Introduction
Glioma a primary tumor of the central nervous system
that is associated with the highest levels of morbidity and
mortality, and accounts for ~45% of intracranial malignant tumors
(1). Glioma exhibits increased
levels of invasive growth, and is prone to invasion and metastasis
(2,3). At present, it is impossible to
achieve total removal of the tumor, and following resection, poor
prognosis and high recurrence rates are common (3). Due to its resistance to radiotherapy
and chemotherapy, the 5-year survival rate of glioma is <5%.
Glioma cannot be completely cured and available treatment options
are also a concern (4,5).
IQ motif containing GTPase activating protein 1
(IQGAP1) was initially cloned in 1994. It is a 189-kDa scaffolding
protein, which belongs to the IQGAP family, alongside its homologs
IQGAP2 and IQGAP3 (6). IQGAP1
contains several protein-interacting domains, including one
polyproline-binding domain, one calponin homology domain, one
Ras-GTPase-activating protein (GAP)-related domain and four
calmodulin-binding motifs (7,8).
Numerous proteins, including the GTP-bound forms of Ras-related C3
botulinum toxin substrate 1 (Rac1) and cell division cycle 42
(Cdc42), and the GDP-bound form of GTPases, are able to bind to
IQGAP1 (9,10). IQGAP1 increases the levels of
active Rac1 in cells (11).
Notably, despite one domain of IQGAP1 that presents sequence
similarity to GAP, IQGAP1 does not possess GAP activity (12). In addition, IQGAP1 binds directly
to E-cadherin (13,14); a previous study reported that
IQGAP1 is associated with the modulation of several cellular
functions and various signaling pathways (7). Notably, numerous IQGAP1 binding
partners have well-defined roles in tumorigenesis (15). Overexpression of IQGAP1 has been
demonstrated to increase proliferation of MCF-7 cells (16) and reduce E-cadherin-mediated
adhesion (14). Conversely,
silencing of IQGAP1 inhibits the invasion of HO-8910PM ovarian
cancer cells (17), and small
interfering (si)RNA-induced knockdown of IQGAP1 reduces the
migration of U87MG human glioblastoma cells (18). These previous studies indicated
that IQGAP1 expression levels are frequently altered in neoplasia;
therefore, IQGAP1 has been proposed as an oncogene (16).
The present study explored the effects of IQGAP1 on
the proliferation and metastasis of U251 and U373 glioma cell
lines. IQGAP1 was shown to be more highly expressed in glioma
tissues compared with in normal tissues. Conversely, IQGAP1-siRNA
significantly inhibited cell proliferation. Furthermore, cell
adhesion, migration and invasion were markedly suppressed following
IQGAP1-siRNA transfection. The expression levels of matrix
metalloproteinase (MMP)2, Snail, MMP9, fibronectin 1 (FN1) and
Twist were suppressed, and E-cadherin was upregulated. The present
study provided insight into the function of IQGAP1 in glioma cell
biology, and indicated that it may be considered an oncogene and a
novel target for glioma treatment.
Materials and methods
Tissue samples
Human normal brain tissues (n=21) and glioma tissues
(n=26) were provided by the Department of Neurosurgery, Wuhan
General Hospital of Guangzhou Command (Wuhan, China). Sample
acquisitions were approved by the Medical Ethics Committee,
Hospital. All samples were cut into small pieces, and were quickly
placed in cryotubes with liquid nitrogen. All glioma tissues were
histopathologically confirmed, and the pathological diagnosis and
classification was determined in accordance with the World Health
Organization classification of tumors of the nervous system
(19). Normal brain tissues were
depressurized resection specimens. The current study was approved
by the ethics committee of Wuhan General Hospital of Guangzhou
Command (Wuhan, China).
Cell culture
The following five human glioma cell lines: U251,
T98G, SHG44, U87 and U373 were obtained from the Shanghai Cell
Bank, Chinese Academy of Science (Shanghai, China). All cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE
Healthcare, Logan, UT, USA), supplemented with 100 µg/ml
streptomycin, 10% heat-inactivated fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 100 U/ml
penicillin. The cells were cultured in an incubator (Thermo Fisher
Scientific, Inc.) at 37°C in an atmosphere containing 100% humidity
and 5% CO2.
siRNA transfection
U251 and U373 cells in the logarithmic growth phase
were collected, trypsin-digested, counted and plated into 6-well
culture plates (1 ml/well, 5×105 cells/ml). The cells
were then seeded in antibiotic-free medium the day prior to
transfection. Subsequently, cells were transfected with 400 nmol/l
IQGAP1-siRNA or negative control (NC) siRNA (Shanghai GenePharma
Co., Ltd., Shanghai, China) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For IQGAP1-siRNA, RNA oligomers were
synthesized in the sense and anti-sense directions, containing 19
nucleotides, corresponding to human IQGAP1 at nucleotides
1,620–1,642 (5′-GGCACAUGCAGAGAAUAAU-3′). After 24, 48 and 72 h, the
cells were collected; cells were also collected at 0 h as a control
for the proliferation assay. Cells collected 48 h post-transfection
were used to conduct the subsequent cell adhesion assay, Transwell
assay, reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analysis.
Cell growth and proliferation
assay
Cell Counting kit (CCK)-8 (Shanghai Tongren
Pharmaceutical Co., Ltd., Shanghai, China) was used to assess the
effects of IQGAP1-siRNA on the viability of U251 and U373 cells.
Briefly, after transfcetion for 0, 24, 48 and 72 h, CCK-8 reagent
[1:10 (v/v) per 100 µl medium] was added to each well and incubated
for 4 h at 37°C. Following incubation, optical density (OD) of the
supernatants was determined at 450 nm using a microplate reader.
Experiments were performed at least three times, each in
triplicate. Cell viability was exhibited by the OD value. Cell
proliferation rate (%) was calculated as follows: (OD value of NC
or IQGAP1-siRNA group/OD value of control group) ×100.
Adhesion assays
Adhesion assays, as well as Transwell assays, were
performed to detect the effects of IQGAP1-siRNA on the metastasis
of U251 and U373 cells. Suspensions of transfected cells were added
to a 12-well plate (1×105 cells/well), incubated for 1 h
at 37°C and 5% CO2, and centrifuged at 1,000 × g for 5
min at room temperature. Subsequently, the supernatant was
discarded, the cells were washed twice with PBS, and the adherent
cells were fixed with 4% methanol for 15 min and stained with
crystal violet for 30 min at room temperature. The number of
adherent cells from three random fields was counted and images were
captured under a microscope (x200 magnification).
Transwell assay
Cell migration and invasion were assessed using
Transwell assays. For the cell migration assay, the various U251
and U373 cell groups were starved in serum-free DMEM for 24 h.
Prior to seeding, cells were digested with 0.25% trypsin (Gibco;
Thermo Fisher Scientific, Inc.), resuspended in DMEM containing 1%
FBS and diluted to 1×105 cells/ml; the lower chamber of
the Transwell contained DMEM with 5% FBS. After a 48 h incubation
at 37°C in the upper chamber of the Transwell in a 24-well plate,
cells were fixed for 10 min using 1 ml/well 4% paraformaldehyde
[JRDUN Biotechnology (Shanghai) Co., Ltd., Shanghai, China],
stained with Giemsa [JRDUN Biotechnology (Shanghai) Co., Ltd.] for
30 min at room temperature and washed three times with 1X PBS.
