-169 |
cagacctcaatgctttgtgcatc | 5510–5532 | P53 | NG_017013 |
| XZ-170 |
tcctagtgaaaactggggctc | 5592–5612 | P53 | NG_017013 |
| XZ-171 |
gttgtgggaccttagcagcttg | 5651–5672 | P53 | NG_017013 |
| XZ-172 |
caagctgctaaggtcccacaac | 5651–5672 | P53 | NG_017013 |
| XZ-173 |
atcgctccaggaaggacaaaggtc | 5678–5701 | P53 | NG_017013 |
| XZ-174 |
gacctttgtccttcctggagcgat | 5678–5701 | P53 | NG_017013 |
| XZ-175 |
ctggtttagcacttctcacttccac | 5761–5785 | P53 | NG_017013 |
| XZ-176 |
gtggaagtgagaagtgctaaaccag | 5761–5785 | P53 | NG_017013 |
| XZ-177 |
ggcagaaatgtaaatgtggagc | 5853–5874 | P53 | NG_017013 |
| XZ-48 |
acaaagaccaggatgagaag | 333–352 | DNMT1 | AF180682 |
| XZ-49 |
cttctcatctttctcgtctc | 576–595 | DNMT1 | AF180682 |
| XZ-52 |
cagaagcgggcaaagaacag | 659–676 | DNMT3a | AF331856 |
| XZ53 |
cttgcgcttgctgatgtagtag | 802–823 | DNMT3a | AF331856 |
| XZ-56 |
cagagtatcaggatgggaag | 965–984 | DNMT3b | AF331857 |
| XZ-57 |
agtttgtctgcagagacctc | 1132–1151 | DNMT3b | AF331857 |
| XZ-60 |
gacctcaactacatggtttac | 175–195 | GAPDH | M33197 |
| XZ-61 |
tgatgggatttccattgatg | 264–283 | GAPDH | M33197 |
| XZ-62 |
aggtgcatgtttgtgcctgtc | 875–895 | P53 | AB082923 |
| XZ-63 |
tcttgcggagattctcttcctc | 916–937 | P53 | AB082923 |
| XZ-64 |
gctgggatgcctttgtggaac | 2039–2059 | BCL2 | M13994 |
| XZ-65 |
tgagcagagtcttcagagacag | 2102–2122 | BCL2 | M13994 |
| XZ-66 |
ccaacatcaccatgcagattatg | 315–337 | VEGF | AY047581 |
| XZ-67 |
Tgctgtaggaagctcatctctc | 369–390 | VEGF | AY047581 |
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was isolated from the cultured cells using
the RNeasy® Mini kit (Qiagen GmbH) according to the
manufacturer's protocol. cDNA was synthesized using a ReverTrace
qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan) using an oligo-dT
primer, and qPCR was then performed using SYBR®-Green
Realtime PCR Master mix (Toyobo Co., Ltd.) with forward and reverse
primers for each gene. The sequences of the qPCR primers are listed
in Table I. To avoid DNA
contamination, at least one primer in each set spanned two exons.
Each qPCR reaction mixture contained 1 µl cDNA, 10 µl
SYBR® Green Real-Time PCR Master Mix (Toyobo Co., Ltd.),
1 µl DNA primer (0.5 µM) and 7 µl ddH2O. The PCR was
performed in a StepOnePlus™ (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the following thermal cycling protocol: 95°C
for 5 min, followed by 40 cycles at 95°C for 15 sec and then at
60°C for 40 sec. The relative expression level of each targeted
gene was normalized to the expression of GAPDH calculated using the
2−ΔΔCq quantification method (17).
