Introduction
Type 2 diabetes mellitus (T2DM), characterized by
hyperglycemia, is one of the most prevalent metabolic disorders.
The International Diabetes Federation estimates that
>400,000,000 diabetic patients are expected by 2030, with over
50% of these being from Asia (1).
Long-term hyperglycemia may lead to macrovascular diseases,
including coronary artery disease, peripheral arterial disease and
stroke, and microvascular complications, including diabetic
nephropathy, neuropathy and retinopathy (2).
The pathogenesis of T2DM arises from the interplay
of genetic, environmental and/or lifestyle factors, which lead to a
decline in insulin sensitivity in the liver, adipose tissues and
skeletal muscles, followed by chronic pancreatic β-cell
dysfunction. Insulin resistance then increases insulin secretion
(hyperinsulinemia) to maintain euglycemia. Subsequently, the
progressive deterioration in insulin sensitivity and a reduction in
pancreatic insulin secretion generate a state of relative insulin
deficiency, resulting in chronic hyperglycemia and the onset of
T2DM (3).
MicroRNAs (miRNAs) are endogenously expressed,
evolutionarily conserved, small single-stranded, non-coding RNA
molecules of 21–23 nucleotides, which function as regulators of
gene expression by partially base-pairing to the 3′ untranslated
regions of their target mRNAs and destabilizing or inhibiting their
translation (4). The latest
estimates revealed that the human genome encodes >1,600 miRNA
precursors, which can generate >2,000 mature miRNAs (www.mirbase.org), which control ~50% of all mammalian
protein-coding genes (5) and are
involved in the biological processes of cell development,
differentiation, metabolism, immunity, apoptosis and proliferation
(4). The dysregulated expression
of miRNAs in various tissues has been associated with a variety of
diseases, including cancer (6,7),
T2DM (8) and its complications
(9).
Serum or plasma miRNAs derived from various
tissues/organs are released by several cellular release mechanisms.
For example, mature miRNAs can bind to RNA-binding proteins or
lipoproteins, or are loaded inside microvesicles or exosomes when
they are to be released (10,11).
These circulating miRNAs may then be delivered to recipient cells,
where they can regulate the translation of target genes, suggesting
that serum or plasma miRNAs can serve as extracellular
communicators (12). Furthermore,
miRNAs levels in serum are stable, reproducible and consistent
among individuals of the same ethnic background (10). The specific serum miRNA expression
profile constitutes the fingerprint of a physiological or disease
condition (10). Evidence from rat
models shows that miRNA expression profiles in different tissues
(pancreas, liver, adipose and skeletal muscle) share high
similarity with those in blood samples (8). Therefore, circulating miRNAs are
suggested as unique biomarkers, which are reflective and predictive
of metabolic health and disorder (8). Furthermore, circulating miRNAs as
novel biomarkers for DM and diabetic complications have been
assessed in different studies (11,13).
For example, Zampetaki et al (14) revealed distinct profiles of serum
miRNAs between patients with T2DM when compared with non-DM
patients in a Bruneck cohort using miRNAs microarray technology.
Similar findings were reported in Singapore by Karolina et
al (15). Previous studies
have also shown that certain specific serum miRNAs arre
differentially expressed in patients with T2DM, compared with
normal individuals, in China using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis (16–19). Furthermore, studies have shown that
the majority of these candidate miRNAs are involved in regulating
insulin secretion, insulin resistance, glucose homeostasis and/or
lipid metabolism implicated in pathology of T2DM (8,20–22).
Therefore, differentially expressed miRNAs in the
blood may be suitable biomarkers for predicting T2DM or associated
complications. However, miRNAs and their role in the etiology and
pathogenesis of T2DM remain to be fully elucidated. Furthermore,
inconsistent results have been obtained from different studies of
T2DM-associated miRNAs, which may be due to ethnic variance of
samples, different inclusion/exclusion criteria or different
methods of miRNA analysis. A previous investigation revealed an
ethnicity-specific miRNA profile of T2DM (23). Although RT-qPCR analysis is
generally used to identify T2DM-associated miRNAs, certain studies
have used high-throughput and microarray profiling, particulary
those investigating Chinese cohorts. Increased knowledge of the
circulating miRNA profiles of Chinese patients with T2DM can
further contribute to current understanding of the development of
T2DM with regards to different ethnic origins. Therefore, in the
present study, an miRNA RT-qPCR array, combining the advantages of
microarray and qPCR technology, was used to investigate differences
in serum miRNA expression profiles between patients with T2DM and
healthy subjects in Chinese cohorts. A total of seven potential
miRNA biomarkers were identified in the patients with T2DM from the
Chinese population. These miRNAs potentially regulated 97 T2DM
candidate genes, which were enriched in several Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathways, including insulin,
adipocytokine and T2DM pathways, elucidating the pathogenesis of
T2DM.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Guangzhou University
of Chinese Medicine (Guangzhou, China). All participants provided
signed written informed consent prior to experiments.
Participants
A total of 10 patients with T2DM, comprising six
women and four men aged 48–66 years old (58.2±7.7 years), were
recruited from the First Affiliated Hospital of Guangzhou
University of Chinese Medicine between October 2013 to December
2013. All patients were diagnosed by the criteria of the American
Diabetes Association (24).
Patients were excluded if they presented with severe diabetic
complications, including stroke and/or other diseases in addition
to T2DM, including infectious or inflammatory diseases, psychiatric
conditions, serious somatic diseases or dyslipidemia. In addition,
five healthy subjects, comprising three women and two mean aged
51–61 years old (56.4±3.7 years), were recruited as a control group
through local advertisement. The healthy subjects were free of any
endocrine diseases, including T2DM, and met the exclusion criteria
for diabetes, which was then confirmed by Professor Ming Hong (The
First Affiliated Hospital of Guangdong Pharmaceutical University)
based on medical examination. These individuals were also excluded
if they were overweight/obese, presented with a family history of
diabetes or were on long-term medication.
Serum sample collection
Each participant, following a period of fasting
between 7:00 a.m. and 9:00 a.m., had whole venous blood (>3 ml)
collected in a vacuum tube sans anti-coagulants. The samples were
stored in a 4°C refrigerator for 1 h to allow complete blood
coagulation. Subsequently, the yellow supernatant (serum) was
centrifuged at 6,640 × g for 10 min at 4°C to remove residual
cellular components. Every 250 µl of serum was then packed in a
frozen storage tube of RNase-free medium (Corning Incorporated,
Corning, NY, USA) and stored at −80°C prior to use. The whole
procedure was completed within 2 h following blood sampling.
RNA isolation
Total RNA, including miRNA, was isolated from serum
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. The
concentration and purity of the RNA samples were determined using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific,
Inc.). RNA integrity was evaluated by denaturing agarose gel
electrophoresis. RNA samples with a met 260/280 value >1.7 and
an RNA concentration (20 µl) >60 µg/µl were used for the miRNA
RT-qPCR array.
miRNA RT-qPCR array
For each sample, ~20–25 ng of total RNA containing
miRNA was reverse transcribed into cDNA using the MicroRNA Reverse
Transcription kit and the RT Primer Pools (Exiqon A/S, Vedbaek,
Denmark) according to the manufacturer's protocol. The resulting
cDNA served as a template for miRNA qPCR analysis in an ABI
PRISM7900 system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the miRCURY LNA™ Universal RT microRNA PCR
system, Ready-to-use Serum/Plasma Focus Human Panel I (Exiqon A/S;
cat. no. 203886), which detected 372 human mature miRNAs in the
serum samples from the 10 T2DM patients and five healthy subjects.
Specifically, the resulting cDNA template was diluted 110 times in
nuclease free water. The 10 µl reaction volume contained 5 µl
SYBR®-Green master mix, 1 µl PCR primer mix (Exiqon A/S)
and 4 µl diluted cDNA template. The amplification profile was
denatured at 95°C for 10 min, followed by 38 cycles of 95°C for 10
sec and 60°C for 60 sec. Melting curve analyses were performed at
the end of the PCR cycles. All procedures were performed according
to the manufacturer's protocol.
Determination of differentially
expressed miRNAs and cluster analysis
The raw quantification cycle (Cq) values were
obtained with the software supplied with the real-time qPCR
instrument. The data was further analyzed with GenEx qPCR (Exiqon
A/S) and SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) analysis
software. Briefly, the threshold value was set in the exponential
amplification phase of the PCR. The Cq values were determined by
the numbers of PCR cycles and threshold values. Undetectable data
were assigned a default Cq value of 38. The Cq values were
normalized by the delta Cq method with the housekeeping gene,
SNORD38B, which had a stable Cq value in the serum of two groups.