Subsequently, the Transwell chamber was wiped carefully to remove
non-migrated cells using a cotton swab. The chambers were then
visualized under a microscope [Olympus (China) Co., Ltd., Shanghai,
China; ×200 magnification] and the number of cells was counted.
For the cell invasion assay, prior to assessment,
the Transwell chamber (pore size, 8 µm; 24-wells; Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany) was washed with 1X PBS for 5
min, and the inserts were coated with 80 µl Matrigel (1:2 dilution;
BD Biosciences, Franklin Lakes, NJ, USA). The cells
(1×105 cells/ml density) were then added to the upper
Transwell chamber in 0.5 ml serum-free medium; whereas 0.75 ml
complete medium containing 10% FBS was added to the lower chamber
as a chemoattractant. Subsequently, the cells were incubated at
37°C for 48 h. Cells with the ability to pass through the filter
were fixed and stained with 1 ml 0.5% crystal violet for 30 min.
Finally, the number of invasive cells in five randomly selected
high power fields was counted under a microscope.
RT-qPCR assay
The mRNA expression levels of IQGAP1 were detected
in U251 and U373 cells, and glioma and normal tissues, using
RT-qPCR. Total RNA was isolated from the samples using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and the obtained mRNA was detected by 0.8% agarose gel
electrophoresis. cDNA was synthesized from ~5 µg RNA using AMV
reverse transcriptase (Fermentas; Thermo Fisher Scientific, Inc.)
with a reaction mixture of 1 µl forward primer, 1 µl reverse
primer, 12.5 µl 2X Supermix, 2 µl cDNA and 8.5µl ddH2O. qPCR
reactions were performed in a 25 µl total volume using
SYBR® Green 10X Supermix (Takara Bio, Inc., Otsu, Japan)
on a Roche Light Cycler® 480II system (Roche
Diagnostics, Basel, Switzerland). Primer Express Software v3
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to
design IQGAP1 primer pairs. GAPDH was used as an internal control.
The PCR cycling conditions were as follows: 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and 60°C for 45 sec; one
cycle at 95°C for 15 sec, 60°C for 1 min; one cycle at 95°C for 15
sec and 60°C for 15 sec. The sequences of the IQGAP1 primer pairs
were as follows: IQGAP1, forward 5′-CAGAGACGTGCTATCCGTGATG-3′,
reverse 5′-CTCCGCTGATTCCGAATATCCC-3′; and GAPDH, forward
5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The size of the amplified IQGAP1
product was 209 bp. The relative expression levels in the various
groups were calculated using the ΔΔCq method by
normalizing to the mRNA expression levels of GAPDH (20). All PCR reactions were performed in
triplicate.
Western blot analysis
The association between IQGAP1 and tumor-related
protein expression was determined by western blot analysis.
Transfected cells were harvested, washed twice with PBS, lysed in
ice-cold radioimmunoprecipitation assay buffer (Beyotime Institute
of Biotechnology, Shanghai, China) containing 0.01% protease and
phosphatase inhibitor (Sigma-Aldrich; Merck Millipore), and were
incubated on ice for 30 min. The lysates were then centrifuged at
12,000 × g for 10 min at 4°C. Proteins in the supernatant were
obtained and quantified using the bicinchoninic acid protein
quantification kit (Thermo Fisher Scientific, Inc.). Protein
samples (20–30 µg) were then separated by 10% SDS-PAGE, and were
electrophoretically transferred to a polyvinylidene fluoride
membrane (Merck Millipore). The membranes were blocked with 5%
bovine serum albumin (Beyotime Institute of Biotechnology) in
PBS-0.1% Tween, and were incubated with primary antibodies against
IQGAP1 (cat. no. ab133490; 1:1,000 dilution; Abcam, Cambridge, UK),
FN1 (cat. no. ab32419; 1:1,000 dilution; Abcam), Snail (cat. no.
3879s; 1:1,000 dilution; Cell Signaling Technology, Inc., Danvers,
MA, USA), Twist (cat. no. ab175430; 1:500 dilution; Abcam), MMP2
(cat. no. ab92536; 1:1,000 dilution; Abcam), MMP9 (cat. no.
ab119906; 1:500 dilution; Abcam), E-cadherin (cat. no. 14472;
1:1,000 dilution; Cell Signaling Technology, Inc.) and GAPDH (cat.
no. 5174; 1:1,500 dilution; Cell Signaling Technology, Inc.). Blots
were then incubated for 1 h at 37°C with goat anti-mouse or
anti-rabbit secondary antibodies (cat. nos. A0192 and A0208,
respectively; 1:3,000 dilution; Beyotime Institute of
Biotechnology) and intensities were measured using enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.).
Statistical analysis
All data are presented as the mean ± standard
deviation. All statistical analyses were conducted using GraphPad
Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Data were analyzed by one-way analysis of variance without
interaction terms, followed by Dunnett's or Duncan's test for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
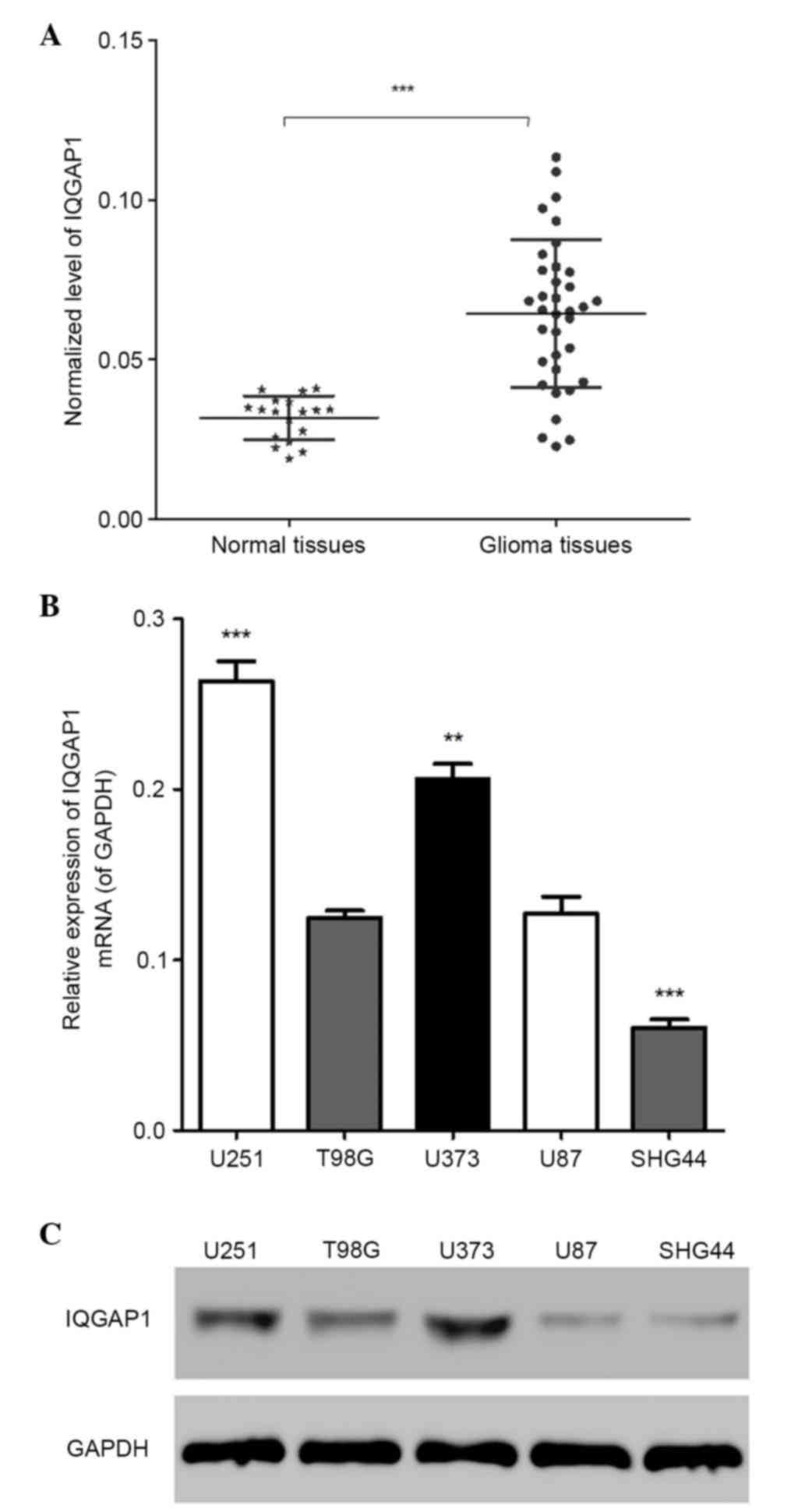
IQGAP1 is highly expressed in glioma
tissues and cell lines
In order to determine the expression levels of
IQGAP1 in normal human brain tissues and glioma tissues, an RT-qPCR
assay was performed; the results indicated that the mRNA expression
levels of IQGAP1 were significantly higher in glioma tissues
compared with in normal tissues (Fig.