Statistical analysis
All data were analyzed using descriptive one-way
analysis of variance using the SPSS (version 16.0) software package
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference. All data are
presented as the mean ± standard deviation.
Results
Antiproliferative effects of
20(S)-ginsenoside Rg3 are more marked, compared with those of
20(R)-ginsenoside Rg3
Ginsenoside Rg3 can exist as either the R type or S
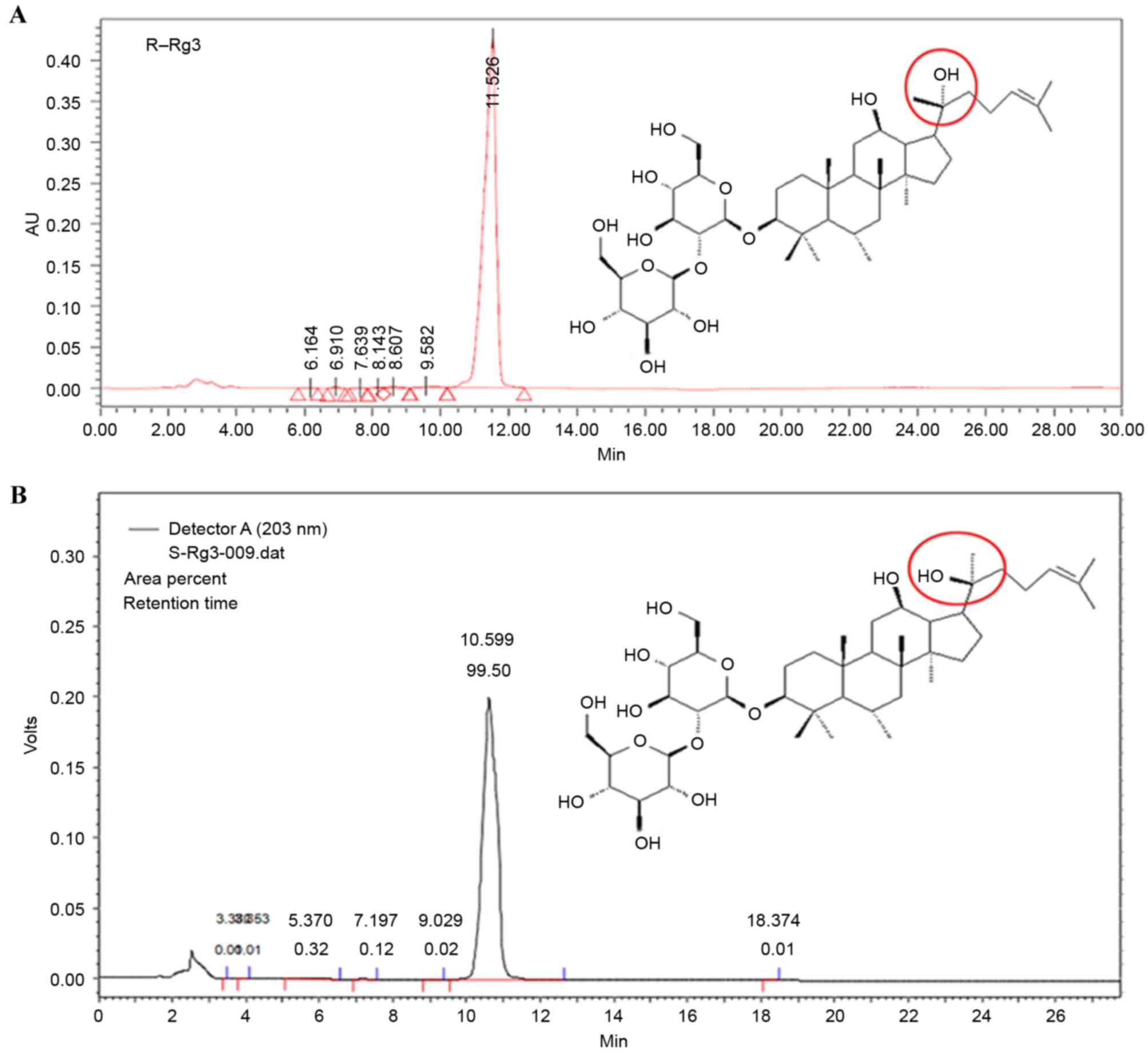
type according to its asymmetric carbon atom (Fig. 1). It has been reported that the
antiproliferative effects of the different ginsenoside Rg3 epimers
are cell line-dependent. Wu et al (18) showed that the tumor inhibition rate
achieved by 20(R)-ginsenoside Rg3 was significantly higher,
compared with that by 20(S)-ginsenoside Rg3 in hepatoma
(H22)-bearing mice. Kim et al (19) found that the antiproliferative
effect of 20(S)-ginsenoside Rg3 was higher, compared with that of
20(R)-ginsenoside Rg3 in A549 cells at the same concentration. Park
et al (20) reported that
20(S)-ginsenoside Rg3 reduced human gastric cancer cellular
proliferation, whereas 20(R)-ginsenoside Rg3 had no effect. Cheong
et al (21) showed that
20(S)-ginsenoside Rg3 had higher cytotoxic potency, compared with
the 20(R) epimer in HepG2 cells.
The present study confirmed the antiproliferative
effects of 20(R)-ginsenoside Rg3 and 20(S)-ginsenoside Rg3 on the
growth of HepG2 cells using a CCK8 assay, which offers higher
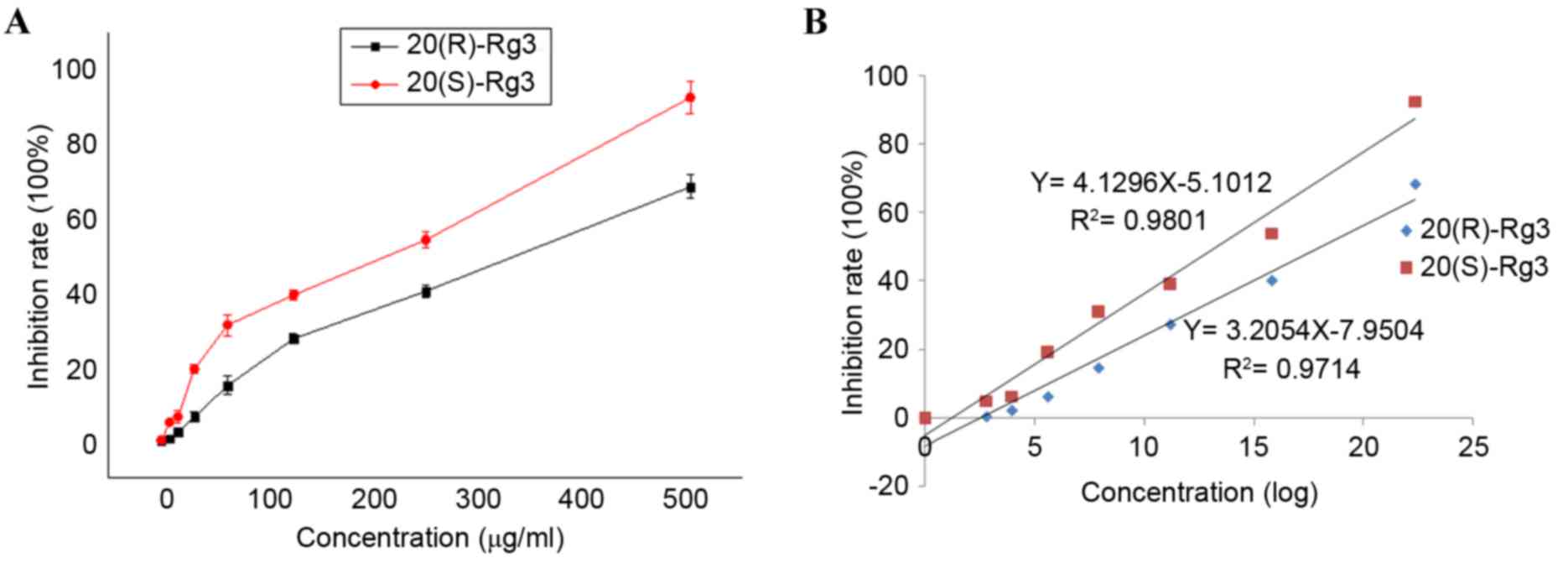
sensitivity, compared with an MTT assay. The results (Fig. 2A) showed that treatment of cells
with 20(S)-ginsenoside Rg3 over a concentration range of 15.63–500
µg/ml resulted in the inhibition of cellular proliferation, and the
antiproliferative effect of 20(R)-ginsenoside Rg3 was observed over
a concentration range of 31.25–500 µg/ml. Thus, the two epimers
inhibited cellular proliferation in a concentration-dependent
manner. As shown in Fig. 2B, the
linear regression equation for 20(S)-ginsenoside Rg3 was
Y=4.1296X-5.1012 (R2=0.9801) and the linear regression
equation for 20(R)-ginsenoside Rg3 was Y=3.2054X-7.9504
(R2=0.9714). From these values, the LD50 of
20(S)-ginsenoside Rg3 was 178.03 µg/ml, whereas that of
20(R)-ginsenoside Rg3 was 326.84 µg/ml. Therefore, treatment with
20(S)-ginsenoside Rg3 resulted in more marked inhibition of cell
growth, compared with treatment with 20(R)-ginsenoside Rg3. The
doses selected for the treatment of cells in the subsequent
experiments were 326.84 µg/ml 20(R)-ginsenoside Rg3 or 178.03 µg/ml
20(S)-ginsenoside Rg3.
