Introduction
Spinal cord injury (SCI) is an ever-increasing
challenge and a devastating neurologic event affecting patients and
their families (1). A myriad of
neurologic and medical sequelae can occur subsequent to the
original insult, including the long-term loss of sensory and motor
functions, amongst other complications (1). Chronic pain is an important problem
following SCI and is a major impediment to effective rehabilitation
(2). It has been shown that
chronic pain following SCI has a high prevalence of at least 80%,
which in at least 40% of individuals manifests as persistent
neuropathic pain (NP) (3).
NP can be categorized as peripheral and central
pain, and is often described as paroxysmal, stabbing, burning,
pulsing, electric shock-like, pricking or tingling, and a
spontaneous or evoked unpleasant abnormal sensation (4). Current treatments use a variety of
surgical, pharmacological, physical and psychological strategies
for the management of NP following SCI (2). However, evidence of effective
therapeutic approaches in use remains to be fully elucidated.
Therefore, an improved understanding of the mechanisms underlying
NP in patients with SCI and the identification of effective
treatment strategies are required.
In previous years, the identification of specific
molecular alterations involved in NP syndromes has become a major
priority in investigations of SCI (5). Several biological alterations have
been implicated in the mechanisms of NP, including cellular
interactions, ion channel expression, extracellular proteins and
epigenetic effects (6). For
example, N-type voltage-dependent Ca2+ channels, which
are expressed in non-excitable microglial cells in mice, have been
demonstrated to contribute to the pathophysiology of NP (7). In addition, Chen et al
(8) demonstrated that astrocytic
connexin-43 enhances spinal cord synaptic transmission and
maintains late-phase NP in mice via the release of chemokines.
Nesic et al (9) performed
DNA microarray analysis and showed that a number of genes with
increased expression were significantly associated with astrocytic
activation and inflammation in the spinal cords of rats, which
developed central NP. Vicuña et al (10) revealed that the serine protease
inhibitor, serpinA3N can attenuate NP by inhibiting T cell-derived
leukocyte elastase in mice, and demonstrated crosstalk between T
cells and neurons in the modulation of NP. However, investigations
of NP following SCI have predominantly been performed in animal
models and the exact molecular mechanisms of persistent NP remain
to be elucidated.
Microarray data provide a global assessment of gene
expression signatures, which may provide insights into the
pathophysiology of disease (11).
Previous gene expression profiling studies of NP following SCI have
been performed predominantly in animal models (12,13).
However, the gene expression profiling of NP in human whole blood
has not been reported. In the present study, the microarray data of
GSE69901 was downloaded from the publicly available Gene Expression
Omnibus (GEO) database and analyzed. Differentially expressed genes
(DEGs) were screened in the peripheral blood mononuclear cells
(PBMCs) of samples from patients with SCI and intractable NP, and
compared with those from patients with SCI without pain. This was
followed by functional enrichment analysis and construction of a
protein-protein interaction (PPI) network. A transcriptional
regulation network was also constructed and functional gene
clustering was performed. The aim of the present study was to
further investigate the molecular mechanisms underlying NP
following SCI, and to identify additional potential pathways and
genes associated with the pathogenesis of NP.
Materials and methods
Microarray data
The GEO (http://www.ncbi.nlm.nih.gov/geo/) is an international
public repository, which archives and freely distributes
high-throughput microarray and next-generation sequencing
functional genomic data deposited by the scientific community
(14). In addition to serving as a
public archive, the GEO database provides available tools to assist
users in identifying, analyzing and visualizing data associated
with their specific interests (14). In the present study, the GSE69901
microarray data, deposited by Adıgüzel et al on 15th June
2015, was retrieved from the publicly available GEO database
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69901).
As shown in the description of the GSE69901 series in the GEO
database, the PBMCs were collected from whole blood samples from 12
patients with intractable NP and 13 patients in the control group
(without pain). All patients had complete SCI with a level of
injury above T5. Data were generated using the platform of the
GPL15207 (PrimeView) Affymetrix Human Gene Expression Array. In the
present study, the 25 samples were used for the subsequent
analysis, comprising the 12 PBMC samples from patients with NP and
13 PBMC samples from patients without pain.
Data preprocessing and differential
expression analysis
The raw data (Series Matrix files) were downloaded.
According to the annotation information on the GPL15207 platform,
the probe symbols were transformed into gene symbols. Gene
expression values were averaged using the aggregate function in R
(version 3.3.1, https://www.r-project.org/) when multiple probe sets
mapped to a same gene symbol. Missing values of probes were imputed
using the k-nearest-neighbor algorithm (15) present in the input package
(16) in R. In addition, quartile
data normalization was performed using the Bioconductor
preprocessCore package (version 1.28.0., http://bioconductor.org/packages/release/bioc/html/preprocessCore.html)
(17).
A t-test in the limma package (version 3.22.7,
http://www.bioconductor.org/packages/3.0/bioc/html/limma.html)
was performed to identify DEGs in the specimens from the patients
with NP, compared with the controls. An absolute value of
log2-fold change (log2FC)>1 and adjusted
P-value of <0.05 were used to determine significant differential
expression.
Functional enrichment analysis
TargetMine is an integrated database enabling
complicated searches, which are difficult to perform using existing
comparable tools and assists in efficient target prioritization
(18). In order to analyze the
identified upregulated and downregulated genes at functional
levels, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses were performed using
TargetMine (http://targetmine.nibio.go.jp/). GO terms or pathways
with a P-value <0.05 were considered to be significantly
enriched.
PPI network construction
The online database resource Search Tool for the
Retrieval of Interacting Genes (STRING) provides uniquely
comprehensive coverage, and access to predicted and experimental
interaction information (19).