Differences in the delta Cq value between control and T2DM subjects
were compared using Student's t-test (two-tailed). The relative
expression levels (fold-change) of miRNAs between the two groups,
were calculated using 2−(ΔCq of disease group−ΔCq of control
group) (25). The miRNAs
which matched P<0.05 and fold change >3.0 or <0.33 were
defined as differentially expressed miRNAs. Data are presented
asrthe mean ± standard deviation. Cluster analysis for
differentially expressed miRNAs was performed using Multiple
Experiment Viewer 4.9 software (TM4; http://www.jcvi.org/cms/research/software/) (26). The median center method was used to
adjust genes/rows. Hierarchical clustering based on Pearson's
correlation distance metric with average linkage was used to
construct gene and sample trees.
miRNA target prediction and T2DM
candidate gene search
To evaluate the functions of the differentially
expressed miRNAs, miRNA target prediction was performed using the
miRSystem database (version 20150312; http://mirsystem.cgm.ntu.edu.tw/) (27), which integrates the seven target
gene prediction algorithms, Diana-microT (version 4.0), miRanda
(August 2010 release), miRBridge (April 2010 release), PicTar
(March 2007 release), PITA (August 2008 release), RNA22 (version
2.0) and Targetscan (version 6.0), and two experimentally validated
databases, TarBase (version 7.0) and miRecords (November 2010
release). In the present study, only validated genes or
miRNA-target interactions identified by at least three prediction
programs were considered for further analysis. Target prediction
was performed separately for upregulated and downregulated
miRNAs.
To investigate the interactive association between
target genes regulated by differentially expressed miRNAs and
candidate genes for T2DM, the VENNY 1.0 tool (http://bioinfogp.cnb.csic.es/tools/venny_old/index.html)
(28) was used to compare the
lists of predicted targets of those miRNAs with a list of 563
candidate genes for T2DM using Venn diagrams. The list of 563
candidate genes was obtained from T2DM in T-the Text-mined
Hypertension, Obesity and Diabetes candidate gene database (last
updated on January 2th, 2014; http://bws.iis.sinica.edu.tw/THOD/) (29). This database provides lists of
candidate genes for hypertension, obesity and diabetes, and is
regularly updated by text-mining technologies, including a
gene/disease identification system and a disease-gene relation
extraction system, which is used to affirm the association of genes
with the three diseases by domain experts. Furthermore, this
database provides textual evidence of previous literature,
disease-centric protein-protein interaction network, and integrated
gene and single-nucleotide polymorphism information.
Functional annotation analysis of the
predicted targets
The intersected gene list between predicted targets
(upregulated and downregulated miRNAs) and candidate genes for T2DM
were separately submitted to the Database for Annotation,
Visualization, and Integrated Discovery (version 6.7) (30,31),
and the putative targets were annotated using KEGG pathway analysis
(http://david.abcc.ncifcrf.gov/). The
count threshold was set as two genes per annotation term. The
threshold of EASE score, expressed as P-value in the present study
and is a modified Fisher Exact P-value for gene-enrichment
analysis, was set as 0.05. P<0.05 was considered to indicate
increased enrichment in the annotation categories (https://david.ncifcrf.gov/helps/functional_annotation.html,
#summary).
Results
Differential miRNA expression in serum
between patients with T2DM and healthy subjects
An miRNA qPCR array containing 372 human serum
mature miRNAs was used to compare the serum miRNA expression
profiles between 10 patients with T2DM and five healthy subjects. A
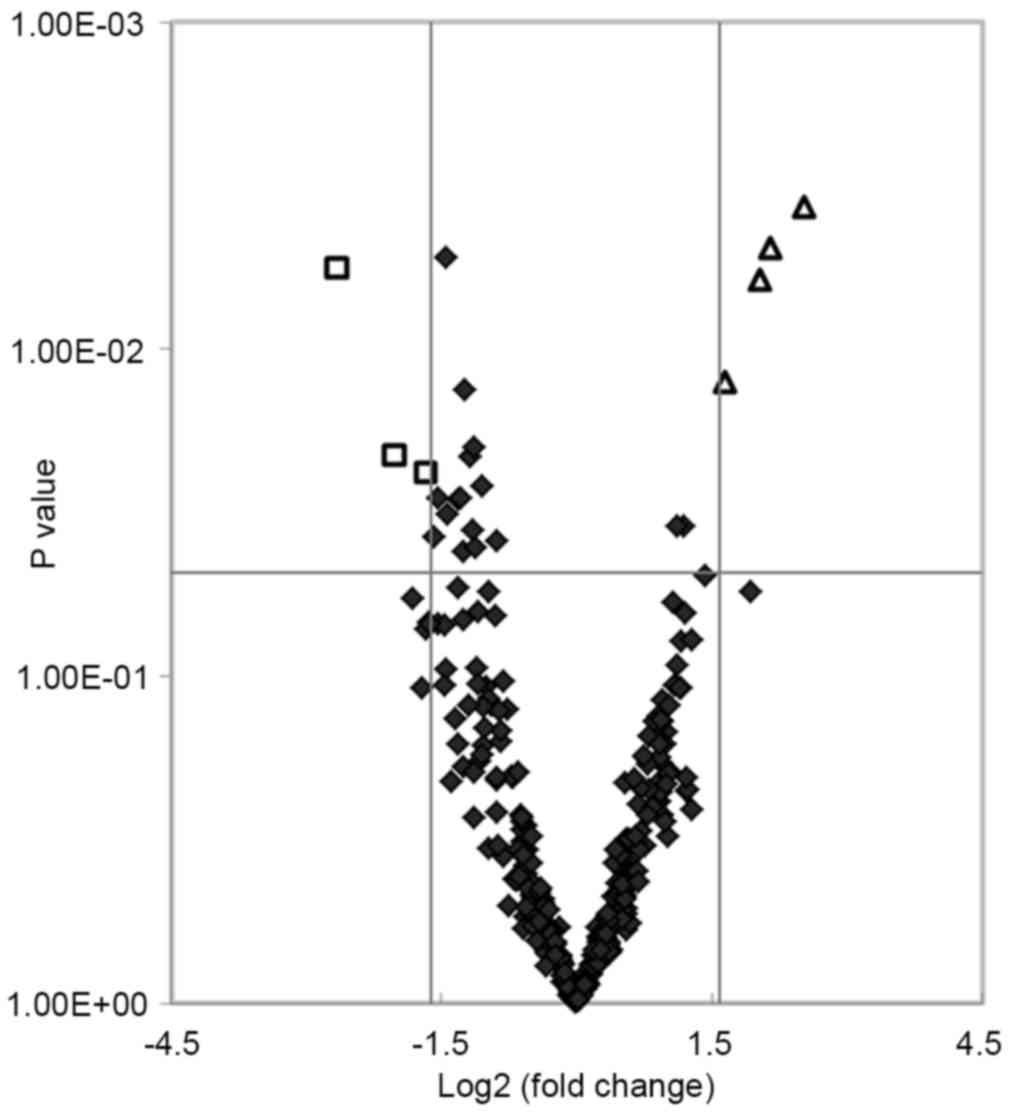
total of 24 miRNAs showed significant differences (P<0.05) in
expression levels between the two groups. Of these, seven miRNAs
matched the fold change >3.0 or <0.33 (shown as red and green
in Fig. 1). The fold changes of
the seven miRNAs are presented in Fig.