Ginsenoside Rg3 induces alterations in
DNA methylation globally and in the promoter regions of specific
genes
The HPLC method used in the present study was
modified from that of Fragou et al (22). In the present study, solvent B was
changed from methanamide to acetonitrile, as acetonitrile was found
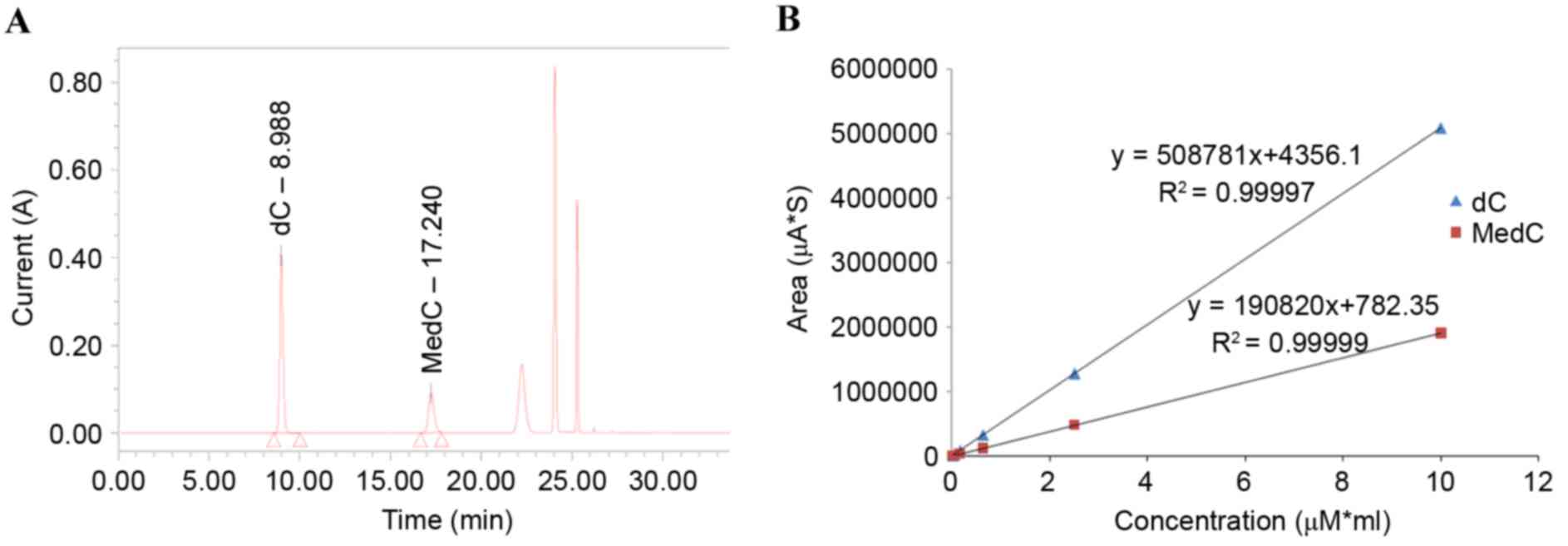
to be more effective. From the HPLC chromatogram of standards, it
was found that the C retention time was ~9 min and the 5-MedC
retention time was ~16 min (Fig.
3A). The linear regression equation for C was Y=508781X+4356.14
(R2=0.99997; Fig. 3B),
and the linear regression equation for MedC was Y=190820X+782.35
(R2=0.99999; Fig.
3B).