Interactions in the STRING database are provided with a confidence
score (19). In the present study,
application of the STRING database (http://string-db.org/) was used to predict PPIs based
on a confidence score >0.4 and other default parameters. The PPI
network was visualized using Cytoscape (version 2.8.2, http://www.cytoscape.org/) (20).
Transcriptional regulation network
construction
The Human Transcriptional Regulation Interactions
database (HTRIdb) is an open-access database (http://www.lbbc.ibb.unesp.br/htri) from which
human experimentally validated interactions among transcription
factors (TFs) and their corresponding target genes can be
extracted. These can be used to construct transcriptional
regulation interaction networks, enabling detailed understanding of
the regulation of biological processes (21). In the present study, the TFs
involved in regulating all DEGs were screened based on the data
derived from the HTRIdb. A transcriptional regulation network was
subsequently constructed and visualized in Cytoscape (20).
Gene clustering
The Gene Cluster with Literature Profile (GenCLip)
(http://www.genclip.com/) is a literature mining
tool for functional clustering of a list of genes with keywords, GO
terms or pathways using the average linkage hierarchical clustering
algorithm (22). In the present
study, all DEGs were analyzed using GenCLip software (version 2.0,
http://ci.smu.edu.cn) to identify functional gene
clusters.
Results
Data preprocessing and identification
of DEGs
Following data preprocessing, 19,419 genes were
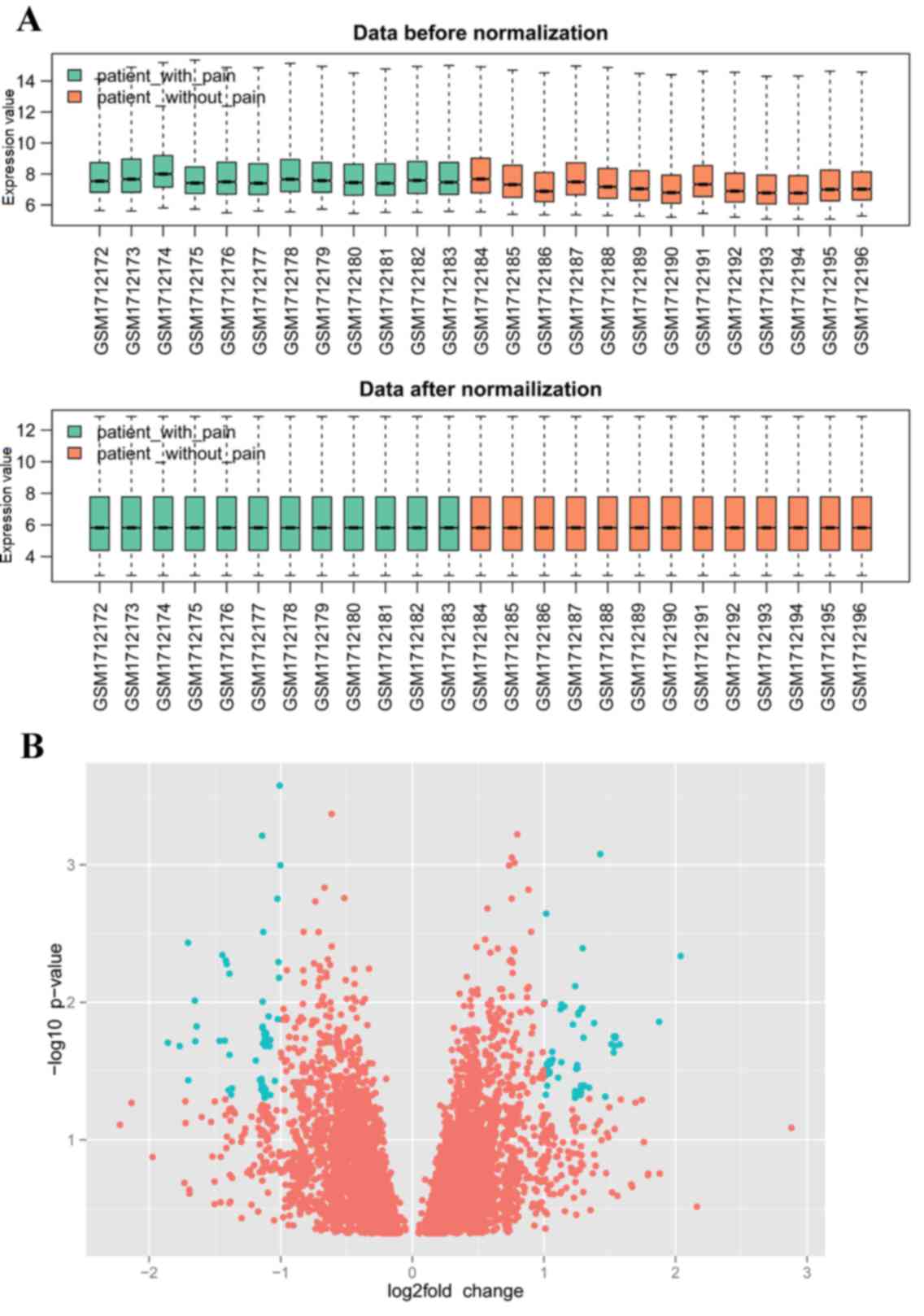
mapped to the probes. The gene expression profiles prior to and
following normalization are shown in Fig. 1A. The results revealed that,
following normalization, the median probe intensity was similar
across all conditions, which reflects the consistency of the
technical quality in the data.