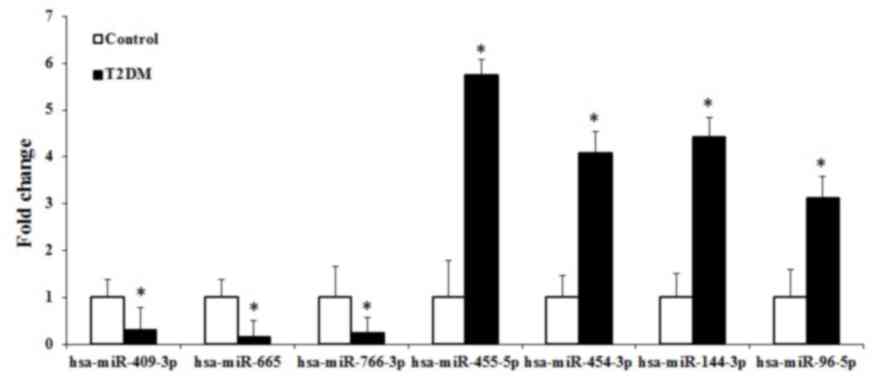
2. The present study found that four miRNAs (hsa-miR-455-5p,
hsa-miR-454-3p, hsa-miR-144-3p and hsa-miR-96-5p) and three miRNAs
(hsa-miR-409-3p, hsa-miR-665 and hsa-miR-766-3p) were upregulated
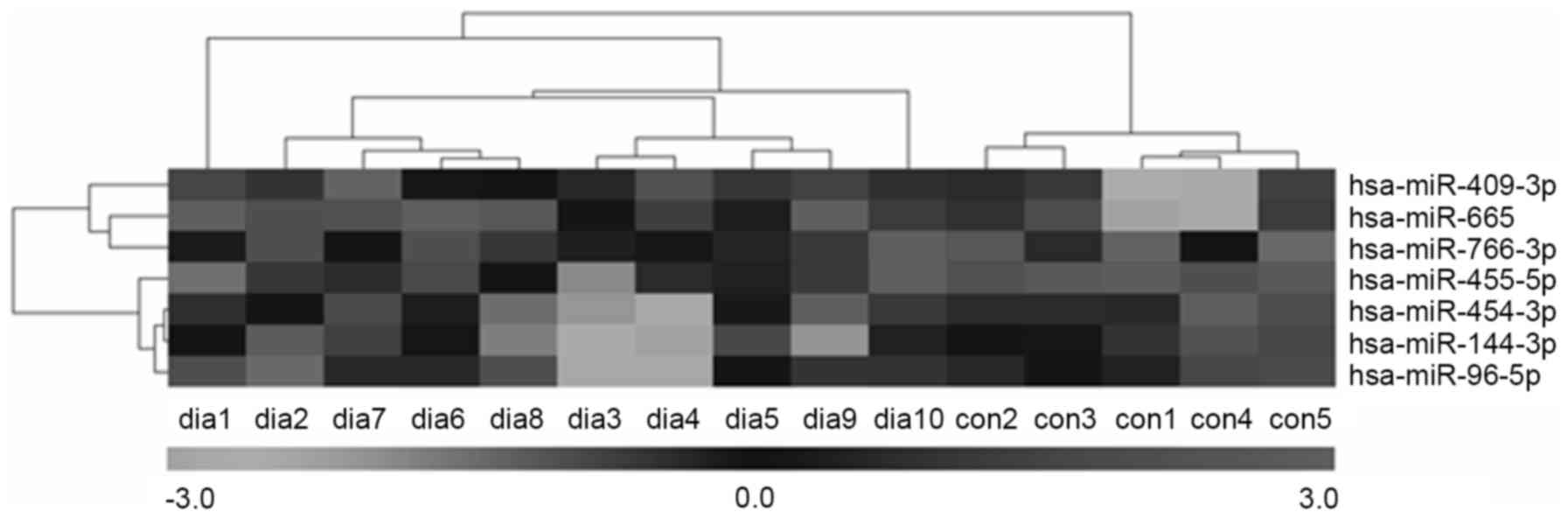
and downregulated, respectively. Furthermore, hierarchical cluster
analysis showed that it was possible to separate patients with T2DM
and control subjects into similar categories via the seven miRNAs,
as all patients were clustered together and separated from the
control subjects (Fig. 3).
T2DM candidate genes potentially
regulated by the differentially expressed miRNAs
The predicted target genes of the seven
differentially expressed miRNAs were identified using the miRSystem
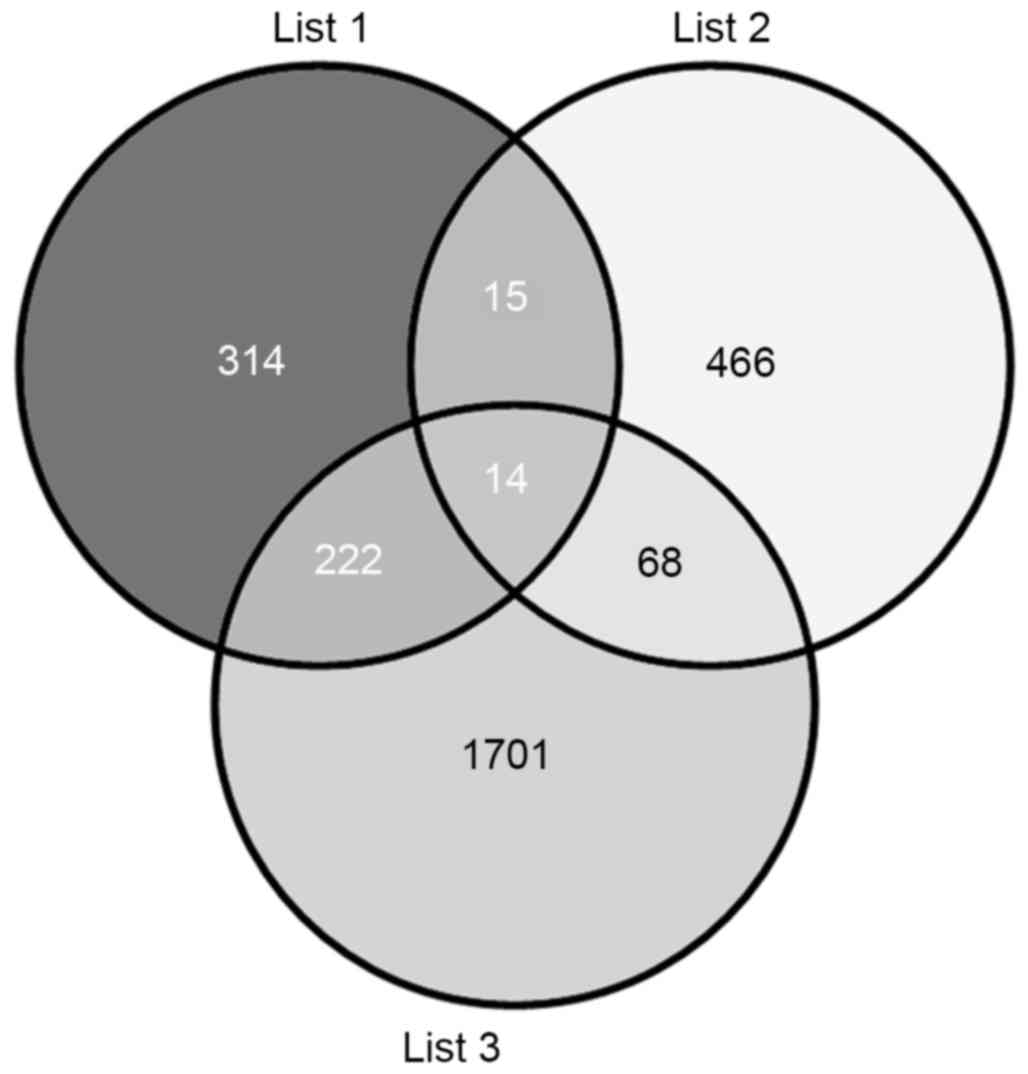
database. The results indicated 2,005 (list 3) and 565 (list 1)
putative target genes for the upregulated and downregulated miRNAs,
respectively (Fig. 4). The VENNY
tool (29) was then used to
analyze the overlaps among the predicted target genes (list 1 and
list 3), and T2DM candidate genes (list 2). The Venn diagrams
showed that, of the 563 T2DM candidate genes, a total of 97 T2DM
candidate genes were regulated by the differentially expressed
miRNAs identified in the present study, with 29 and 82 genes being
predicted by the downregulated and upregulated miRNAs,
respectively, and 14 by predicted by both (Fig. 4).