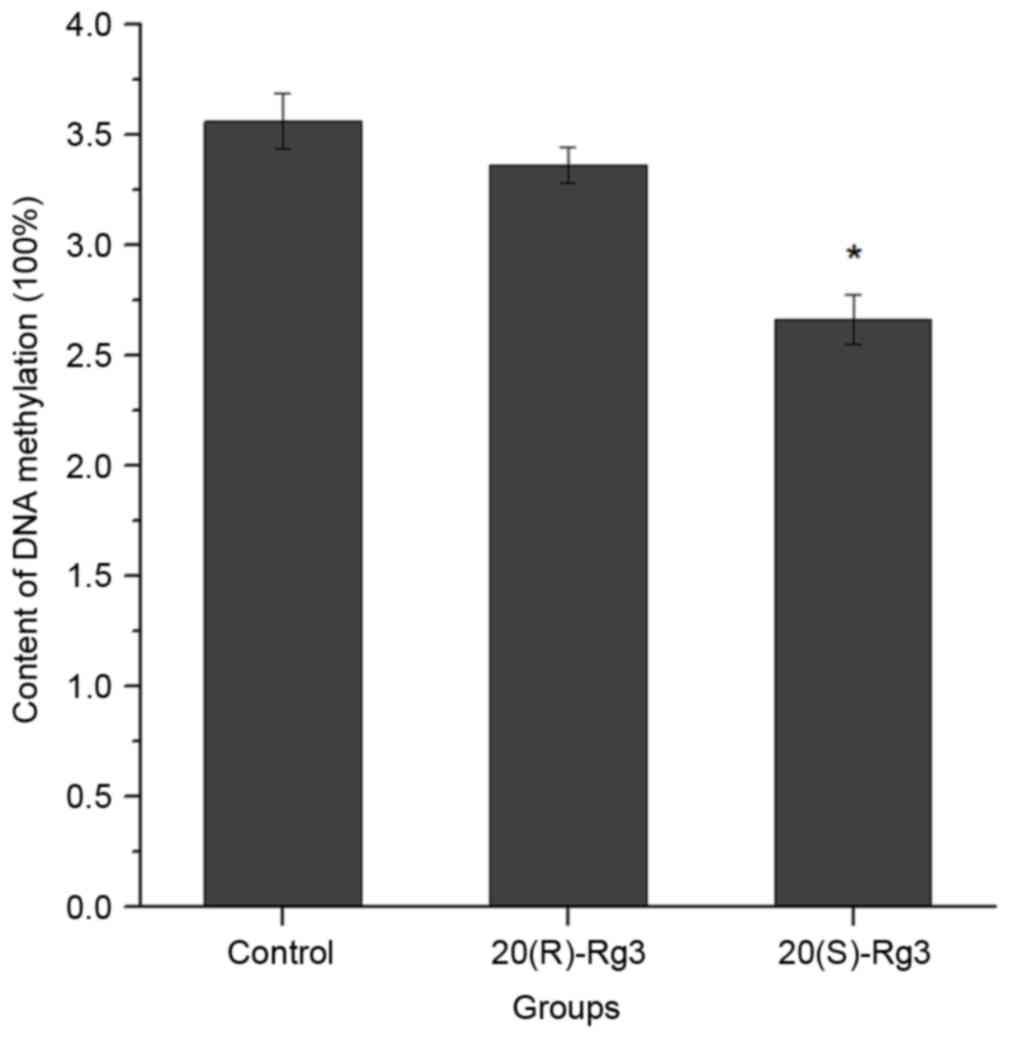
The alterations in DNA methylation in the untreated
HepG2 cells and HepG2 cells treated with ginsenoside Rg3 are shown
in Fig. 4. The data showed that
the average DNA methylation was 3.56±0.125% in the untreated cells,
3.36±0.082% in the 20(R)-ginsenoside Rg3-treated cells (P=0.115,
vs. untreated group) and 2.66±0.111% in 20(S)-ginsenoside
Rg3-treated cells (P=0.0115). The difference between DNA
methylation in the 20(R)-ginsenoside Rg3 and 20(S)-ginsenoside Rg3
groups was also statistically significant (P=0.0327).
20(R)-ginsenoside Rg3 is known to induce the global
hypermethylation of DNA; therefore, the present study examined
whether 20(R)-ginsenoside Rg3 affected the methylation of the
promoter region of a specific gene. Zhou et al (23) showed that ginsenoside Rg3 can
inhibit HepG2 cell proliferation by downregulating the expression
of VEGF. Yuan et al (24)
reported that the protein expression of anti-apoptotic Bcl2 was
downregulated and the protein expression of pro-apoptotic P53 was
upregulated by 20(S)-ginsenoside Rg3 in HT-29 colon cancer cells.
The present study obtained similar results, which showed that P53
was upregulated, and BCL2 and VEGF were downregulated by treatment
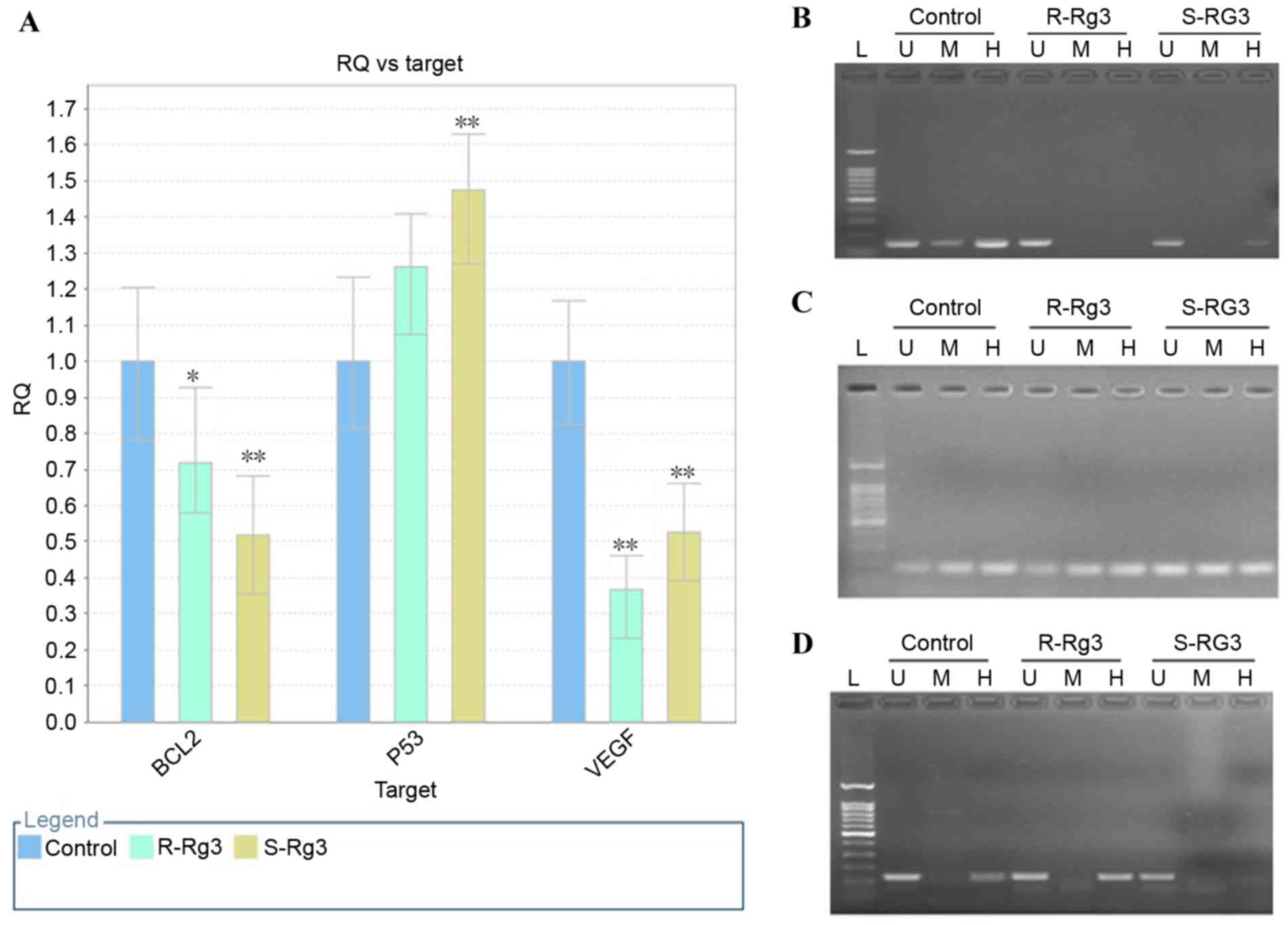
of the HepG2 cells with ginsenoside Rg3 (Fig. 5A). Therefore, the present study
examined whether the altered gene expression levels were associated
with the methylation status of the promoter. The primers used are
listed in Table I.