1A) (n=3, P<0.01). The mRNA and protein expression levels of
IQGAP1 in the following five glioma cell lines: U251, T98G, SHG44,
U87 and U373, were detected by RT-qPCR and western blotting,
respectively. Notably, IQGAP1 was more highly expressed in U251 and
U373 cell lines compared with the others (Fig. 1B and C) (n=3, P<0.01). These
findings suggest that IQGAP1 is highly expressed in glioma tissues
and cell lines. U251 and U373 cell lines were used to conduct the
subsequent assays.
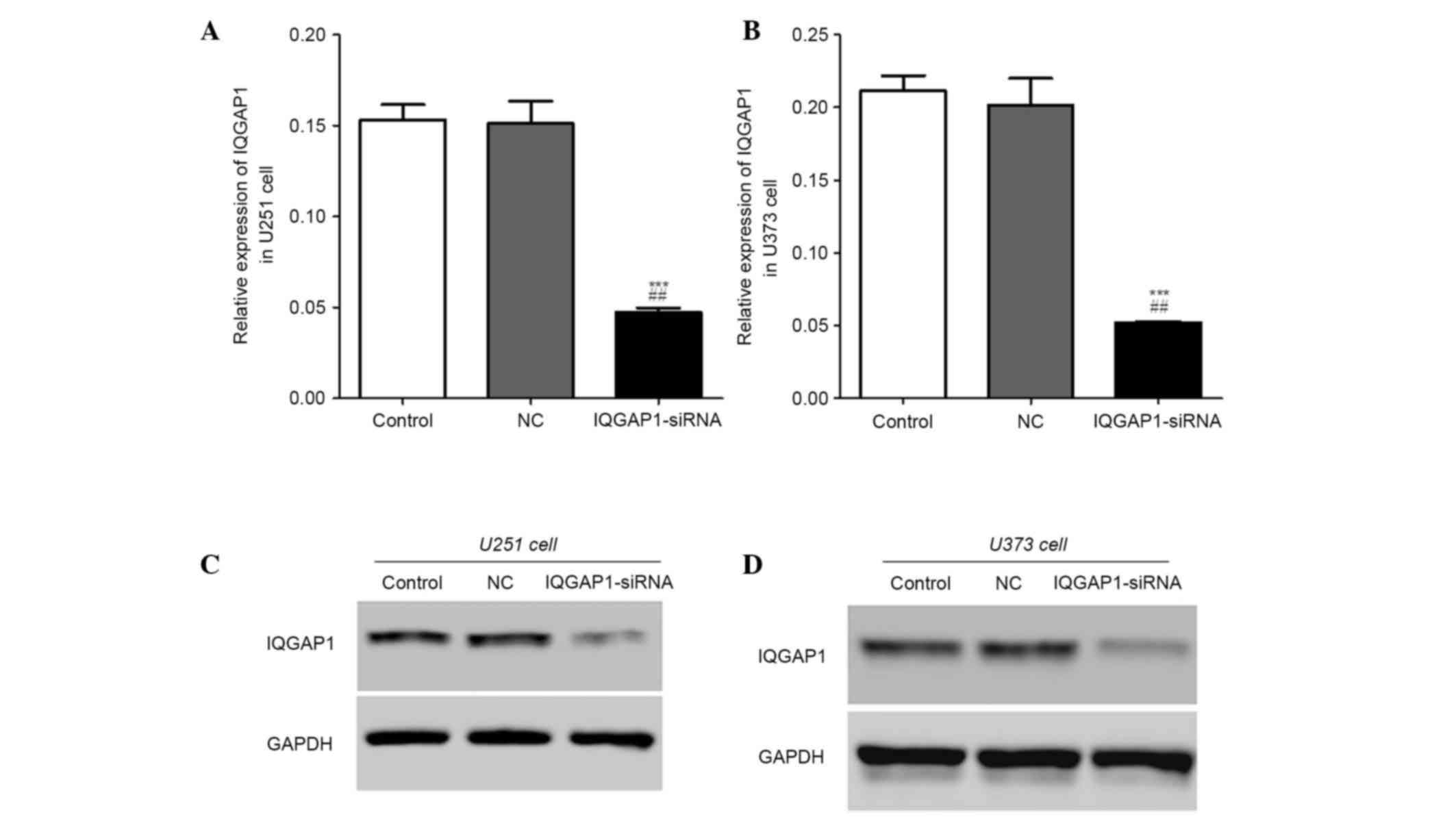
IQGAP1 expression in U251 and U373
cells post-transfection
In order to detect whether IQGAP1-siRNA and NC-siRNA
were transfected successfully, RT-qPCR and western blotting were
performed. As shown in Fig. 2A and
B, in the IQGAP1-siRNA group, the relative mRNA expression
levels of IQGAP1 were markedly decreased in U251 and U373 cells.
Similarly, the protein expression levels of IQGAP1 were
downregulated in the IQGAP1-siRNA U251 and U373 cell groups
(Fig. 2C and D). These results
indicate that IQGAP1 expression was successfully knocked down.