Potential functions of the
differentially expressed miRNAs
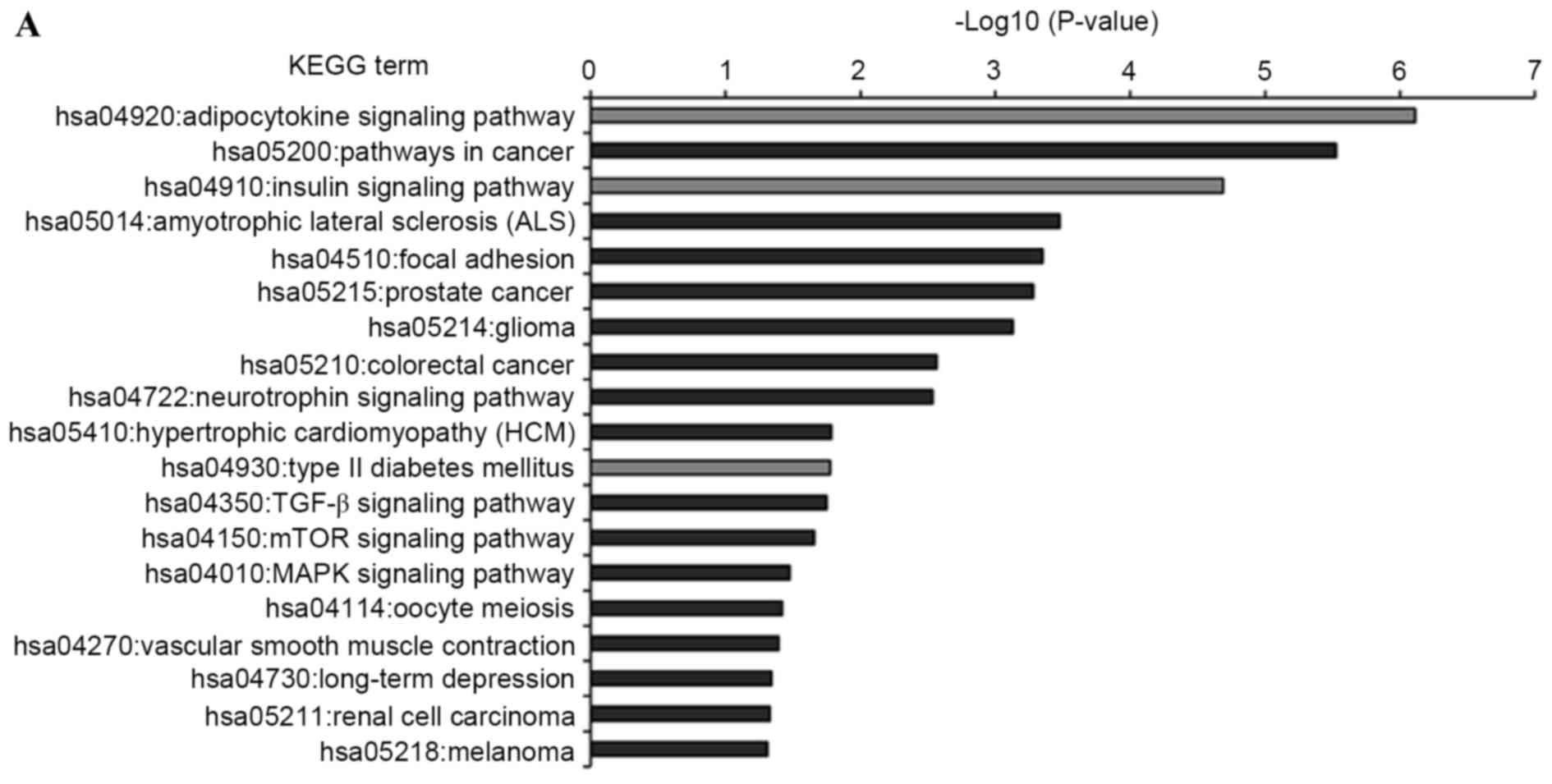
KEGG pathway analysis was introduced to annotate the
T2DM candidate genes predicted by the differentially expressed
miRNAs. The results showed that the T2DM candidate genes predicted
by upregulated and downregulated miRNAs were enriched in 19 and 13
KEGG pathways, respectively (Fig.
5). Furthermore, eight KEGG pathways were shared by both, which
comprised insulin, adipocytokine, mammalian target of rapamycin
(mTOR), long-term depression, hypertrophic cardiomyopathy (HCM),
cancer (melanoma and glioma) and focal adhesion signaling pathways.
Of the 24 enriched pathways, three KEGG pathways were directly
associated with the pathomechanism of T2DM. These were T2DM,
insulin and adipocytokine signaling pathways, which included 19
T2DM candidate genes, which were regulated by the differentially
expressed miRNAs. The interactions between these T2DM targets and
the seven differentially expressed miRNAs are shown in Fig. 6. Of the 19 targets, seven targets
were regulated by downregulated miRNAs, 12 targets were regulated
by upregulated miRNAs, and three targets were regulated by both,
which were RPS6KB1, SHC1 and peroxisome proliferator-activated
receptor γ, coactivator 1α (PPARGC1A). Considering the fold-changes
of the miRNAs for these three targets, RPS6KB1 and PPARGC1A were
potentially repressed and SHC1 was overexpressed by corresponding
miRNAs (Figs. 2 and 7).
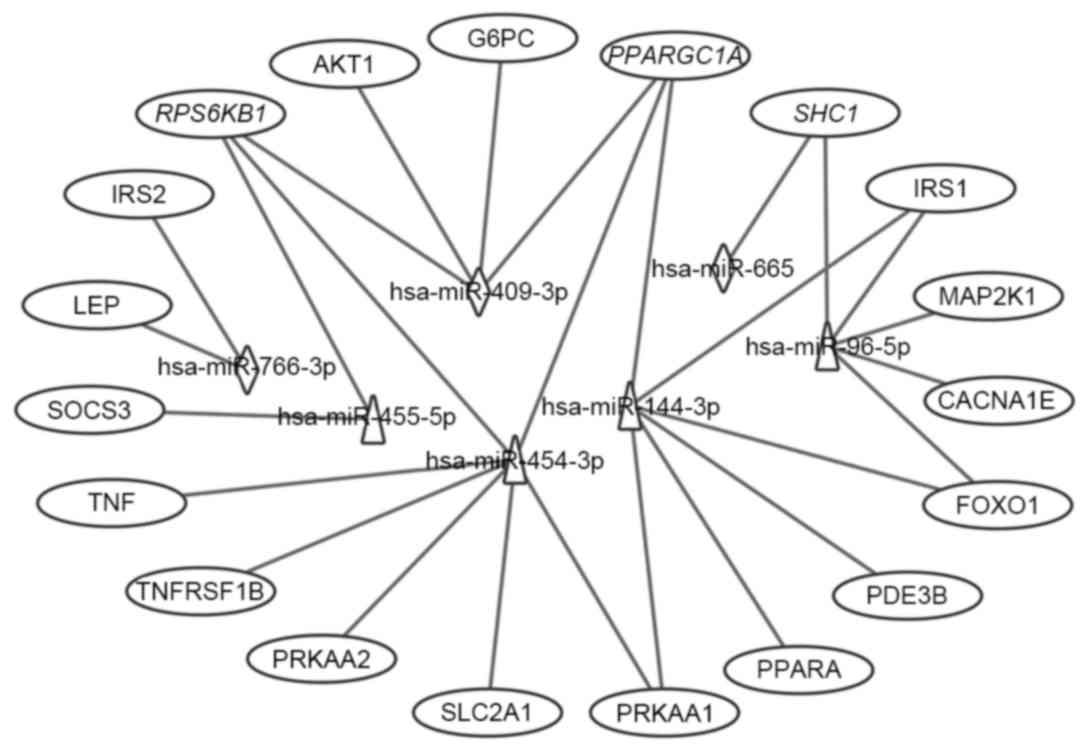 | Figure 6.Interaction networks of the
differential miRNAs and their T2DM target genes associated with
type II diabetes mellitus, insulin and adipocytokine signaling
pathways. Triangles represent upregulated miRNAs; diamonds
represent downregulated miRNAs and ovals represent target genes for
T2DM. Italics (RPS6KB1, PPARGC1A and SHC1) indicate genes targeted
by downregulated and upregulated miRNAs. miR, microRNA; T2DM, type
2 diabetes mellitus; CACNA1E, calcium voltage-gated channel subunit
α1 E; TNFRSF1B, tumor necrosis factor α receptor superfamily,
member 1B; IRS, insulin receptor substrate; SOCS3, suppressor of
cytokine signaling 3; PPARGC1A, peroxisome proliferator-activated
receptor γ, coactivator 1α; PPARA, peroxisome
proliferator-activated receptor α; PRKAA1, protein kinase,
AMP-activated, α1; AKT1, v-akt murine thymoma viral oncogene
homolog 1; SLC2A1, solute carrier family 2, member 1; G6PC,
glucose-6-phosphatase, catalytic subunit; LEP, leptin; SHC1, Src
homology 2 domain containing, transforming protein 1; MAP2K1,
mitogen-activated protein kinase kinase 1; RPS6KB1, ribosomal
protein S6 kinase, 70 kDa, polypeptide 1; PDE3B. phosphodiesterase
3B; FOXO1, forkhead box O1. |
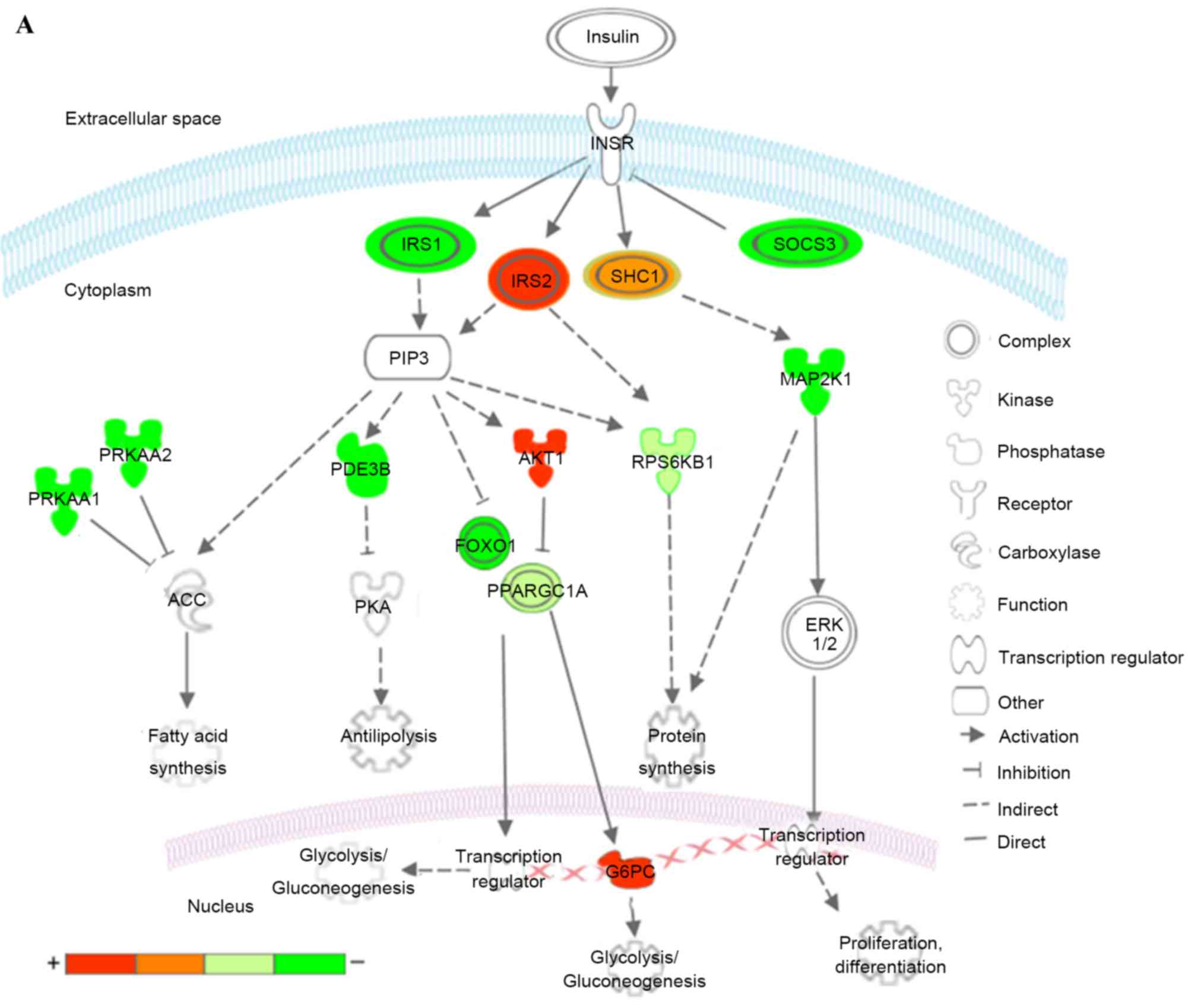 | Figure 7.Kyoto Encyclopedia of Genes and
Genomes pathways associated with T2DM and T2DM candidate genes
regulated by the differentially expressed miRNAs. (A) Insulin
signaling pathway (B) Pathways of adipocytokine signaling and type
II diabetes mellitus. Red represents overexpressed genes targeted
by downregulated miRNAs, and green represents repressed genes
targeted by upregulated miRNAs. Orange and pea green represent
genes targeted by both downregulated and upregulated miRNAs,
although the former was primarily overexpressed and the latter was
repressed. miR, microRNA; T2DM, type 2 diabetes mellitus; CACNA1E,
calcium voltage-gated channel subunit α1 E; KIR, inward rectifying
potassium channel; SUR1, sulfonylurea receptor 1; GLUT2, glucose
transporter 2; TNFα, tumor necrosis factor α; TNFRSF1B, receptor
superfamily, member 1B; IRS, insulin receptor substrate; INSR, INS
receptor; SOCS3, suppressor of cytokine signaling 3; PPARGC1A,
peroxisome proliferator-activated receptor γ, coactivator 1α;
PPARA, peroxisome proliferator-activated receptor α; PRKAA1/2,
protein kinase, AMP-activated, α1 and 2; AKT1, v-akt murine thymoma