The results of the MspI/HpaII PCR
analysis of DNA methylation alterations in BCL2, VEGF and P53 genes
induced by ginsenoside Rg3 treatment are shown in Fig. 5. Alterations in DNA methylation at
the CCGG sequence (2371–2374 bp) were identified in the promoter of
BCL2 induced by ginsenoside Rg3, compared with the untreated
control. As shown in Fig. 5B, no
significant amplification band was detected in the
20(R)-ginsenoside Rg3-treated sample digested by HpaII or
MspI, or in the 20(S)-ginsenoside Rg3-treated sample
digested by MspI. However, a weak specific band was observed
in the 20(S)-ginsenoside Rg3-treated sample digested by
HpaII. These data indicated that the cytosine demethylation
at this site was induced by ginsenoside Rg3, and that
20(R)-ginsenoside Rg3 had a higher potential to induce methylation
at this site, compared with 20(S)-ginsenoside Rg3. Alterations in
DNA methylation at the CCGG sequence (1,981–1,984 bp) in the VEGF
promoter are shown in Fig. 5C. No
significant differences were found in the amplification profiles
among the three groups, which indicated that the DNA methylation
status was not altered following ginsenoside Rg3 treatment.
Alterations in DNA methylation at the CCGG sequence (5,641–5,644
bp) in the P53 promoter are shown in Fig. 5D, and an amplification band was
found in the untreated control and 20(R)-ginsenoside Rg3-treated
sample digested by HpaII, however, no band was observed for
the 20(S)-ginsenoside Rg3-treated sample digested by HpaII.
These data indicated that DNA methylation was decreased upon
treatment with 20(S)-ginsenoside Rg3 at this site, whereas the DNA
methylation status remained unchanged (Table II). If the band patterns remained
unchanged, compared with those of the untreated control group, it
was scored 0. If the patterns showed that cytosine methylation was
increased, it was scored 1. If the patterns showed that cytosine
methylation was decreased, it was scored 2. The results of the
MspI/HpaII PCR analysis showed that the percentages
of CCGG sites remaining unchanged were 52 and 64% following
treatment with 20(R)-ginsenoside Rg3 and 20(S)-ginsenoside Rg3,
respectively. The percentages of methylated bands were 24 and 16%
in the 20(R)-ginsenoside Rg3 and 20(S)-ginsenoside Rg3-treated
groups, whereas the percentages of demethylated bands were 24 and
20% in the 20(R)-ginsenoside Rg3- and 20(S)-ginsenoside Rg3-treated
group, respectively. From the data in Table II, it was found that the level of
DNA methylation was decreased at the promoter region of P53 and
BCL2 in cells treated with 20(S)-ginsenoside Rg3, and that the gene
expression of P53 was increased, whereas that of BCL2 was
decreased. The level of DNA methylation was increased in the
promoter region of VEGF and decreased in that of P53 upon treatment
with 20(R)-ginsenoside Rg3. In addition, the gene expression of
VEGF was decreased, whereas that of P53 was increased. These
results indicated that gene expression was not only associated with
the level of DNA methylation, but was also associated with specific
sites in the promoter region of the gene.
 | Table II.MspI/HpaII polymerase
chain reaction assay. |
Table II.
MspI/HpaII polymerase
chain reaction assay.