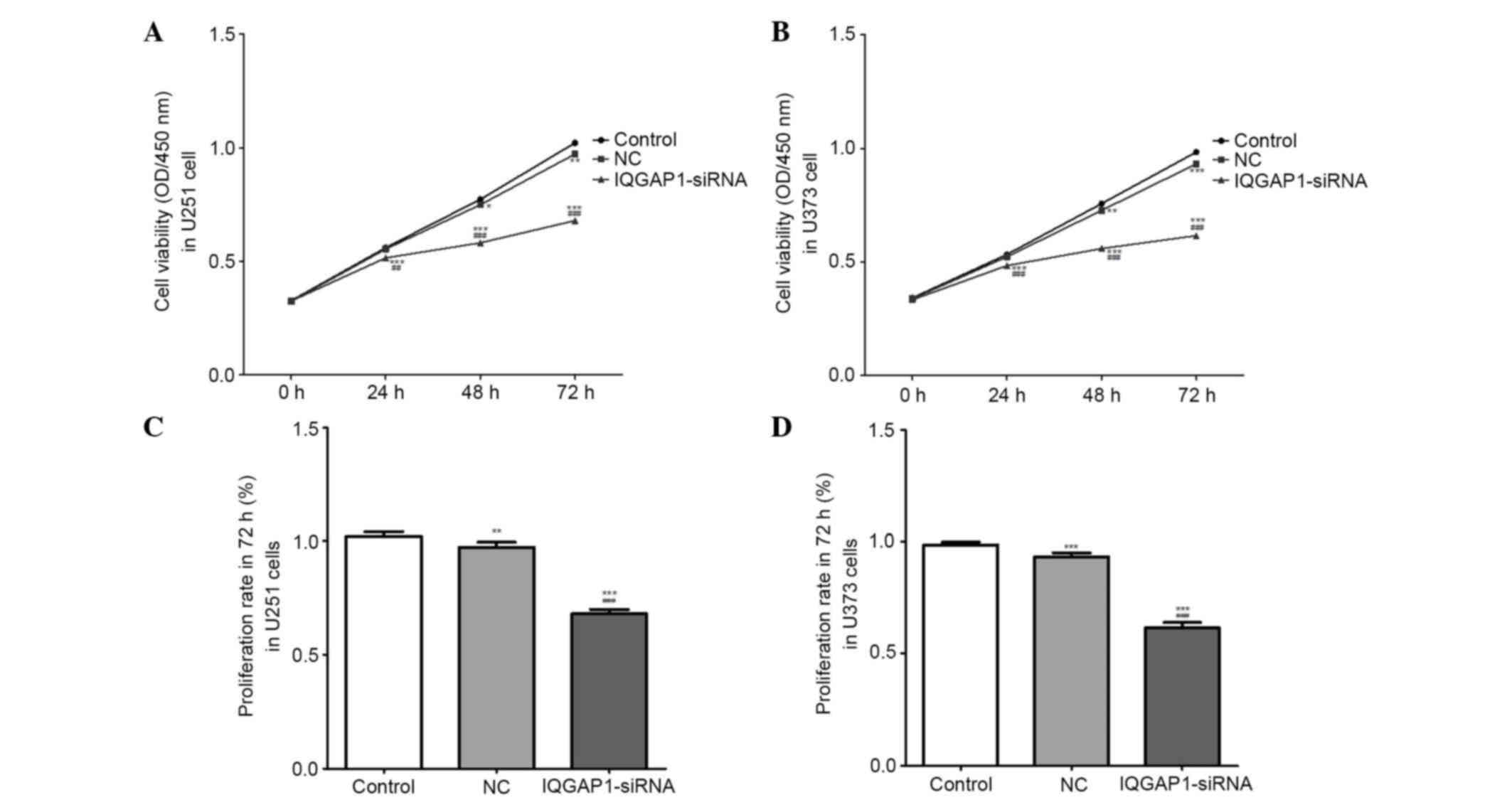
IQGAP1-siRNA inhibits cell
viability
Cell viability (OD, 450 nm) and proliferation rates
were measured using the CCK-8 assay. Following transfection with
IQGAP1-siRNA or NC-siRNA for 24, 48 or 72 h, the viability of U251
and U373 cells was markedly reduced in the IQGAP1-siRNA groups in a
time-dependent manner (n=3, P<0.01) (Fig. 3A and B). Post-transfection for 72
h, the proliferation rates of U251 and U373 cells in the
IQGAP1-siRNA group were significantly reduced compared with that of
the controls (n=3, P<0.01) (Fig. 3C
and D). These results indicate that IQGAP1-siRNA inhibits cell
viability and proliferation.
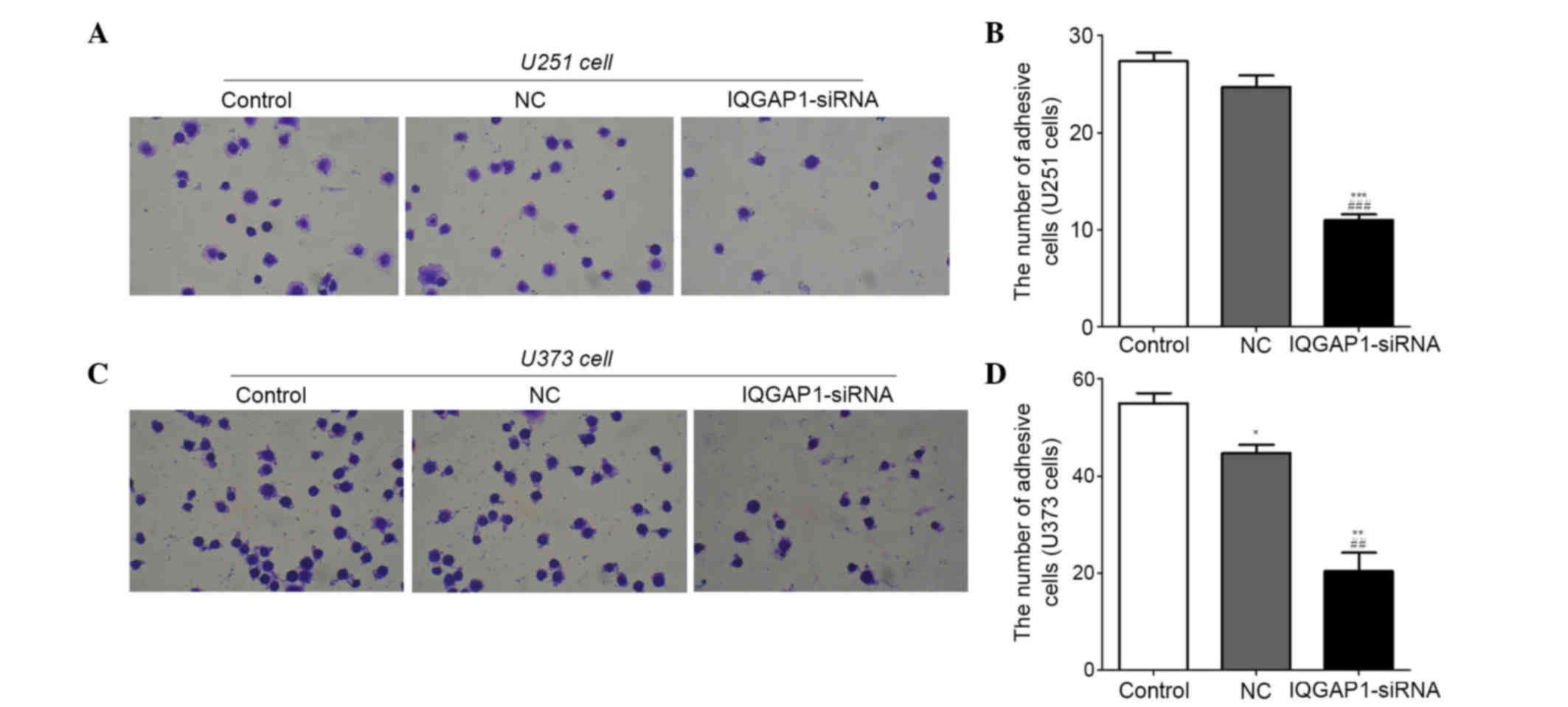
IQGAP1-siRNA inhibits adhesion of U251
and U373 cells
Cell metastasis serves a vital role in cancer
progression. In order to determine whether IQGAP1-siRNA had an
effect on glioma cell metastasis, cell adhesion assays (Fig. 4) and Transwell assays (Figs. 5 and 6) were performed in U251 and U373 cells.