viral oncogene homolog 1; GTP-1, glutamate pyruvate transaminase-1;
SLC2A1, solute carrier family 2, member 1; G6PC,
glucose-6-phosphatase, catalytic subunit; LEP, leptin; LEPR, leptin
receptor; ADIPO, adiponectinp; ADIPOR, adiponectin receptor; SHC1,
Src homology 2 domain containing, transforming protein 1; MAP2K1,
mitogen-activated protein kinase kinase 1; RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; ACC, acetyl-CoA
carboxylase; PDE3B, phosphodiesterase 3B; FOXO1, forkhead box O1;
ERK, extracellular signal-regulated kinase. |
In the insulin signaling pathway (Fig. 7A), upregulated miRNAs inhibited
insulin receptor substrate (IRS) 1, further reduced antilipolysis
via phosphodiesterase 3B, (PDE3B; cGMP-inhibited) and protein
synthesis via ribosomal protein S6 kinase, 70 kDa, polypeptide 1
(RPS6KB1), increased lipogenesis via protein kinase, AMP-activated,
α1 and 2 catalytic subunits (PRKAA1 and 2), and affected
glycolysis/gluconeogenesis via forkhead box O1 (FOXO1) and
PPARGC1A. The upregulated miRNAs decreased cell proliferation,
differentiation and protein synthesis via Src homology 2 domain
containing, transforming protein 1 (SHC1) and mitogen-activated
protein kinase kinase 1 (MAP2K1). By contrast, the downregulated
miRNAs predominantly affected glycolysis/gluconeogenesis via IRS2,
v-akt murine thymoma viral oncogene homolog 1 (AKT1), PPARGC1A and
glucose-6-phosphatase, catalytic subunit (G6PC). The downregulated
miRNAs also increased protein synthesis via SHC1 and RPS6KB1.
Combining these two aspects, patients with T2DM exhibited insulin
resistance, characteristic of a metabolic disorder of glucose and
lipid homeostasis.
In the adipocytokine and T2DM signaling pathways
(Fig. 7B), upregulated miRNAs
affected the insulin signaling pathway via IRS1, tumor necrosis
factor α (TNFα), tumor necrosis factor receptor superfamily, member
1B (TNFRSF1B) and suppressor of cytokine signaling 3 (SOCS3),
reduced mitochondrion β-oxidation via PPARGC1A and peroxisome
proliferator-activated receptor α (PPARA), and decreased glucose
uptake via PRKAA1/2 and solute carrier family 2, member 1 (SLC2A1).
In addition, upregulated miRNAs inhibited insulin secretion via
calcium voltage-gated channel subunit α1 E (CACNA1E), a voltage
dependent R type calcium channel. The downregulated miRNAs
increased mitochondrion β-oxidation via PPARGC1A and leptin (LEP),
and increase gluconeogenesis via G6PC.
Discussion
In the present study, a qPCR array, which included
372 human mature miRNAs, was used to examine differences in serum
miRNA expression profiles between patients with T2DM and healthy
subjects. A total of seven differentially expressed miRNAs were
identified, which improved stratification of the two groups. Target
gene prediction indicated that 97 T2DM candidate genes were
regulated by these miRNAs. KEGG functional annotation showed that
these T2DM candidate genes were significantly enriched in the
insulin, adipocytokine and T2DM signaling pathways, as well as
several other pathways.
As the pathogenesis of T2DM remains to be fully
elucidated, biomarkers used for the early detection and
identification of at risk individuals have the potential to improve
patient quality of life by providing improved management. The
levels of serum miRNAs derived from various tissues/organs are
stable, reproducible and consistent among individuals of the same
ethnic origin. Additionally, the expression profiles of serum
miRNAs better reflect underlying pathological/physiologic processes
(10,32) and have been used extensively in
various types of cancer (6,7) and
metabolic syndromes (15),
including T2DM (8,11). Furthermore, accumulating evidence
has identified several serum miRNAs, which regulate insulin
signaling, glucose and lipid metabolism, as implicated in T2DM
pathology (8,20–22).
Thus, serum miRNAs may serve as novel biomarkers for T2DM and also
assist in explaining its pathogenesis.