In the original analysis by Adıgüzel et al,
only 16 DEGs, including nine upregulated and seven downregulated
genes, were obtained with a cut-off degree of a 4-fold change.
However, the present study identified a total of 131 DEGs in the
PBMC samples from patients with intractable NP, compared with
controls under the specific thresholds of |log2 FC|>1
and adjusted P-value <0.05, including 70 upregulated and 61
downregulated genes. A volcano plot was applied to visualize the
genes identified (Fig. 1B). In
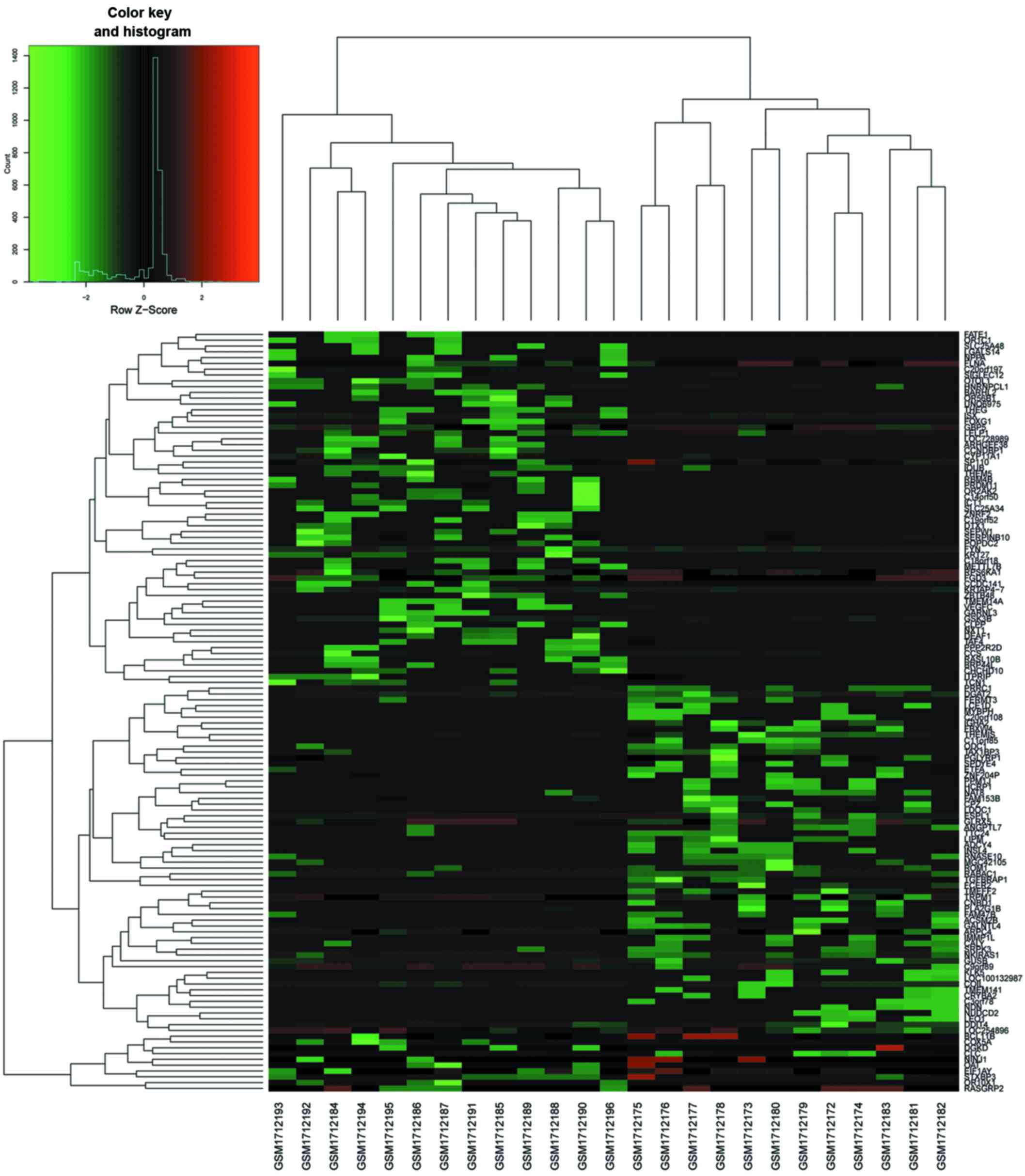
addition, a heat map of gene expression values was constructed
(Fig. 2). The color patterns in
the heat map indicate the variability of gene expression between
the groups of SCI with NP and without NP.
GO and pathway enrichment analysis. The upregulated
and downregulated genes were enriched in different GO terms and
pathways. The results showed that the upregulated genes were
enriched in categories of 39 biological processes (BPs), four
molecular functions (MFs), four cellular components (CCs) and four
KEGG pathways. By contrast, the downregulated genes were enriched
in GO terms of 15 BPs, one CC, nine MFs and nine KEGG pathways. The
top five GO terms in each category and the results of the KEGG
pathway enrichment analysis are shown in Tables I and II, respectively.
 | Table I.Top five GO terms in each category
enriched by upregulated and downregulated genes. |
Table I.