The results indicated that the number of adhesive U251 and U373
cells was significantly decreased in the IQGAP1-siRNA groups
(Fig. 4A and C). In the U251
groups, the number of adhesive cells was 27±2, 25±2 and 11±1 in the
control, NC and IQGAP-siRNA groups, respectively (Fig. 4B). In the U373 groups, the number
of adhesive cells was 55±4, 45±3 and 20±7 in the control, NC and
IQGAP1-siRNA groups, respectively (Fig. 4D). These results suggest that
IQGAP1-siRNA may inhibit the adhesion of U251 and U373 cells.
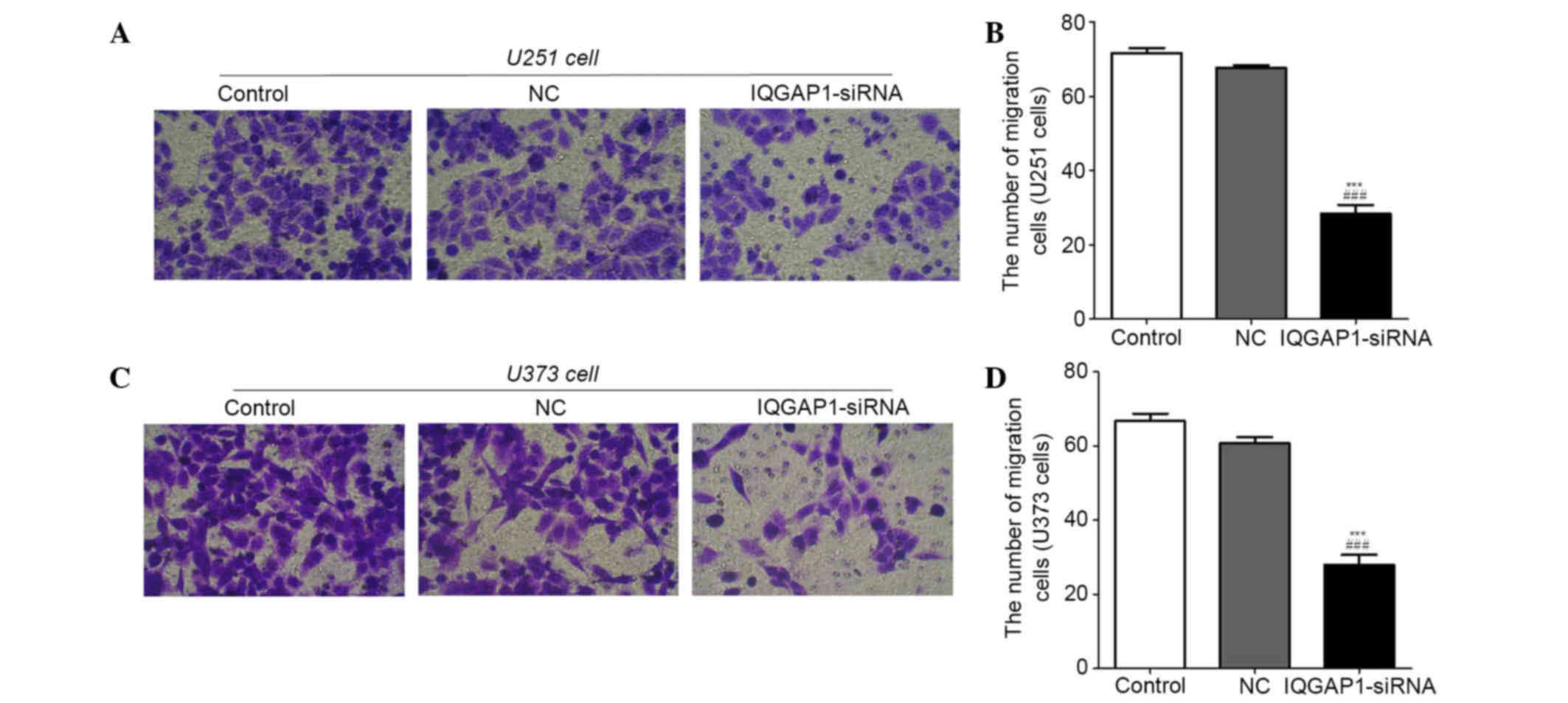
IQGAP1-siRNA inhibits migration of
U251 and U373 cells
A Transwell migration assay was performed to explore
the effects of IQGAP1-siRNA on U251 and U373 cells. As shown in
Fig. 5A and C, transfection with
IQGAP1-siRNA markedly suppressed the migratory ability of cells
compared with in the NC and control groups. The number of migratory
cells in the control, NC and IQGAP1-siRNA U251 groups was 72±3,
68±1 and 33±4, respectively (Fig.
5B), and was 67±4, 61±3 and 28±5, respectively in the U373 cell
groups (Fig. 5D). These results
indicate that IQGAP1-siRNA inhibits the migration of U251 and U373
cells.