Several studies have assessed the differences in
serum or plasma miRNA expression between patients with T2DM and
healthy non-diabetic subjects. Using miRNA microarray profiling
confirmed by qPCR, Zampetaki et al (14) first identified low plasma levels of
miR-15a, miR-29b, miR-126 and miR-223, and high levels of miR-28-3p
in patients with T2DM, compared with non-diabetic individuals in
Bruneck, Italy. Karolina et al (15) found upregulation in the levels of
miR-27a, miR-150, miR-192, miR-320a and miR-375 in the blood and
exosomes of patients with T2DM, compared with healthy controls in
Singapore. Using qPCR analysis of specific miRNAs, Kong et
al (18) also found that seven
candidate miRNAs (miR-9, miR-29a, miR-30d, miR-34a, miR-124a,
miR-146a and miR-375) were significantly upregulated in serum from
patients newly diagnosed with T2DM, compared with T2DM-susceptible
individuals and normal glucose tolerance in a Chinese cohort. These
pioneering studies demonstrated the potential of miRNAs as
biomarkers for T2DM, although with mixed results. In the present
study, seven differently expressed miRNAs were identified. The
upregulation of miR-144-3p has been supported in several other
reports. Wang et al (23)
found that a higher expression of miR-144 in plasma was
significantly associated with T2DM in Sweden. Similar results were
reported by Yang et al (33), and Zhu and Leung (34) showed that the upregulation of
circulating miR-144 may be a potential biomarker for T2DM in a
meta-analysis of controlled profiling studies. Karolina et
al (8) found that miR-144 was
significantly increased in blood samples from a T2DM rat model. In
addition, the upregulation of miR-144 was shared among patients
with T1DM, T2DM and gestational diabetes mellitus in peripheral
blood mononuclear cells, and expression was higher in muscles of
patients with T2DM, compared with healthy individuals (35). Compared with the findings of the
present study, low expression levels of miR-96 were reported by
Yang et al (19) in the
serum of patients with T2DM, compared with normal glucose tolerance
controls. To the best of our knowledge, none of the other
differentially expressed miRNAs identified in the present study
have been reported in previous studies associated with T2DM. Thus,
the present study may have identified novel dysregulated miRNAs in
patients with T2DM, compared with control individuals, at least in
the Chinese population examined.
The present study also predicted the T2DM candidate
genes, which were potentially regulated by the seven differential
miRNAs. Of the 563 T2DM candidate genes, 97 genes were identified
(Fig. 4), which may be important
in explaining the role of these miRNAs in the pathogenesis of T2DM.
KEGG functional annotation of these targets showed that several
pathways were potentially modulated by these upregulated and/or
downregulated miRNAs (Fig. 5). The
majority of these pathways have been previously associated with
T2DM, including insulin and adipocytokine signaling pathways, T2DM,
pathways in cancer, focal adhesion, and hypertrophic cardiomyopathy
(as described below). These findings may provide novel insights
into the complex molecular mechanisms involved in T2DM.
Relative insulin deficiency and insulin resistance
are important characteristics in the development of T2DM
pathogenesis. In the present study, three signaling pathways
(insulin, adipocytokine and T2DM) showed marked enrichment with the
19 T2DM candidate genes modulated by the downregulated and
upregulated miRNAs (Figs. 6 and
7), which have been implicated in
insulin secretion and function in T2DM. For insulin secretion, the
upregulation of miR-96-5p represses CACNA1E and then results in
impaired insulin secretion. Similar reports have shown that miR-96
negatively regulates insulin exocytosis by granuphilin/SLP4
(20). Dysregulated insulin and
adipocytokine signaling pathways can affect glucose, lipid, and
protein metabolism, which result in insulin resistance.
Specifically, these identified miRNAs may dysregulate the
glycolysis/gluconeogenesis process via the targeting of IRS1, IRS2,
FOXO1, PPARGC1A, AKT1 and G6PC, and repress glucose uptake via
PRKAA1/2 and SLC2A1. They may also dysregulate the process of
lipogenesis via the targeting of IRS1, IRS2, PDE3B and PRKAA1/2,
and inhibit mitochondrial β-oxidation via PPARA and PPARGC1A. In
addition, these miRNAs may dysregulate protein synthesis processes
via the targeting of IRS1, IRS2, SHC1, MAP2K1 and RPS6KB1. In
addition, TNFα, TNFRSF1B and LEP indirectly affect insulin
signaling pathways and lipolysis processes. Karolina et al
(8) experimentally demonstrated
that IRS1 is the target of miR-144, and that increased circulating
levels of miR-144 are correlated with downregulation of its
predicted target, IRS1, at the mRNA and protein levels. Similar
results were reported by Yang et al (33). Furthermore, Jeong et al
(36) and Wang et al
(37) revealed that IRS1 is also
the target of miR-96. FOXO1 was experimentally demonstrated in
several investigations (38,39)
to serve as the target of miR-96. None of the other interactions of
the target-miRNAs identified in the present study have been
reported previously. Of note, previous reports have shown that
several miRNAs identified in the present study were involved in
carbohydrate and lipid metabolism. Hu et al (40) and Ramírez et al (41) revealed that miR-144 regulates
cholesterol metabolism and plasma levels of high-density
lipoprotein, and promotes pro-inflammatory cytokine production. Fu
et al (42) found that
miR-144 regulates carbohydrate and lipid metabolism by inhibiting
isocitrate dehydrogenase 2, which acts as key enzyme of the
tricarboxylic acid cycle. Similar reports have also demonstrated
functions of miR-96, which controls selective high-density
lipoprotein cholesterol and cholesteryl ester uptake, and regulates
endogenous lipid synthesis (22,43,44).
In addition, Milagro et al (45) found that the expression of miR-766
is correlated with weight loss. Therefore, these miRNAs may be able
to regulate lipid metabolism through the insulin, adipocytokine and
T2DM pathways. Additionally, the dysregulation of carbohydrate and
lipid metabolism modulated by the identified miRNAs may be an
important pathogenic mechanism of T2DM.
Evidence of an association between DM and cancer has
been sugested, although without a definitive conclusion. Previous
reports have shown that DM and insulin resistance are risk factors
for gastric, hepatocellular and prostate cancer (46). In addition, breast cancer, colon
cancer (47), melanoma (48), renal cell carcinoma (49) and pancreatic cancer (50) have been implicated in the
progression of T2DM. In the present study, and in agreement with
the previous studies, several signaling pathways were found to be
involved. Previous studies have also shown that these miRNAs are
associated with increased risk of cancer. For example, miR-144-3p
exerts antitumor effects in glioblastoma (51), and is a diagnostic marker for
breast cancer (52), follicular
thyroid cancer (53), laryngeal
carcinoma (54) and papillary
thyroid carcinoma (55). Similar
results also revealed an association between miRNA-96-5p and
several types of cancer, including breast cancer (56), colorectal carcinoma (57), epithelial ovarian cancer (58), pancreatic carcinoma (59) and prostate cancer (60). miR-454-3p can enhance cellular
radiosensitivity in renal carcinoma cells by inhibiting the
expression of BTG anti-proliferation factor 1 (61). miR-455-5p can promote melanoma
growth and metastasis through inhibition of the tumor suppressor
gene, cytoplasmic polyadenylation element binding protein 1
(62). In additiob, miR-455-5p was
identified as a molecular signature associated with anaplastic
large cell lymphoma (63), basal
cell carcinoma (64), endometrial
serous adenocarcinomas (65) and
laryngeal cancer (66). miR-409-3p
suppresses the invasion and metastasis of colorectal (67) and bladder cancer (68), but promotes the tumorigenesis of
human prostate cancer (69) and
gastric cancer (70). Furthermore,
plasma miR-409-3p serves as a promising biomarker for the early
detection of breast cancer (71)
and colorectal cancer (72). The
downregulation of miR-665 may be closely associated with the
invasive metastatic and chemoresistance of gastric signet ring cell
carcinoma (73). However, no
report has shown an association between miR-766-3p and cancer.
Taken together, the findings of the present study corroborated with
previous studies, which linked T2DM and cancer. It is possible that
a number of the patients with T2DM in the present study were at
risk of cancer.
In the present study, the predicted target genes
were also significantly enriched in the focal adhesion and
hypertrophic cardiomyopathy pathways. Wang et al (74) found that the focal adhesion pathway
is significantly dysregulated in the progression of T2DM by
assessing differentially expressed genes between human pancreatic
islets with T2DM and normal islets. Similar results were reported
in female visceral and subcutaneous adipose, and in male visceral
adipose and skeletal muscle of patients with T2DM (75). In terms of HCM pathway, asymmetric
left ventricular hypertrophy and impairment in diastolic function
were important characteristics of HCM. Dinh et al (76) and Shigematsu et al (77) found that insulin resistance and
glycemic abnormalities were associated with the deterioration of
left ventricular diastolic function. Okayama et al (78) also revealed that the presence of
obstructive coronary stenosis and the magnitude of left ventricular
hypertrophy were associated with the presence of diabetes,
triglyceride levels and estimated glomerular filtration rate. In
addition, the results of previous studies have shown that the mTOR
signaling pathway, also enriched in the present study, is
implicated in left ventricular remodeling, myocardial infarction
and hypertrophic cardiomyopathy (79,80).
Therefore, the findings of the present study suggested that the
abnormal pathway of focal adhesion may be a pathological feature of
T2DM, and that aberrant expression of miRNAs may also induce
diabetic cardiomyopathy by targets implicated in the HCM
pathway.