Top five GO terms in each category
enriched by upregulated and downregulated genes.
| Gene change | Ontology | ID | Pathway | P-value | n | Genes |
|---|
| Upregulated | BP | GO:0030168 | Platelet
activation | 5.16E-05 | 6 | DGKD, FLNA, FYN,
RASGRP2, STXBP3, VEGFC |
|
| BP | GO:0001775 | Cell
activation | 0.002258 | 7 | DGKD, FLNA, FYN,
RASGRP2, RPS6KA1, STXBP3, VEGFC |
|
| BP | GO:0007596 | Blood
coagulation | 0.003756 | 6 | DGKD, FLNA, FYN,
RASGRP2, STXBP3, VEGFC |
|
| BP | GO:0050817 | Coagulation | 0.003864 | 6 | DGKD, FLNA, FYN,
RASGRP2, STXBP3, VEGFC |
|
| BP | GO:0007599 | Hemostasis | 0.0039 | 6 | DGKD, FLNA, FYN,
RASGRP2, STXBP3, VEGFC |
|
| CC | GO:0005759 | Mitochondrial
matrix | 0.001476 | 5 | CLPP, CYP11A1,
ICT1, OAT, THEM5 |
|
| CC | GO:0044429 | Mitochondrial
part | 0.002033 | 7 | CHCHD10, CLPP,
COX5A, CYP11A1, ICT1, OAT, THEM5 |
|
| CC | GO:0005739 | Mitochondrion | 0.008474 | 8 | CHCHD10, CLPP,
COX5A, CYP11A1, ICT1, MPC1, OAT, THEM5 |
|
| CC | GO:0031091 | Platelet α
granule | 0.013661 | 2 | STXBP3, VEGFC |
|
| MF | GO:0019992 | Diacylglycerol
binding | 2.45E-04 | 2 | DGKD, RASGRP2 |
|
| MF | GO:0001948 | Glycoprotein
binding | 0.020786 | 2 | FLNA, FYN |
| Downregulated | BP | GO:0050830 | Defense response to
Gram-positive bacterium | 0.006167 | 2 | PGLYRP1,
PLA2G1B |
|
| BP | GO:0010518 | Positive regulation
of phospholipase activity | 0.0173 | 2 | ADCY4, PLA2G1B |
|
| BP | GO:0060401 | Cytosolic calcium
ion transport | 0.020834 | 2 | PLA2G1B, TRPM1 |
|
| BP | GO:0060402 | Calcium ion
transport into cytosol | 0.020834 | 2 | PLA2G1B, TRPM1 |
|
| BP | GO:0010517 | Regulation of
phospholipase activity | 0.021295 | 2 | ADCY4, PLA2G1B |
|
| CC | GO:0015629 | Actin
cytoskeleton | 0.038255 | 3 | ARPC4, FERMT3,
TAX1BP3 |
|
| MF | GO:0005178 | Integrin
binding | 0.015317 | 2 | FCER2, FERMT3 |
|
| MF | GO:0016829 | Lyase activity | 0.025162 | 2 | ADCY4, ODC1 |
|
| MF | GO:0004175 | Endopeptidase
activity | 0.028922 | 3 | CLC, ESPL1,
KLK5 |
|
| MF | GO:0004197 | Cysteine-type
endopeptidase activity | 0.029558 | 2 | CLC, ESPL1 |
|
| MF | GO:0016747 | Transferase
activity, transferring acyl groups other than amino-acyl
groups | 0.033704 | 2 | DGAT2, NAT8 |
 | Table II.Enriched pathways of the upregulated
and downregulated genes. |
Table II.
Enriched pathways of the upregulated
and downregulated genes.
| Gene change | ID | Pathway | P-value | n | Genes |
|---|
| Upregulated | hsa04510 | Focal adhesion | 0.005135 | 4 | FLNA, FYN, GSK3B,
VEGFC |
|
| hsa03015 | mRNA surveillance
pathway | 0.039026 | 2 | NXT1, PPP2R2D |
|
| hsa04740 | Olfactory
transduction | 0.048956 | 4 | OR10X1, OR1C1,
OR2AK2, OR56B1 |
|
| hsa04660 | T cell receptor
signaling pathway | 0.049678 | 2 | FYN, GSK3B |
| Downregulated | hsa04975 | Fat digestion and
absorption | 0.00297 | 2 | DGAT2, PLA2G1B |
|
| hsa04972 | Pancreatic
secretion | 0.015504 | 2 | ADCY4, PLA2G1B |
|
| hsa04114 | Oocyte meiosis | 0.021098 | 2 | ADCY4, ESPL1 |
|
| hsa04270 | Vascular smooth
muscle contraction | 0.023984 | 2 | ADCY4, PLA2G1B |
|
| hsa01100 | Metabolic
pathways | 0.024489 | 6 | ACSM2B, DGAT2,
GALNT18, GUSB, ODC1, PLA2G1B |
|
| hsa04611 | Platelet
activation | 0.027808 | 2 | ADCY4, FERMT3 |
|
| hsa00531 | Glycosaminoglycan
degradation | 0.037737 | 1 | GUSB |
|
| hsa03060 | Protein export | 0.045511 | 1 | IMMP1L |
|
| hsa00592 | α-linolenic acid
metabolism | 0.049376 | 1 | PLA2G1B |
The results revealed that the upregulated genes were
predominantly involved in GO terms associated with platelet
activation, cell activation, blood coagulation and mitochondrial
function. The upregulated genes were also enriched in four
pathways, which included focal adhesion and the T cell receptor
signaling pathway. The downregulated genes were involved with
cytosolic calcium ion transport and the positive regulation of
phospholipase activity. Additionally, the downregulated genes were
involved in pathways, including fat digestion and absorption,
pancreatic secretion and vascular smooth muscle contraction.