|
| Methylation
score |
|---|
|
|
|
|---|
| Gene | Primers used | Position of CCGG
site (bp) | R-Rg3 | S-Rg3 |
|---|
| Bcl2 | XZ-82/XZ-83 | 1701–1704;
1715–1718 | 0 | 0 |
| Bcl2 | XZ-84/XZ-85 | 1903–1906 | 0 | 0 |
| Bcl2 | XZ-86/XZ-87 | 1977–1980;
1982–1985; 2000–2003 | 1 | 1 |
| Bcl2 | XZ-88/XZ-89 | 2141–2144 | 0 | 0 |
| Bcl2 | XZ-90/XZ-91 | 2176–2179;
2181–2184; 2198–2199; 2225–2228 | 0 | 2 |
| Bcl2 | XZ-94/XZ-95 | 2371–2374 | 2 | 0 |
| Bcl2 | XZ-96/XZ-97 | 2673–2676;
2689–2692 | 0 | 0 |
| Bcl2 | XZ-98/XZ-99 | 2844–2947 | 0 | 0 |
| Bcl2 | XZ-100/XZ-101 | 2946–2949 | 1 | 2 |
| Bcl2 | XZ-102/XZ-103 | 3095–3098 | 0 | 0 |
| Bcl2 | XZ-104/XZ-105 | 3163–3166 | 2 | 0 |
| Bcl2 total |
|
| 1: 18% | 1: 9% |
|
|
|
| 2: 18% | 2: 18% |
| VEGF | XZ-106/XZ-107 | 1435–1438 | 2 | 1 |
| VEGF | XZ-110/XZ-111 | 1981–1984 | 0 | 0 |
| VEGF | XZ-112/XZ-113 | 2120–2123 | 1 | 0 |
| VEGF | XZ-114/XZ115 | 2225–2228 | 0 | 0 |
| VEGF | XZ-116/XZ-117 | 2275–2278;
2285–2288; 2299–2302 | 1 | 1 |
| VEGF | XZ-118/XZ-119 | 2384–2385 | 1 | 0 |
| VEGF | XZ-120/XZ-121 | 2512–2515 | 2 | 2 |
| VEGF total |
|
| 1: 42% | 1: 29% |
|
|
|
| 2: 29% | 2: 14% |
| P53 | XZ-162/XZ163 | 4928–4931 | 0 | 1 |
| P53 | XZ-164/XZ-165 | 5108–5111 | 2 | 2 |
| P53 | XZ-168/XZ-169 | 5476–5479 | 1 | 0 |
| P53 | XZ-170/XZ-171 | 5641–5644 | 0 | 2 |
| P53 | XZ-172/XZ-173 | 5675–5678 | 2 | 0 |
| P53 | XZ-174/XZ-175 | 5717–5720 | 0 | 0 |
| P53 | XZ-176-XZ-177 | 5831–5834 | 0 | 0 |
| P53 total |
|
| 1: 14% | 1: 14% |
|
|
|
| 2: 29% | 2: 29% |
| Overall total |
|
| 1: 24% | 1: 16% |
|
|
|
| 2: 24% | 2: 20% |
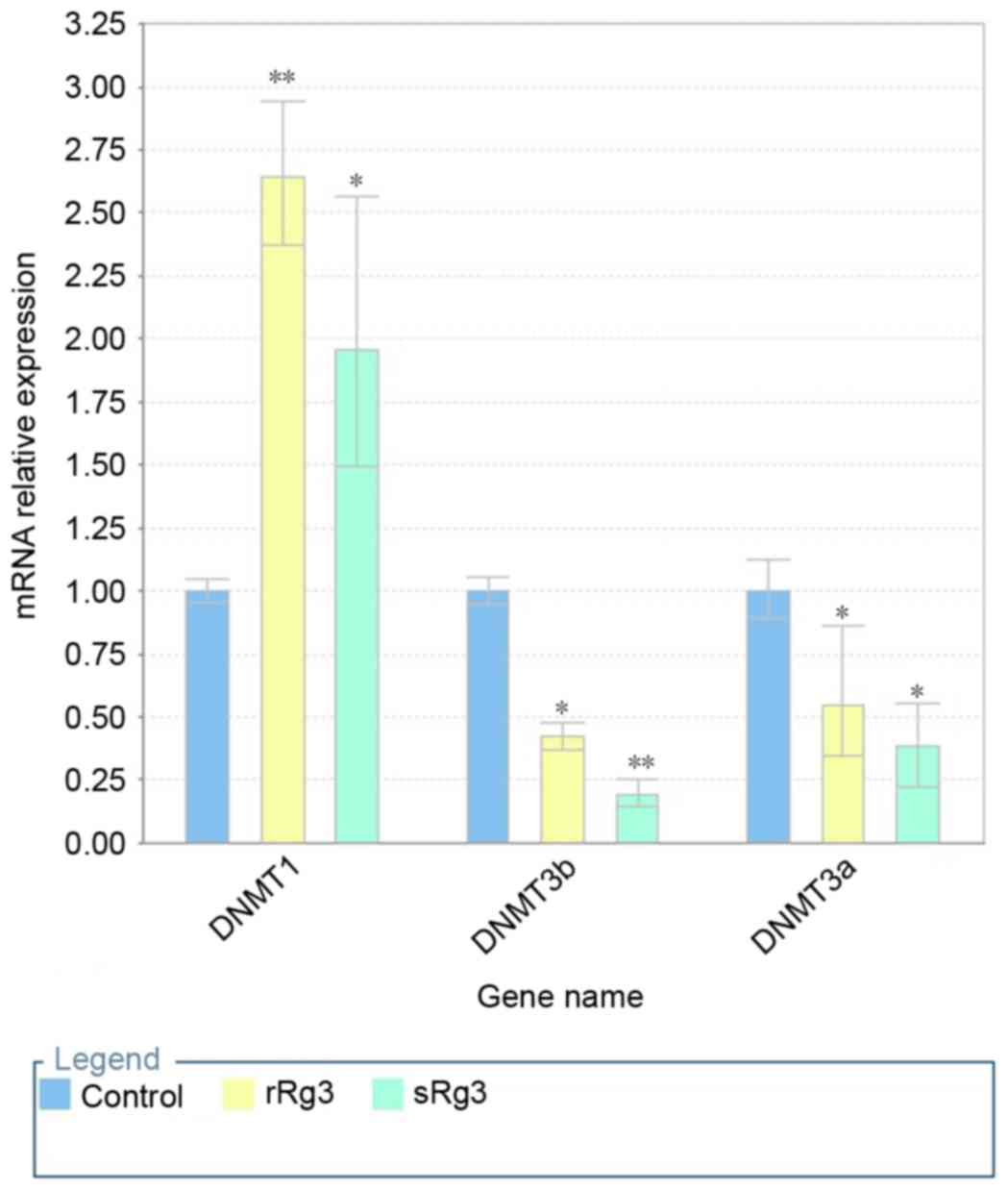
DNMT1 is upregulated, and DNMT3a and
DNMT3b are downregulated by ginsenoside Rg3
The expression levels of DNMTs are closely linked to
DNA methylation patterns. To investigate the molecular mechanism of
cytosine methylation induced by ginsenoside Rg3, the mRNA levels of
DNMT1, DNMT3a and DNAMT3b were measured using RT-qPCR analysis
following the treatment of HepG2 cells with ginsenoside Rg3 for 24
h. As shown in Fig. 6, the
exposure of HepG2 cells to ginsenosides Rg3 significantly increased
the mRNA expression of DNMT1, compared with that in the untreated
cells (P<0.001). However, the mRNA expression levels of DNMT3a
and DNMT3b showed significant decreases following exposure to
ginsenoside Rg3 (P<0.001), compared with the untreated cells,
particularly the mRNA level of DNMT3a, which was reduced to
<0.25% of that in the untreated cells. Therefore,
20(S)-ginsenoside Rg3 treatment differentially regulated the
expression of DNMT, possibly resulting in a decrease of global
demethylation.