In conclusion, the present study identified seven
differentially expressed miRNAs by using an miRNA qPCR array. These
miRNAs clearly discriminated patients with T2DM from healthy
subjects and offer potential as suitable biomarkers for T2DM by
assessing for abnormal expression. In addition, target gene
prediction revealed that a total of 97 T2DM candidate genes may be
regulated by these differential miRNAs. The results of the present
study were concordant with those of previous reports, to a certain
extent, in that several biological pathways previously implicated
in T2DM were potentially modulated by the seven miRNAs, including
insulin and adipocytokine signaling pathways, T2DM and several
cancer-associated pathways. Taken together, the results of the
present study may provide novel insight into the possibility that
circulating miRNAs can be used as potential biomarkers for T2DM,
which assists in improving current understanding of the
pathomechanism and biological pathways underlying T2DM.
Acknowledgements
The authors would like to thank Dr Zhang-Zhi Zhu and
Dr Sai-Mei Li of the First Affiliated Hospital of Guangzhou
University of Chinese Medicine for support in participant
recruitment, and Li-Ping Zhang of the Art Department of Guangdong
Light Industry School, China, for assisting with figures. The
authors would also like to thank John Lees (Monell Chemical Senses
Center, Philadelphia, PA, USA), for language editing. This study
was supported by the National Natural Science Foundation of China
(grant no. 81102703 to Professor Ze-Min Yang), the Science and
Technology Planning Project of Guangdong Province of China (grant
no. 2013A032500005 to Professor Ze-Min Yang), the Administration of
Traditional Chinese Medicine of Guangdong Province of China (grant
no. 20123001 to Professor Wei-Wen Chen), the Special Funds from
Central Finance of China in Support of the Development of Local
Colleges and Universities in 2013 (grant no. 338 to Professor
Wei-Wen Chen), the Natural Science Foundation for Fostering of
Guangdong Pharmaceutical University of China (grant no.
GYFYLH201303 to Professor Ze-Min Yang), and the South China Chinese
Medicine Collaborative Innovation Center (grant no. A1-AFD01514A05
to Professor Wei-Wen Chen). Dr Long-Hui Chen received support from
the China Scholarship Council as a joint PhD student at the
University of Pennsylvania, USA.
References
|
1
|
Whiting DR, Guariguata L, Weil C and Shaw
J: IDF diabetes atlas: Global estimates of the prevalence of
diabetes for 2011 and 2030. Diabetes Res Clin Pract. 94:311–321.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fowler MJ: Microvascular and macrovascular
complications of diabetes. Clinical Diabetes. 26:77–82. 2008.
View Article : Google Scholar
|
|
3
|
Ferrannini E, Gastaldelli A and Iozzo P:
Pathophysiology of prediabetes. Med Clin North Am. 95:327–339,
vii-viii. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
6
|
Rossing M, Borup R, Henao R, Winther O,
Vikesaa J, Niazi O, Godballe C, Krogdahl A, Glud M, Hjort-Sørensen
C, et al: Down-regulation of microRNAs controlling tumourigenic
factors in follicular thyroid carcinoma. J Mol Endocrinol.
48:11–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karolina DS, Armugam A, Tavintharan S,
Wong MT, Lim SC, Sum CF and Jeyaseelan K: MicroRNA 144 impairs
insulin signaling by inhibiting the expression of insulin receptor
substrate 1 in type 2 diabetes mellitus. PLoS One. 6:e228392011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kantharidis P, Wang B, Carew RM and Lan
HY: Diabetes complications: The microRNA perspective. Diabetes.
60:1832–1837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guay C and Regazzi R: Circulating
microRNAs as novel biomarkers for diabetes mellitus. Nat Rev
Endocrinol. 9:513–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chien HY, Lee TP, Chen CY, Chiu YH, Lin
YC, Lee LS and Li WC: Circulating microRNA as a diagnostic marker
in populations with type 2 diabetes mellitus and diabetic
complications. J Chin Med Assoc. 78:204–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zampetaki A, Kiechl S, Drozdov I, Willeit
P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E,
et al: Plasma microRNA profiling reveals loss of endothelial
miR-126 and other microRNAs in type 2 diabetes. Circ Res.
107:810–887. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karolina DS, Tavintharan S, Armugam A,
Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF and Jeyaseelan K:
Circulating miRNA profiles in patients with metabolic syndrome. J
Clin Endocrinol Metab. 97:E2271–E2276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang T, Lv C, Li L, Chen S, Liu S, Wang C
and Su B: Plasma miR-126 is a potential biomarker for early
prediction of type 2 diabetes mellitus in susceptible individuals.
Biomed Res Int. 2013:7616172013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rong Y, Bao W, Shan Z, Liu J, Yu X, Xia S,
Gao H, Wang X, Yao P, Hu FB and Liu L: Increased microRNA-146a
levels in plasma of patients with newly diagnosed type 2 diabetes
mellitus. PLoS One. 8:e732722013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao
Y, Dong Q, Pang Z, Guan Q, Gao L, et al: Significance of serum
microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A
clinical study. Acta Diabetol. 48:61–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Z, Chen H, Si H, Li X, Ding X, Sheng
Q, Chen P and Zhang H: Serum miR-23a, a potential biomarker for
diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol.
51:823–831. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen H, Lan HY, Roukos DH and Cho WC:
Application of microRNAs in diabetes mellitus. J Endocrinol.
222:R1–R10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dehwah MA, Xu A and Huang Q: MicroRNAs and
type 2diabetes/obesity. J Genet Genomics. 39:11–18. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeon TI, Esquejo RM, Roqueta-Rivera M,
Phelan PE, Moon YA, Govindarajan SS, Esau CC and Osborne TF: An
SREBP-responsive microRNA operon contributes to a regulatory loop
for intracellular lipid homeostasis. Cell Metab. 18:51–61. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Sundquist J, Zöller B, Memon AA,
Palmér K, Sundquist K and Bennet L: Determination of 14 Circulating
microRNAs in Swedes and Iraqis with and without Diabetes Mellitus
Type 2. PLoS One. 9:e867922014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
American Diabetes Association, . Economic
costs of diabetes in the U.S. in 2012. Diabetes Care. 36:1033–1046.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saeed AI, Sharov V, White J, Li J, Liang
W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et
al: TM4: A free, open-source system for microarray data management
and analysis. Biotechniques. 34:374–378. 2003.PubMed/NCBI
|
|
27
|
Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK,
Lai LC and Chuang EY: miRSystem: An integrated system for
characterizing enriched functions and pathways of microRNA targets.
PLoS One. 7:e423902012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oliveros JC; VENNY. An interactive tool
for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.htmlNovember
20–2013.
|
|
29
|
Dai HJ, Wu JC, Tsai RT, Pan WH and Hsu WL:
T-HOD: A literature-based candidate gene database for hypertension,
obesity and diabetes. Database (Oxford). 2013:bas0612013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gilad S, Meiri E, Yogev Y, Benjamin S,
Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed
N, et al: Serum microRNAs are promising novel biomarkers. PLoS One.
3:e31482008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang S, Zhao J, Chen Y and Lei M:
Biomarkers associated with ischemic stroke in diabetes mellitus
patients. Cardiovasc Toxicol. 16:213–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu H and Leung SW: Identification of
microRNA biomarkers in type 2 diabetes: A meta-analysis of
controlled profiling studies. Diabetologia. 58:900–911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Collares CV, Evangelista AF, Xavier DJ,
Rassi DM, Arns T, Foss-Freitas MC, Foss MC, Puthier D,
Sakamoto-Hojo ET, Passos GA and Donadi EA: Identifying common and
specific microRNAs expressed in peripheral blood mononuclear cell
of type 1, type 2, and gestational diabetes mellitus patients. BMC
Res Notes. 6:4912013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeong HJ, Park SY, Yang WM and Lee W: The
induction of miR-96 by mitochondrial dysfunction causes impaired
glycogen synthesis through translational repression of IRS-1 in
SK-Hep1 cells. Biochem Biophys Res Commun. 434:503–508. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Luo H, Li Y, Chen T, Wu S and Yang
L: hsa-miR-96 up-regulates MAP4K1 and IRS1 and may function as a
promising diagnostic marker in human bladder urothelial carcinomas.