PPI network analysis
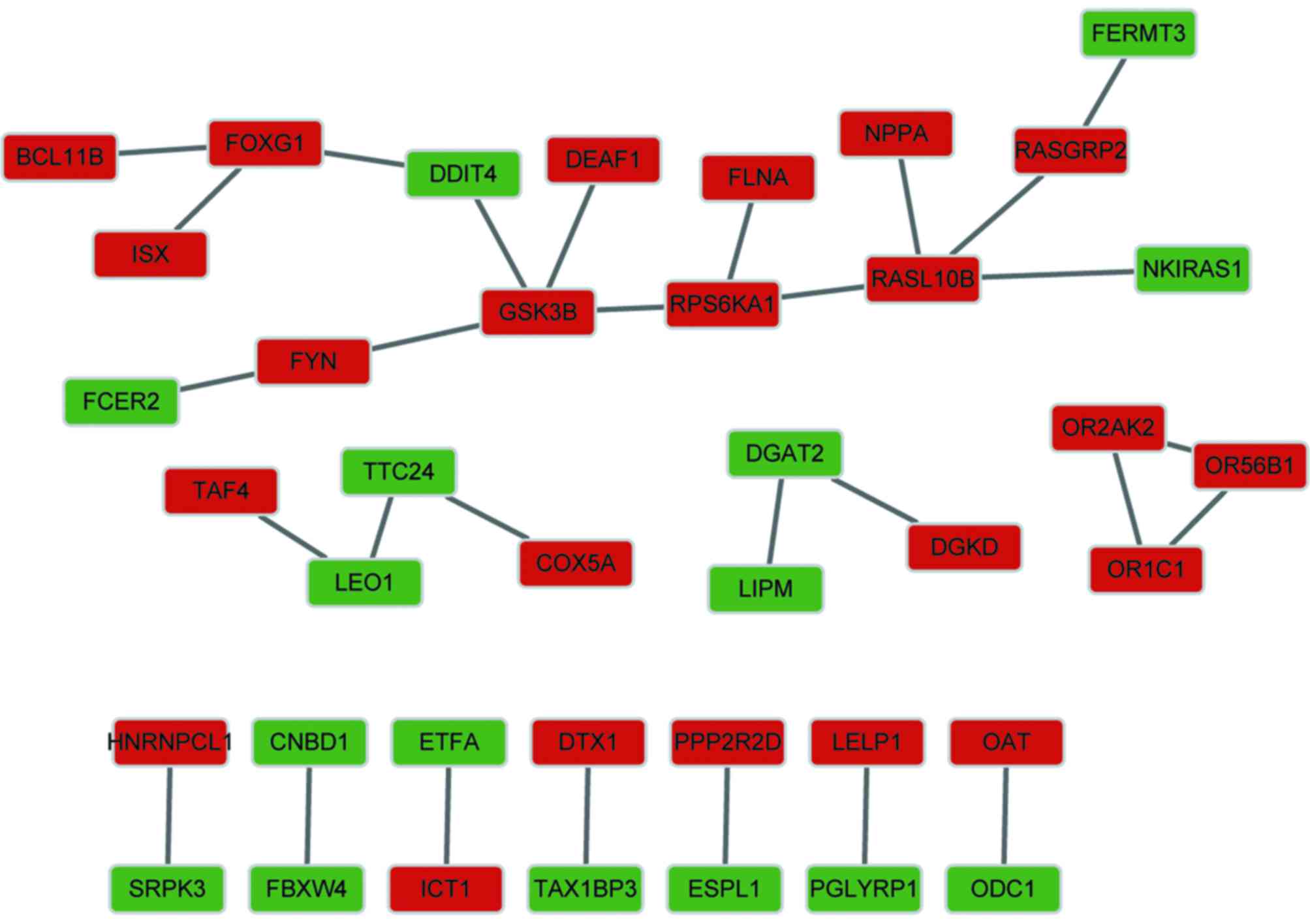
As shown in Fig. 3,
the constructed PPI network contained 39 nodes and 29 interactions,
consisting of 23 upregulated genes and 16 downregulated genes. The
results revealed that the PPI network had a loose structure.
However, glycogen synthase kinase 3β (GSK3B) and RAS-like, family
10, member B (RASL10B) had higher node degrees.