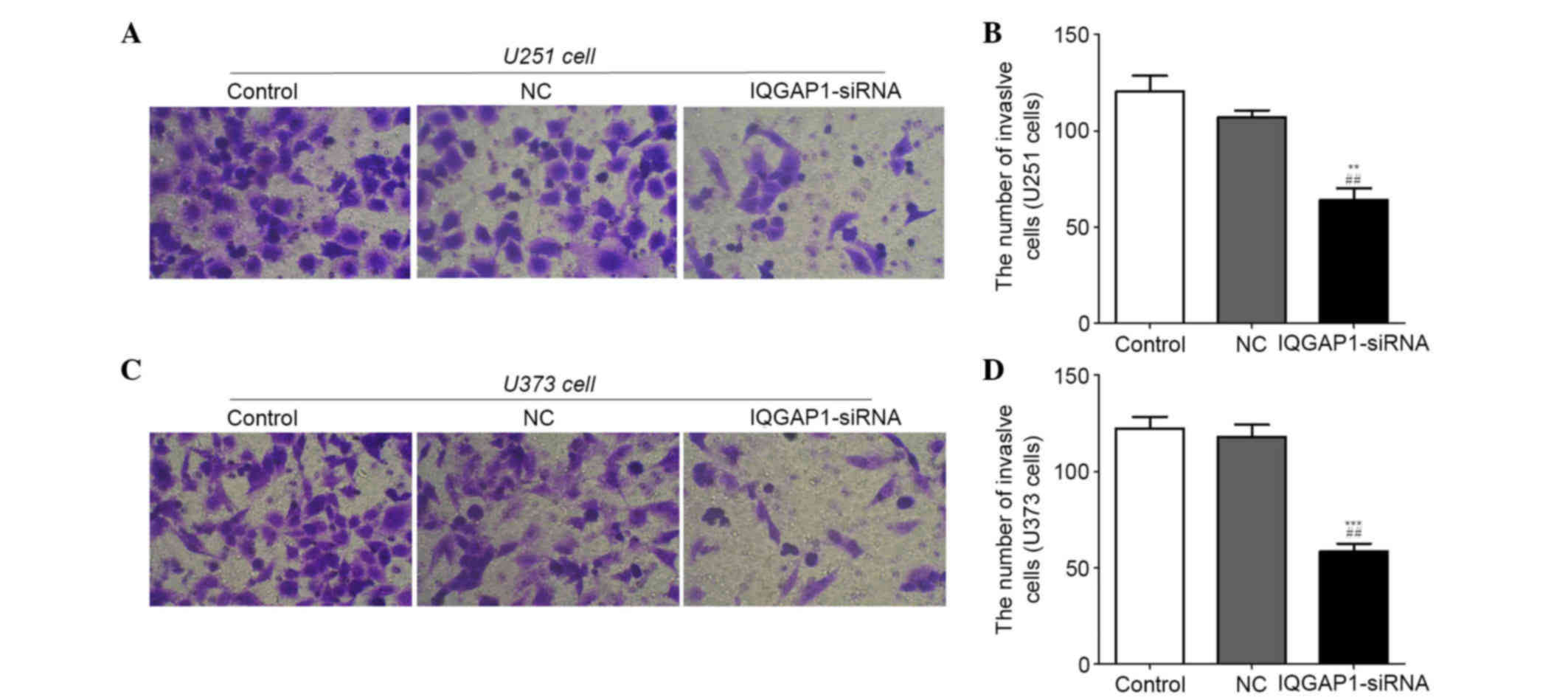
IQGAP1-siRNA inhibits invasion of U251
and U373 cells
A Transwell invasion assay was used to determine the
effects of IQGAP1-siRNA on U251 and U373 cells. As shown in
Fig. 6A and C, IQGAP1-siRNA
markedly suppressed the invasive ability of cells compared with in
the NC and control groups. The number of invasive U251 cells in the
control, NC and IQGAP1-siRNA groups was 120±14, 107±6 and 64±11,
respectively (Fig. 6B), and was
122±11, 118±11 and 58±7, respectively in the U373 cell groups
(Fig. 6D). These results indicate
that IQGAP1-siRNA may inhibit invasion of U251 and U373 cells.
IQGAP1-siRNA regulates the expression
of tumor suppressor genes and oncogenes
In order to further determine the mechanism by which
IQGAP1-siRNA inhibits glioma cell proliferation and metastasis, the
mRNA and protein expression levels of several tumor-associated
genes were detected in U251 and U373 cells by RT-qPCR and western
blotting, respectively. As presented in Fig. 7A and B, in U251 and U373 cells, the
mRNA expression levels of MMP2, Snail, MMP9, FN1 and Twist were
decreased, whereas E-cadherin was increased in the IQGAP1-siRNA
groups, compared with in the controls. Similarly, the protein
levels corresponded with the mRNA levels, which indicated that with
the exception of E-cadherin, all other proteins were downregulated
by IQGAP1-siRNA (Fig. 7C and
D).
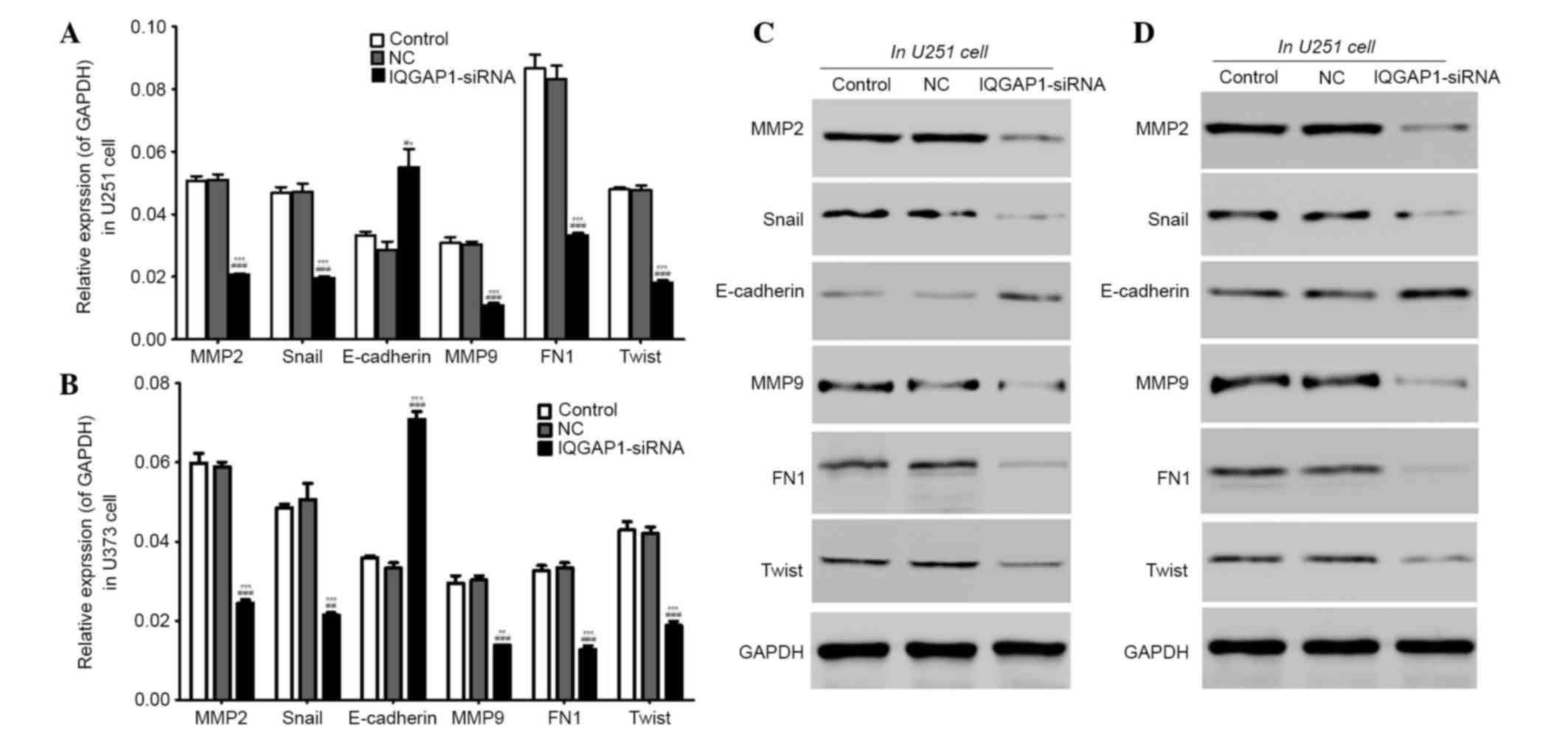 | Figure 7.IQGAP1-siRNA regulated the expression
of tumor suppressor genes and oncogenes. (A and B) The mRNA
expression levels of MMP2, Snail, MMP9, FN1 and Twist were
decreased, whereas E-cadherin was increased, in the IQGAP1-siRNA
groups compared with in the control groups. (C and D) The protein
expression levels of MMP2, Snail, MMP9, FN1 and Twist were
downregulated, whereas E-cadherin was upregulated. n=3. Data are
presented as the mean ± standard deviation. **P<0.01,
***P<0.001 vs. the control group, ##P<0.01,
###P<0.001 vs. the NC group. IQGAP1, IQ motif
containing GTPase activating protein 1; siRNA, small interfering
RNA; NC, negative control; MMP, matrix metalloproteinase; FN1,
fibronectin 1. |
Discussion
It has previously been revealed that IQGAP1 RNA and
protein expression is increased in various human malignancies
(15). Although an increasing
number of studies has aimed to elucidate the expression of IQGAP1
in various types of cancer, including lung cancer (21), endometrial cancer (22), ovarian cancer (23), gastric cancer (24), colon cancer (25), hepatic carcinoma (26), breast cancer (27) and even glioma (28), the molecular roles of IQGAP1 remain
to be understood and the correlation between clinical outcomes and
IQGAP1 expression in human malignant tumors require further
investigation. The present study indicated that IQGAP1-siRNA
inhibited the proliferation and metastasis of U251 and U373 glioma
cells. Furthermore, several tumor suppressor genes and oncogenes
were modulated following IQGAP1-siRNA transfection.