Discussion
Several reports have demonstrated that ginsenoside
Rg3 has pharmacological actions through regulating gene expression
(25). In the present study, the
CCK8 data showed that the ability of 20(S)-ginsenoside Rg3 to
inhibit HepG2 cell growth was more marked, compared with that of
20(R)-ginsenoside Rg3. The HPLC assay also demonstrated that
20(S)-ginsenoside Rg3 induced a more marked reduction in global DNA
methylation, compared with 20(R)-ginsenoside Rg3. DNA
hypomethylation has been reported in hepatocellular carcinoma and
hypomethylation is associated with DNA damage, the promotion of
carcinogenesis and activation of tumor suppressor genes (16). Thus, DNA hypomethylation induced by
ginsenoside Rg3, particularly 20(S)-ginsenoside Rg3, may be
responsible for the inhibition of HepG2 cell proliferation.
The present study further investigated the effect of
ginsenoside Rg3 on the methylation of the promoter regions of
specific genes. In total, three genes were selected, which are
closely associated with tumor angiogenesis, growth and apoptosis.
P53 is important in apoptosis, genomic stability and the inhibition
of angiogenesis, and its expression is increased by ginsenoside-Rg3
treatment (26). VEGF stimulates
vasculogenesis and angiogenesis. The overexpression of VEGF can
cause vascular disease and the development of cancer. A decrease in
its expression is induced by 20-ginsenoside Rg3 (23). Bcl2 inhibits cell apoptosis and is
classified as an oncogene. The expression was increased by
20-ginsenoside Rg3 (5).
Classically, hypermethylation of the promoter region is associated
with gene silencing, and hypomethylation of the promoter region is
correlated with gene activation. In the present study study, it was
found that hypomethylation of the promoter region of P53 was
consistent with gene activation. DNA methylation of the promoter
regions of BCL2 were unchanged or decreased, however, the
methylated sites of the promoter region and the gene expression
were altered, suggesting that the DNA methylation level and the
specific methylated site of the promoter region may be important
for gene expression.
DNMTs are responsible for continued and de
novo DNA methylation. DNMT1 maintains the DNA methylation
status by maintaining the transfer of methyl groups to the newly
synthesized DNA strands during DNA replication. DNMT3a and DNMT3b
introduce the novel methylated cytosine site to the unmodified
cytosine residues. Oh et al (27) reported that the DNA methylation and
expression of DNMT1, DNMT3a and DNMT3b were higher in
hepatocellular carcinoma, compared with those in normal liver
tissue. In the present study, it was found that treatment with
ginsenoside Rg3 increased the relative mRNA levels of DNMT1, but
significantly reduced the levels of DNMT3a and DNMT3b. Therefore,
low expression levels of DNMT3a and DNMT3b may account for
decreased methylation, although high levels of DNMT1 expressed
during DNA replication maintained the methylation pattern.
20(S)-Ginsenoside Rg3 and 20(R)-ginsenoside Rg3 have
exhibited stereo-specific effects in several studies. For example,
Kim et al (19) showed that
20(R)-ginsenoside Rg3 regulates the expression of multiple genes
during the initiation of the transforming growth factor-β1-induced
epithelial-mesenchymal transition and suppresses lung cancer
development, whereas 20(S)-ginsenoside Rg3 does not. Park et
al (20) showed that
20(S)-ginsenoside Rg3 has more potent anticancer activity, compared
with 20(R)-ginsenoside Rg3 in inducing human gastric cancer cell
apoptosis. In the present study, 20(S)-ginsenoside Rg3 exhibited
more potent antiproliferative effects on HepG2 cells and induced a
more marked reduction in the methylation of global DNA and CpG
islands, compared with 20(R)-ginsenoside Rg3, and reduced the
expression levels of DNMT3a and DNMT3b. Thus, the different
ginsenoside Rg3 epimers exhibit different stereo-specific effects
in different types of cell, and selection of the appropriate epimer
is required for treating different diseases.
In conclusion, data obtained in the present study
indicated that ginsenoside Rg3 inhibited HepG2 cell proliferation
in a dose-dependent manner, induced a reduction in the methylation
of global genomic DNA and, for a promoter region of a specific
gene, upregulated the expression of DNMT1 and downregulated the
expression of DNMT3a and DNMT3b. In addition, 20(S)-ginsenoside Rg3
exhibited superior biological activity, compared with
20(S)-ginsenoside Rg3. These data indicated that 20(S)-ginsenoside
Rg3 may offer increased potential, compared with 20(R)-ginsenoside
Rg3, as an adjuvant treatment for hepatocellular carcinoma.
Acknowledgements
The authors would like to thank the National Natural
Science Foundation of China (grant no. 31200953) and the China
Postdoctoral Science Foundation Funded Project (grant no.
2013M530977) for financial support.
References
|
1
|
Shan X, Fu YS, Aziz F, Wang XQ, Yan Q and
Liu JW: Ginsenoside Rg3 inhibits melanoma cell proliferation
through down-regulation of histone deacetylase 3 (HDAC3) and
increase of p53 acetylation. PLoS One. 9:e1154012014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Joo EJ, Chun J, Ha YW, Ko HJ, Xu MY and
Kim YS: Novel roles of ginsenoside Rg3 in apoptosis through
downregulation of epidermal growth factor receptor. Chem Biol
Interact. 233:25–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shan X, Tian LL, Zhang YM, Wang XQ, Yan Q
and Liu JW: Ginsenoside Rg3 suppresses FUT4 expression through
inhibiting NF-κB/p65 signaling pathway to promote melanoma cell
death. Int J Oncol. 47:701–709. 2015.PubMed/NCBI
|
|
4
|
Zeng D, Wang J, Kong P, Chang C and Li J
and Li J: Ginsenoside Rg3 inhibits HIF-1α and VEGF expression in
patient with acute leukemia via inhibiting the activation of
PI3K/Akt and ERK1/2 pathways. Int J Clin Exp Pathol. 7:2172–2178.