Transcriptional regulation network
construction and functional gene clustering analysis
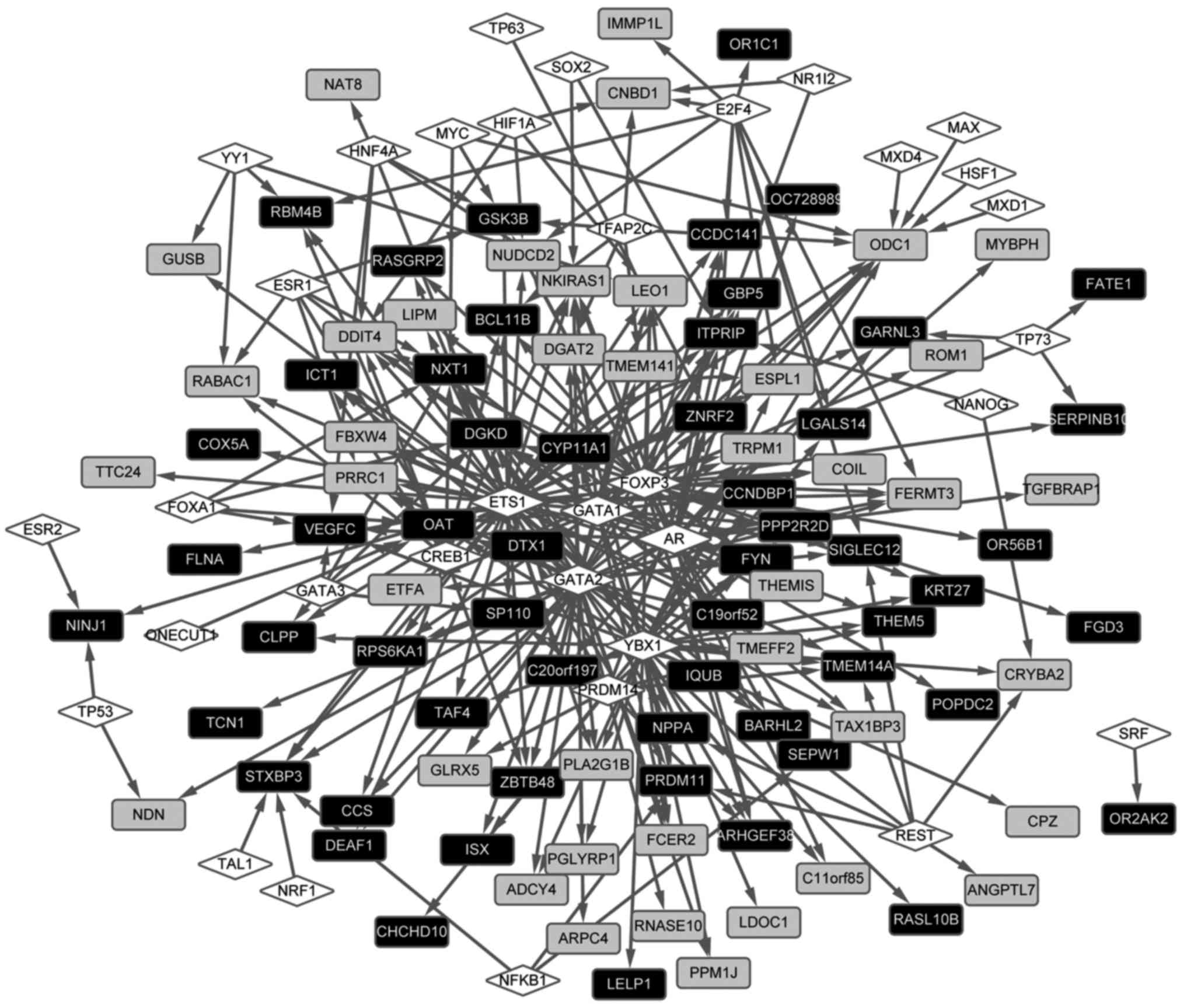
The transcriptional regulation network was
constructed (Fig. 4). The results
revealed that a total of 34 TFs regulated 101 DEGs in the
regulation network, comprising 322 regulation pairs. In addition,
TFs, including androgen receptor, GATA-binding protein 2 and Y
box-binding protein 1, were found to regulate a higher number of
DEGs. Ornithine decarboxylase 1 (ODC1) (downregulated), ornithine
aminotransferase (OAT) (upregulated), diacylglycerol kinase δ 130
kDa (DGKD; upregulated) were identified to be regulated by a
higher number of TFs. It was also found that ODC1 interacted
with OAT in the PPI network.
The present study also analyzed the potential
functions of the identified DEGs using the GeneCLip text-mining
program. The results showed that the DEGs were significantly
involved with two functional gene clusters, namely the function of
mitochondrial membrane and DNA binding (Table III). In addition, the
GSK3B, OAT and ODC1 DEGs were clearly
clustered into the subgroup associated with the function of
mitochondrial membrane. As shown in Table III, the GSK3B and
ODC1 DEGs were also closely associated with DNA binding.
 | Table III.Results of functional gene clustering
analysis. |
Table III.
Results of functional gene clustering
analysis.
| Function of
cluster | Genes | P-value |
|---|
| Function of
mitochondrial membrane | COX5A, CYP11A1,
GSK3B, IMMP1L, MPC1, OAT, ODC1, ROM1, SLC25A34, SLC25A48,
TMEM14A | 0.001229 |
| DNA binding | BCL11B, CCNDBP1,
CLC, CLPP, COIL, CYP11A1, DDIT4, DEAF1, DTX1, FCER2, FOXG1, GSK3B,
GUSB, LDOC1, LEO1, NDN, NPPA, ODC1, RPS6KA1, SP110, SRPK3,
TAF4 | 0.048263 |
Discussion
NP is a common problem following SCI and is usually
difficult to treat (23). In the
present study, gene expression profiling was performed to further
investigate the molecular mechanisms underlying NP in patients with
SCI. As a result, a total of 70 upregulated and 61 downregulated
genes were identified in the PBMC samples from patients with
intractable NP, compared with the controls. The upregulated genes
and downregulated genes were significantly involved in different GO
terms and pathways, including focal adhesion, the T cell receptor
signaling pathway and mitochondrial function. GSK3B was
identified as a hub protein in the PPI network. In addition,
ODC1, DGKD and OAT were regulated by a higher
number of TFs in the transcriptional regulation network.
GSK3B, OAT and ODC1 were also significantly
enriched in two functional gene clusters, namely the functions of
mitochondrial membrane and DNA binding.
In the present study, a total of 131 DEGs (70
upregulated and 61 downregulated) were identified in the PBMC
samples from patients with SCI and NP, compared with controls under
the specific thresholds. However, in the original analysis by
Adıgüzel et al, only 16 DEGs (nine upregulated and seven
downregulated) were obtained with a cut-off degree of a 4-fold
change. Thus, the results of the present study showed that distinct
genetic features were identified in PBMC samples using different
screening methods with different thresholds.