The expression of IQGAP1 has been explored in
various types of cancer in vivo and in vitro. With
regards to glioma, patients with glioblastoma and negative IQGAP1
expression have been reported to survive for >3 years (28). Studies regarding the effects of
IQGAP1 on cell proliferation are increasing. It has previously been
reported that IQGAP1, as a mitogen-activated protein kinase (MAPK)
scaffold that responds to MAPK signaling activation, may modulate
cellular functions and enhance proliferation (29). Overexpression of IQGAP1 increases
the proliferation of MCF-7 cells (16), whereas knockdown of IQGAP1 with
siRNA inhibits cell proliferation of umbilical vein endothelial
cells, and IQGAP1 is required for vascular endothelial-derived
growth factor-stimulated proliferation (30). Taken together, IQGAP1 expression
appears to be upregulated in various cancer cells, and is involved
in the regulation of cell proliferation. Similarly, in the present
study, knockdown of IQGAP1 with siRNA inhibited the proliferation
of glioma cell lines.
IQGAP1 contains several protein-interacting domains,
including one polyproline-binding domain, one calponin homology
domain, one Ras-GAP-related domain and four calmodulin-binding
motifs (7,8). The domains possess several important
interacting partners, including calmodulin, Rac1, Csc42, Rap1,
β-catenin, E-cadherin and members of the MAPK pathway (7,31).
IQGAP1 regulates various basic cellular activities such as
cell-cell adhesion, cell migration and invasion through the
aforementioned interactions (32).
For example, IQGAP1 binds directly to E-cadherin (13,14)
and overexpression of IQGAP1 reduces E-cadherin-mediated cell
adhesion (14). In ovarian cancer,
it has been reported that overexpression of IQGAP1 is significantly
correlated with poor prognosis, as determined by multivariate
analysis (33). In addition,
IQGAP1 is strongly expressed at the invasion front; furthermore,
this invasion front-associated expression pattern appeared more
frequently in advanced carcinoma, compared with other carcinomas
(32). Taken together, these
findings suggested that IQGAP1 has a function on cancer cell
metastasis, and the existing reports agree with the present study
that transfection of cells with IQGAP1-siRNA may inhibit cell
metastasis.
IQGAP1 may serve a critical role in pathways leading
to cell proliferation and metastasis; however, the precise
mechanism remains unknown. In order to explore this, the present
study indicated that IQGAP1 affects the expression of several
tumor-associated genes. Knockdown of IQGAP1 with siRNA resulted in
upregulation of the tumor suppressor gene E-cadherin, whereas
oncogenes, including MMP2, Snail, MMP9, FN1 and Twist, were
downregulated. Among them, MMP2 and MMP9 are members of the MMP
family (34). A previous study
indicated that human prostate cancer cell invasion was inhibited by
finasteride via MMP2 and MMP9 downregulation (35). Furthermore, it has been suggested
that MMP2 serves a role in the migration of tumor cells (36). Snai1, E-cadherin and Twist have
vital roles in the epithelial-mesenchymal transition (EMT) process
(36,37), which is closely associated with
tumor cell metastasis (38,39).
Furthermore, Snai1 and E-cadherin regulate EMT to initiate the
metastasis of several types of tumor cell (40). Twist is known to be a highly
conserved basic helix-loop-helix protein transcription factor,
which promotes EMT and may have prognostic significance in
endometrial cancers (41). As for
FN1, U94 alters gene expression of angiopoietin-like 4 and FN1
resulting in inhibition of tumorigenesis of PC3 prostate cancer
cells (42). These results
indicated that IQGAP1 knockdown-mediated inhibition of cell
proliferation and metastasis in glioma cell lines may be associated
with the expression of these tumor-associated genes. Further
studies are required to determine the functional roles of IQGAP1 in
cancer.
In conclusion, IQGAP1 was highly expressed in glioma
tissues and cell lines. The present study indicated that
IQGAP1-siRNA inhibited proliferation and metastasis of U251 and
U373 glioma cell lines. Furthermore, the expression levels of
several tumor suppressor genes and oncogenes were modulated. These
results provide evidence regarding the functional roles of IQGAP1
in cancer.
Acknowledgements
The present study was supported by grants from the
Wuhan Youth Science and Technology Fund (grant no.
2014070404010224).
References
|
1
|
Shirai K and Chakravarti A: Towards
personalized therapy for patients with glioblastoma. Expert Rev
Anticancer Ther. 11:1935–1944. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vranic A: New developments in surgery of
malignant gliomas. Radiol Oncol. 45:159–165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ware ML, Hirose Y, Scheithauer BW, Yeh RF,
Mayo MC, Smith JS, Chang S, Cha S, Tihan T and Feuerstein BG:
Genetic aberrations in gliomatosis cerebri. Neurosurgery.
60:150–158; discussion 158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mawrin C: Molecular genetic alterations in
gliomatosis cerebri: What can we learn about the origin and course
of the disease? Acta Neuropathol. 110:527–536. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patil CG, Eboli P and Hu J: Management of
multifocal and multicentric gliomas. Neurosurg Clin N Am.
23:343–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weissbach L, Settlemean J, Kalady MF,
Snijders AJ, Murthy AE, Yan YX and Bernards A: Identification of a
human rasGAP-related protein containing calmodulin-binding motifs.