2014.PubMed/NCBI
|
|
5
|
Luo Y, Zhang P, Zeng HQ, Lou SF and Wang
DX: Ginsenoside Rg3 induces apoptosis in human multiple myeloma
cells via the activation of Bcl-2-associated X protein. Mol Med
Rep. 12:3557–3562. 2015.PubMed/NCBI
|
|
6
|
Ge X, Zhen F, Yang B, Yang X, Cai J, Zhang
C, Zhang S, Cao Y, Ma J, Cheng H and Sun X: Ginsenoside Rg3
enhances radiosensitization of hypoxic oesophageal cancer cell
lines through vascular endothelial growth factor and hypoxia
inducible factor 1α. J Int Med Res. 42:628–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim JH, Cho SY, Lee JH, Jeong SM, Yoon IS,
Lee BH, Lee JH, Pyo MK, Lee SM, Chung JM, et al: Neuroprotective
effects of ginsenoside Rg3 against homocysteine-induced
excitotoxicity in rat hippocampus. Brain Res. 1136:190–199. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoon SJ, Park JY, Choi S, Lee JB, Jung H,
Kim TD, Yoon SR, Choi I, Shim S and Park YJ: Ginsenoside Rg3
regulates S-nitrosylation of the NLRP3 inflammasome via suppression
of iNOS. Biochem Biophys Res Commun. 463:1184–1189. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paska AV and Hudler P: Aberrant
methylation patterns in cancer: A clinical view. Biochem Med
(Zagreb). 25:161–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Poke FS, Qadi A and Holloway AF: Reversing
aberrant methylation patterns in cancer. Curr Med Chem.
17:1246–1254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Price RJ, Lillycrop KA and Burdge GC:
Folic acid supplementation in vitro induces cell type-specific
changes in BRCA1 and BRCA 2 mRNA expression, but does not alter DNA
methylation of their promoters or DNA repair. Nutr Res. 35:532–544.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gadgil MS, Joshi KS, Naik SS, Pandit AN,
Otiv SR and Patwardhan BK: Association of homocysteine with global
DNA methylation in vegetarian Indian pregnant women and neonatal
birth anthropometrics. J Matern Fetal Neonatal Med. 27:1749–1753.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sie KK, Li J, Ly A, Sohn KJ, Croxford R
and Kim YI: Effect of maternal and postweaning folic acid
supplementation on global and gene-specific DNA methylation in the
liver of the rat offspring. Mol Nutr Food Res. 57:677–685. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sceusi EL, Loose DS and Wray CJ: Clinical
implications of DNA methylation in hepatocellular carcinoma. HPB
(Oxford). 13:369–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Komatsu Y, Waku T, Iwasaki N, Ono W,
Yamaguchi C and Yanagisawa J: Global analysis of DNA methylation in
early-stage liver fibrosis. BMC Med Genomics. 5:52012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nishida N and Kudo M: Alteration of
epigenetic profile in human hepatocellular carcinoma and its
clinical implications. Liver Cancer. 3:417–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu R, Ru Q, Chen L, Ma B and Li C:
Stereospecificity of ginsenoside Rg3 in the promotion of cellular
immunity in hepatoma H22-bearing mice. J Food Sci. 79:H1430–H1435.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim YJ, Choi WI, Jeon BN, Choi KC, Kim K,
Kim TJ, Ham J, Jang HJ, Kang KS and Ko H: Stereospecific effects of
ginsenoside 20-Rg3 inhibits TGF-β1-induced epithelial-mesenchymal
transition and suppresses lung cancer migration, invasion and
anoikis resistance. Toxicology. 322:23–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Park EH, Kim YJ, Yamabe N, Park SH, Kim
HK, Jang HJ, Kim JH, Cheon GJ, Ham J and Kang KS: Stereospecific
anticancer effects of ginsenoside Rg3 epimers isolated from
heat-processed American ginseng on human gastric cancer cell. J
Ginseng Res. 38:22–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheong JH, Kim H, Hong MJ, Yang MH, Kim
JW, Yoo H, Yang H, Park JH, Sung SH, Kim HP and Kim J:
Stereoisomer-specific anticancer activities of ginsenoside Rg3 and
Rh2 in HepG2 cells: Disparity in cytotoxicity and
autophagy-inducing effects due to 20(S)-epimers. Biol Pharm Bull.
38:102–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fragou D, Zanos P, Kouidou S, Njau S,
Kitchen I, Bailey A and Kovatsi L: Effect of chronic heroin and
cocaine administration on global DNA methylation in brain and
liver. Toxicol Lett. 218:260–265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou B, Wang J and Yan Z: Ginsenoside Rg3
attenuates hepatoma VEGF overexpression after hepatic artery
embolization in an orthotopic transplantation hepatocellular
carcinoma rat model. Onco Targets Ther. 7:1945–1954. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan HD, Quan HY, Zhang Y, Kim SH and
Chung SH: 20(S)-Ginsenoside Rg3-induced apoptosis in HT-29 colon
cancer cells is associated with AMPK signaling pathway. Mol Med
Rep. 3:825–831. 2010.PubMed/NCBI
|
|
25
|
Lü JM, Yao Q and Chen C: Ginseng
compounds: An update on their molecular mechanisms and medical
applications. Curr Vasc Pharmacol. 7:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang F, Li M, Wu X, Hu Y, Cao Y, Wang X,
Xiang S, Li H, Jiang L, Tan Z, et al: 20(S)-ginsenoside Rg3
promotes senescence and apoptosis in gallbladder cancer cells via
the p53 pathway. Drug Des Devel Ther. 9:3969–3987. 2015.PubMed/NCBI
|
|
27
|
Oh BK, Kim H, Park HJ, Shim YH, Choi J,
Park C and Park YN: DNA methyltransferase expression and DNA
methylation in human hepatocellular carcinoma and their
clinicopathological correlation. Int J Mol Med. 20:65–73.
2007.PubMed/NCBI
|