Previous evidence has led to an increased awareness
of the contribution of immune and inflammatory systems to NP, and
has revealed the roles of immune cells and inflammatory mediators
(24,25). In addition, in the original
analysis by Adıgüzel et al, KEGG pathway analysis revealed
that 14.6% of the DEGs identified were enriched in nodes of the
immune system. Signals through the T cell receptor are shown to be
significant for the initiation of T helper cell activation
(26). Calvo et al
(25) suggested that T cell
involvement in the course of NP and modulation of the T lymphocyte
phenotype may be one method for the management of NP. In accordance
with the previous study, the present study found that DEGs were
significantly enriched in the T cell receptor signaling pathway.
Focal adhesion kinase is critical to cellular functions, including
the migration, proliferation and survival of anchorage-dependent
cells (27). Hua and Cabot
(28) indicated that the immune
system used the cell migration mechanisms not only to fight
pathogens, but also to control pain within the injured issue. In
accordance with the previous studies, the results of the present
study showed that the DEGs were involved in the focal adhesion
pathway. Taken together, the present study indicated that the focal
adhesion and T cell receptor signaling pathways may be crucial for
the development of NP following SCI. Genes, which were involved in
these two pathways and active in lymphocytes may also be crucial in
the molecular mechanisms of NP.
The protein encoded by GSK3B is a
serine-threonine kinase, which belongs to the GSK subfamily
(29). In previous years,
GSK3 has been implicated in a number of roles in the immune
system (30). Beurel et al
(30) demonstrated that GSK3 is an
important regulator of the balance between the production of pro-
and anti-inflammatory cytokines in the peripheral and central
nervous system. T cell differentiation, proliferation and survival
were found to be affected by GSK3 (30). In the present study, GSK3B
was a hub protein in the PPI network. In addition, it was found
that GSK3B was significantly linked to the focal adhesion
and T cell receptor signaling pathways (Table II). As discussed above, genes
involved in the focal adhesion and T cell receptor signaling
pathways may be crucial for the development of NP following SCI. In
this context, the present study indicated that active GSK3B
in lymphocytes may be significant in the mechanisms underlying NP
in patients with SCI, involved in the focal adhesion and T cell
receptor signaling pathways. Furthermore, GSK3B may be a novel and
effective pharmacological target for the treatment of NP following
SCI, in accordance with a previous study, which showed that a
GSK3-specific inhibitor, AR-A014418, produced marked
antihyperalgesic effects in mice with NP (31). However, additional experimental
verification is required to confirm this result.
ODC1 encodes the rate-limiting enzyme of the
polyamine biosynthesis pathway, which catalyzes ornithine to
putrescine, a type of polyamine (32). Rivat et al (33) investigated the ability of a
polyamine-deficient diet to prevent long-lasting pain
hypersensitivity associated with tissue injury in rats, and found
that the diet markedly reduced long-lasting hyperalgesia. In the
present study, ODC1 was associated with metabolic pathways
and was concerned with the function of mitochondrial membrane. This
indicated that ODC1 may be crucial in PBMCs in the
mechanisms of NP following SCI. By contrast, OAT encodes the
mitochondrial enzyme, ornithine aminotransferase, which is a key
enzyme in the pathway converting arginine and ornithine into the
major excitatory and inhibitory neurotransmitters, glutamate and
gamma-aminobutyric acid (34).
Studies have demonstrated that the balance of chloride ions,
glutamate and gamma-aminobutyric acid distribution are disrupted
following SCI, resulting in neuronal hyperexcitability and chronic
NP (35). In addition, OAT
and ODC1 are involved in ornithine metabolism (32,34).
In accordance with the previous studies, the present study
identified that OAT and ODC1 had interactions in the
PPI network, and were regulated by a higher number of TFs. Taken
together, the results suggested that the interactions of OAT
and ODC1, which were involved in the metabolism process, may
be key in the development and maintenance of chronic NP in patients
with SCI. Further investigations are required to confirm this.
In conclusion, the data obtained in the present
study demonstrated that gene expression profiles were altered in
patients with SCI and NP, compared with the SCI patients without
pain. The focal adhesion and T cell receptor signaling pathways may
be crucial for the development of NP following SCI. GSK3B
was found to be significantly involved in these two pathways, and
may be a novel and effective pharmacological target for the
treatment of NP. In addition, OAT may be key in its
association with metabolic process in the development and
maintenance of chronic NP following SCI, via interacting with
ODC1. Further investigations and experiments with additional
patient cohorts, and delineation of the specific roles of these
genes may lead to an improved understanding of the mechanisms
underlying NP and the development of novel therapeutic options.
Acknowledgements
This study was supported by the Special Fund for
Medical Service of Jilin Finance Department (grant no.
SCZSY201507).
References
|
1
|
Cao HQ and Dong ED: An update on spinal
cord injury research. Neurosci Bull. 29:94–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Finnerup NB and Baastrup C: Spinal cord
injury pain: Mechanisms and management. Curr Pain Headache Rep.
16:207–216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mehta S, Orenczuk K, McIntyre A, Willems
G, Wolfe DL, Hsieh JT, Short C, Loh E and Teasell RW: SCIRE
Research Team: Neuropathic pain post spinal cord injury part 1:
Systematic review of physical and behavioral treatment. Top Spinal
Cord Inj Rehabil. 19:61–77. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Finnerup NB and Jensen TS: Spinal cord
injury pain-mechanisms and treatment. Eur J Neurol. 11:73–82. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Von Schack D, Agostino MJ, Murray BS, Li
Y, Reddy PS, Chen J, Choe SE, Strassle BW, Li C, Bates B, et al:
Dynamic changes in the microRNA expression profile reveal multiple
regulatory mechanisms in the spinal nerve ligation model of
neuropathic pain. PLoS One. 6:e176702011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schomberg D, Miranpuri G, Duellman T,
Crowell A, Vemuganti R and Resnick D: Spinal cord injury induced
neuropathic pain: Molecular targets and therapeutic approaches.