J Biol Chem. 269:20517–20521. 1994.PubMed/NCBI
|
|
7
|
Brown MD and Sacks DB: IQGAP1 in cellular
signaling: Bridging the GAP. Trends Cell Biol. 16:242–249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joyal JL, Annan RS, Ho YD, Huddleston ME,
Carr SA, Hart MJ and Sacks DB: Calmodulin modulates the interaction
between IQGAP1 and CDC42. Identification of IQGAP1 by
nanoelectrospray tandem mass spectrometry. J Biol Chem.
272:15419–15425. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hart MJ, Callow MG, Souza B and Polakis P:
IQGAP1, a calmodulin-binding protein with a rasGAP-related domain,
is a potential effector for cdc42Hs. EMBO J. 15:2997–3005.
1996.PubMed/NCBI
|
|
11
|
Mataraza JM, Briggs MW, Li Z, Entwistle A,
Ridley AJ and Sacks DB: IQGAP1 promotes cell motility and invasion.
J Biol Chem. 278:41237–41245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z, Kim SH, Higgins JM, Brenner MB and
Sacks DB: IQGAP1 and calmodulin modulate E-cadherin function. J
Biol Chem. 274:37885–37892. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kuroda S, Fukata M, Nakagawa M, Fujii K,
Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al:
Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in
regulation of Ecadherin-mediated cell-cell adhesion. Science.
281:832–835. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumourigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong PX, Jia N, Xu ZJ, Liu YT, Li DJ and
Feng YJ: Silencing of IQGAP1 by shRNA inhibits the invasion of
ovarian carcinoma HO-8910PM cells in vitro. J Exp Clin Cancer Res.
27:772008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu B, Shi B, Jarzynka MJ, Yiin JJ,
D'Souza-Schorey C and Cheng SY: ADP-ribosylation factor 6 regulates
glioma cell invasion through the IQ-domain GTPase-activating
protein 1-Rac1-mediated pathway. Cancer Res. 69:794–801. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervoussystem. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakamura H, Fujita K, Nakagawa H, Kishi F,
Takeuchi A, Aute I and Kato H: Expression pattern of the scaffold
protein IQGAP1 in lung cancer. Oncol Rep. 13:427–431.
2005.PubMed/NCBI
|
|
22
|
Miyamoto S, Baba H, Kuroda S, Kaibuchi K,
Fukuda T, Maehara Y and Saito T: Changes in E-cadherin associated
with cytoplasmic molecules in well and poorly differentiated
endometrial cancer. Br J Cancer. 83:1168–1175. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong PX, Jian N, Xu ZJ, Liu YT, Li DJ and
Feng YJ: Silencing of IQGAP1 by shRNA inhibits the invasion of
ovarian carcinoma HO-890PM cells in vitro. J Exp Clin Cancer Res.
27:772008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Walch A, Seidl S, Hermannstädter C, Rauser
S, Deplazes J, Langer R, von Weyhern CH, Sarbia M, Busch R, Feith
M, et al: Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin
expression in gastric cancer. Mod Pathol. 21:544–552. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nabeshma K, Shmao Y, Inoue T and Koono M:
Immunohistochemical analysis of IQGAP1 expression in human
colorectal carcinomas: Its overexpression in carcinomas and
association with invasion fronts. Cancer Lett. 176:101–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmidt VA, Chiariello CS, Capilla E,
Miller F and Bahou WF: Development of hepatocellular carcinoma in
Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol.
28:1489–1502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jadesk L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McDonald KL, O'Sullivan MG, Parkinson JF,
Shaw JM, Payne CA, Brewer JM, Young L, Reader DJ, Wheeler HT, Cook
RJ, et al: IQGAP1 and IGFBP2: Valuable biomarkers for determining
prognosis in glioma patients. J Neuropathol Exp Neruol. 66:405–417.
2007. View Article : Google Scholar
|
|
29
|
Ussar S and Voss T: MEK1 and MEK2,
different regulators of the G1/S transition. J Biol Chem.
279:43861–43869. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L,
Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA,
Wang N, et al: IQGAP1, a novel vascular endothelial growth factor
receptor binding protein, is involved in reactive oxygen
species-dependent endothelial migration and proliferation. Circ
Res. 95:276–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Owen D, Campbell LJ, Littlefield K, Evetts
KA, Li Z, Sacks DB, Lowe PN and Mott HR: The IQGAP1-Rac1 and
IQGAP1-Cdc42 interactions: Interfaces differ between the complexes.
J Biol Chem. 283:1692–1704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayashi H, Nabeshima K, Aoki M, Hamasaki
M, Enatsu S, Yamauchi Y, Yamashita Y and Iwasaki H: Overexpression
of IQGAP1 in advanced colorectal cancer correlates with poor
prognosis-critical role in tumor invasion. Int J Cancer.
126:2563–2574. 2010.PubMed/NCBI
|
|
33
|
Dong P, Nabeshima K, Nishimura N, Kawakami
T, Hachisuga T, Kawarabayashi T and Iwasaki H: Overexpression and
diffuse expression pattern of IQGAP1 at invasion fronts are
independent prognostic parameters in ovarian carcinomas. Cancer
Lett. 243:120–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mitra A, Chakrabarti J, Chattopadhyay N
and Chatterjee A: Membrane-associated MMP-2 in human cervical
cancer. J Environ Pathol Toxicol Oncol. 22:93–100. 2003.PubMed/NCBI
|
|
35
|
Moroz A, Delella FK, Almeida R, Lacorte
LM, Fávaro WJ, Deffune E and Felisbino SL: Finasteride inhibits
human prostate cancer cell invasion through MMP2 and MMP9
downregulation. PLoS One. 8:e847572013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ansieau S, Courtois-Cox S, Morel AP and
Puisieux A: Failsafe program escape and EMT: A deleterious
partnership. Semin Cancer Biol. 21:392–396. 2011.PubMed/NCBI
|
|
37
|
Julien S, Puig I, Caretti E, Bonaventure
J, Nelles L, van Roy F, Dargemont C, De Herreros AG, Bellacosa A
and Larue L: Activation of NF-kappaB by Akt upregulates Snail
expression and induces epithelium mesenchyme transition. Oncogene.
26:7445–7456. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tiwari N, Gheldof A, Tatari M and
Christofori G: EMT as the ultimate survival mechanism of cancer
cells. Semin Cancer Biol. 22:194–207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peña C, García JM, Larriba MJ, Barderas R,
Gómez I, Herrera M, García V, Silva J, Domínguez G, Rodríguez R, et
al: SNAI1 expression in colon cancer related with CDH1 and VDR
downregulation in normal adjacent tissue. Oncogene. 28:4375–4385.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kyo S, Sakaguchi J, Ohno S, Mizumoto Y,
Maida Y, Hashimoto M, Nakamura M, Takakura M, Nakajima M, Masutomi
K and Inoue M: High twist expression is involved in infiltrative
endometrial cancer and affects patient survival. Hum Pathol.
37:431–438. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ifon ET, Pang AL, Jonhnson W, Cashman K,
Zimmerman S, Muralidhar S, Chan WY, Casey J and Rosenthal LJ: U94
alters FN1 and ANGPTL4 gene expression and inhibits tumorigenesis
of prostate cancer cell line PC3. Cancer Cell Int. 5:192005.
View Article : Google Scholar : PubMed/NCBI
|