Metab Brain Dis. 30:645–658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saegusa H and Tanabe T: N-type
voltage-dependent Ca2+ channel in non-excitable microglial cells in
mice is involved in the pathophysiology of neuropathic pain.
Biochem Biophys Res Commun. 450:142–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen G, Park CK, Xie RG, Berta T,
Nedergaard M and Ji RR: Connexin-43 induces chemokine release from
spinal cord astrocytes to maintain late-phase neuropathic pain in
mice. Brain. 137:2193–2209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nesic O, Lee J, Johnson KM, Ye Z, Xu GY,
Unabia GC, Wood TG, McAdoo DJ, Westlund KN, Hulsebosch CE and
Regino Perez-Polo J: Transcriptional profiling of spinal cord
injury-induced central neuropathic pain. J Neurochem. 95:998–1014.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vicuña L, Strochlic DE, Latremoliere A,
Bali KK, Simonetti M, Husainie D, Prokosch S, Riva P, Griffin RS,
Njoo C, et al: The serine protease inhibitor SerpinA3N attenuates
neuropathic pain by inhibiting T cell-derived leukocyte elastase.
Nat Med. 21:518–523. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim DS, Figueroa KW, Li KW, Boroujerdi A,
Yolo T and Luo ZD: Profiling of dynamically changed gene expression
in dorsal root ganglia post peripheral nerve injury and a critical
role of injury-induced glial fibrillary acetic protein in
maintenance of pain behaviors. Pain. 143:114–122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim CF and Moalem-Taylor G: Interleukin-17
contributes to neuroinflammation and neuropathic pain following
peripheral nerve injury in mice. J Pain. 12:370–383. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database issue):
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altman NS: An introduction to kernel and
nearest-neighbor nonparametric regression. The American
Statistician. 46:175–185. 2012. View Article : Google Scholar
|
|
16
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: impute: Imputation for microarray data. R package version.
1:2012.
|
|
17
|
López-Romero P, González MA, Callejas S,
Dopazo A and Irizarry RA: Processing of Agilent microRNA array
data. BMC Res Notes. 3:182010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen YA, Tripathi LP and Mizuguchi K:
TargetMine, an integrated data warehouse for candidate gene
prioritisation and target discovery. PLoS One. 6:e178442011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database issue): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bovolenta LA, Acencio ML and Lemke N:
HTRIdb: An open-access database for experimentally verified human
transcriptional regulation interactions. BMC Genomics. 13:4052012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang JH, Zhao LF, Lin P, Su XR, Chen SJ,
Huang LQ, Wang HF, Zhang H, Hu ZF, Yao KT and Huang ZX: GenCLiP
2.0: A web server for functional clustering of genes and
construction of molecular networks based on free terms.
Bioinformatics. 30:2534–2536. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hagen EM and Rekand T: Management of
neuropathic pain associated with spinal cord injury. Pain Ther.
4:51–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moalem G and Tracey DJ: Immune and
inflammatory mechanisms in neuropathic pain. Brain Res Rev.
51:240–264. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calvo M, Dawes JM and Bennett DL: The role
of the immune system in the generation of neuropathic pain. Lancet
Neurol. 11:629–642. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huppa JB, Gleimer M, Sumen C and Davis MM:
Continuous T cell receptor signaling required for synapse
maintenance and full effector potential. Nat Immunol. 4:749–755.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang D, Khoe M, Befekadu M, Chung S,
Takata Y, Ilic D and Bryer-Ash M: Focal adhesion kinase mediates
cell survival via NF-kappaB and ERK signaling pathways. Am J
Physiol Cell Physiol. 292:C1339–C1352. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hua S and Cabot PJ: Mechanisms of
peripheral immune-cell-mediated analgesia in inflammation: Clinical
and therapeutic implications. Trends Pharmacol Sci. 31:427–433.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Galli C, Piemontese M, Lumetti S, Manfredi
E, Macaluso G and Passeri G: GSK3b-inhibitor lithium chloride
enhances activation of Wnt canonical signaling and osteoblast
differentiation on hydrophilic titanium surfaces. Clin Oral
Implants Res. 24:921–927. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beurel E, Michalek SM and Jope RS: Innate
and adaptive immune responses regulated by glycogen synthase
kinase-3 (GSK3). Trends Immunol. 31:24–31. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mazzardo-Martins L, Martins D, Stramosk J,
Cidral-Filho F and Santos A: Glycogen synthase kinase 3-specific
inhibitor AR-A014418 decreases neuropathic pain in mice: Evidence
for the mechanisms of action. Neuroscience. 226:411–420. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hogarty MD, Norris MD, Davis K, Liu X,
Evageliou NF, Hayes CS, Pawel B, Guo R, Zhao H, Sekyere E, et al:
ODC1 is a critical determinant of MYCN oncogenesis and a
therapeutic target in neuroblastoma. Cancer Res. 68:9735–9745.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rivat C, Richebé P, Laboureyras E, Laulin
JP, Havouis R, Noble F, Moulinoux JP and Simonnet G: Polyamine
deficient diet to relieve pain hypersensitivity. Pain. 137:125–137.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bouché N, Lacombe Bt and Fromm H: GABA
signaling: A conserved and ubiquitous mechanism. Trends Cell Biol.
13:607–610. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gwak YS and Hulsebosch CE: GABA and
central neuropathic pain following spinal cord injury.
Neuropharmacology. 60:799–808. 2011. View Article : Google Scholar : PubMed/NCBI
|