Mol Med Rep. 5:260–265. 2012.PubMed/NCBI
|
|
38
|
Yu JJ, Wu YX, Zhao FJ and Xia SJ: miR-96
promotes cell proliferation and clonogenicity by down-regulating of
FOXO1 in prostate cancer cells. Med Oncol. 31:9102014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fendler A, Jung M, Stephan C, Erbersdobler
A, Jung K and Yousef GM: The antiapoptotic function of miR-96 in
prostate cancer by inhibition of FOXO1. PLoS One. 8:e808072013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu YW, Hu YR, Zhao JY, Li SF, Ma X, Wu SG,
Lu JB, Qiu YR, Sha YH, Wang YC, et al: An agomir of miR-144-3p
accelerates plaque formation through impairing reverse cholesterol
transport and promoting pro-inflammatory cytokine production. PLoS
One. 9:e949972014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ramírez CM, Rotllan N, Vlassov AV, Dávalos
A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno
A, et al: Control of cholesterol metabolism and plasma high-density
lipoprotein levels by microRNA-144. Circ Res. 112:1592–1601. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fu X, Huang X, Li P, Chen W and Xia M:
7-Ketocholesterol inhibits isocitrate dehydrogenase 2 expression
and impairs endothelial function via microRNA-144. Free Radic Biol
Med. 71:1–15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si
SY and Hong B: MicroRNAs 185, 96, and 223 repress selective
high-density lipoprotein cholesterol uptake through
posttranscriptional inhibition. Mol Cell Biol. 33:1956–1964. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Meyer JM, Graf GA and van der Westhuyzen
DR: New developments in selective cholesteryl ester uptake. Curr
Opin Lipidol. 24:386–392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Milagro FI, Miranda J, Portillo MP,
Fernandez-Quintela A, Campión J and Martínez JA: High-throughput
sequencing of microRNAs in peripheral blood mononuclear cells:
Identification of potential weight loss biomarkers. PLoS One.
8:e543192013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Best LG, García-Esquinas E, Yeh JL, Yeh F,
Zhang Y, Lee ET, Howard BV, Farley JH, Welty TK, Rhoades DA, et al:
Association of diabetes and cancer mortality in American Indians:
The strong heart study. Cancer Causes Control. 26:1551–1560. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Onitilo AA, Stankowski RV, Berg RL, Engel
JM, Glurich I, Williams GM and Doi SA: Type 2 diabetes mellitus,
glycemic control, and cancer risk. Eur J Cancer Prev. 23:134–140.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qi L, Qi X, Xiong H, Liu Q, Li J, Zhang Y,
Ma X, Wu N, Liu Q and Feng L: Type 2 diabetes mellitus and risk of
malignant melanoma: A systematic review and meta-analysis of cohort
studies. Iran J Public Health. 43:857–866. 2014.PubMed/NCBI
|
|
49
|
Vavallo A, Simone S, Lucarelli G,
Rutigliano M, Galleggiante V, Grandaliano G, Gesualdo L, Campagna
M, Cariello M, Ranieri E, et al: Pre-existing type 2 diabetes
mellitus is an independent risk factor for mortality and
progression in patients with renal cell carcinoma. Medicine
(Baltimore). 93:e1832014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brodovicz KG, Kou TD, Alexander CM,
O'Neill EA, Engel SS, Girman CJ and Goldstein BJ: Impact of
diabetes duration and chronic pancreatitis on the association
between type 2 diabetes and pancreatic cancer risk. Diabetes Obes
Metab. 14:1123–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lan F, Yu H, Hu M, Xia T and Yue X:
miR-144-3p exerts anti-tumor effects in glioblastoma by targeting
c-Met. J Neurochem. 135:274–286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang CW, Wu HC, Terry MB and Santella RM:
microRNA expression in prospectively collected blood as a potential
biomarker of breast cancer risk in the BCFR. Anticancer Res.
35:3969–3977. 2015.PubMed/NCBI
|
|
53
|
Stokowy T, Eszlinger M, Świerniak M,
Fujarewicz K, Jarząb B, Paschke R and Krohn K: Analysis options for
high-throughput sequencing in miRNA expression profiling. BMC Res
Notes. 7:1442014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lu ZM, Lin YF, Jiang L, Chen LS, Luo XN,
Song XH, Chen SH and Zhang SY: Micro-ribonucleic acid expression
profiling and bioinformatic target gene analyses in laryngeal
carcinoma. Onco Targets Ther. 7:525–533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Swierniak M, Wojcicka A, Czetwertynska M,
Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M,
Koperski L, de la Chapelle A and Jazdzewski K: In-depth
characterization of the microRNA transcriptome in normal thyroid
and papillary thyroid carcinoma. J Clin Endocrinol Metab.
98:E1401–E1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Matamala N, Vargas MT, González-Cámpora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor MicroRNA expression
profiling identifies circulating MicroRNAs for early breast cancer
detection. Clin Chem. 61:1098–1096. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang L, Zhu MJ, Ren AM, Wu HF, Han WM, Tan
RY and Tu RQ: A ten-microRNA signature identified from a
genome-wide microRNA expression profiling in human epithelial
ovarian cancer. PLoS One. 9:e964722014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li C, Du X, Tai S, Zhong X, Wang Z, Hu Z,
Zhang L, Kang P, Ji D, Jiang X, et al: GPC1 regulated by miR-96-5p,
rather than miR-182-5p, in inhibition of pancreatic carcinoma cell
proliferation. Int J Mol Sci. 15:6314–6327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Larne O, Martens-Uzunova E, Hagman Z,
Edsjö A, Lippolis G, den Berg MS, Bjartell A, Jenster G and Ceder
Y: miQ-a novel microRNA based diagnostic and prognostic tool for
prostate cancer. Int J Cancer. 132:2867–2875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu X, Ding N, Hu W, He J, Xu S, Pei H, Hua
J, Zhou G and Wang J: Down-regulation of BTG1 by miR-454-3p
enhances cellular radiosensitivity in renal carcinoma cells. Radiat
Oncol. 9:1792014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu C, Iqbal J, Teruya-Feldstein J, Shen
Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino
E, et al: MicroRNA expression profiling identifies molecular
signatures associated with anaplastic large cell lymphoma. Blood.
122:2083–2092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Saito K, Inagaki K, Kamimoto T, Ito Y,
Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H,
et al: MicroRNA-196a is a putative diagnostic biomarker and
therapeutic target for laryngeal cancer. PLoS One. 8:e714802013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bai R, Weng C, Dong H, Li S, Chen G and Xu
Z: MicroRNA-409-3p suppresses colorectal cancer invasion and
metastasis partly by targeting GAB1 expression. Int J Cancer.
137:2310–2322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu
X, Zhu Y, Li S, Zheng X and Xie L: MicroRNA-409-3p inhibits
migration and invasion of bladder cancer cells via targeting c-Met.
Mol Cells. 36:62–88. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/−5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li C, Nie H, Wang M, Su L, Li J, Yu B, Wei
M, Ju J, Yu Y, Yan M, et al: MicroRNA-409-3p regulates cell
proliferation and apoptosis by targeting PHF10 in gastric cancer.
Cancer Lett. 320:189–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cuk K, Zucknick M, Heil J, Madhavan D,
Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, et al:
Circulating microRNAs in plasma as early detection markers for
breast cancer. Int J Cancer. 132:1602–1612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X,
Yu L, Wang L, Wang J, Wu Y, et al: A plasma microRNA panel for
early detection of colorectal cancer. Int J Cancer. 136:152–161.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen J, Sun D, Chu H, Gong Z, Zhang C,
Gong B, Li Y, Li N and Jiang L: Screening of differential microRNA
expression in gastric signet ring cell carcinoma and gastric
adenocarcinoma and target gene prediction. Oncol Rep. 33:2963–2971.
2015.PubMed/NCBI
|
|
74
|
Wang Q, Zhao Z, Shang J and Xia W: Targets
and candidate agents for type 2 diabetes treatment with
computational bioinformatics approach. J Diabetes Res.
2014:7639362014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jain P, Vig S, Datta M, Jindel D, Mathur
AK, Mathur SK and Sharma A: Systems biology approach reveals genome
to phenome correlation in type 2 diabetes. PLoS One. 8:e535222013.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dinh W, Lankisch M, Nickl W, Scheyer D,
Scheffold T, Kramer F, Krahn T, Klein RM, Barroso MC and Füth R:
Insulin resistance and glycemic abnormalities are associated with
deterioration of left ventricular diastolic function: A
cross-sectional study. Cardiovasc Diabetol. 9:632010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shigematsu Y, Hamada M, Nagai T, Nishimura
K, Inoue K, Suzuki J, Ogimoto A and Higaki J: Risk for atrial
fibrillation in patients with hypertrophic cardiomyopathy:
Association with insulin resistance. J Cardiol. 58:18–25. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Okayama S, Soeda T, Kawakami R, Takami Y,
Somekawa S, Ueda T, Sugawara Y, Matsumoto T, Sung JH, Nishida T, et
al: Evaluation of coronary artery disease and cardiac morphology
and function in patients with hypertrophic cardiomyopathy, using
cardiac computed tomography. Heart Vessels. 30:28–35. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Diniz GP, Carneiro-Ramos MS and
Barreto-Chaves ML: Angiotensin type 1 receptor mediates thyroid
hormone-induced cardiomyocyte hypertrophy through the
Akt/GSK-3beta/mTOR signaling pathway. Basic Res Cardiol.
104:653–667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Marin TM, Keith K, Davies B, Conner DA,
Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, et al:
Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of
LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest.
121:1026–1043. 2011. View Article : Google Scholar : PubMed/NCBI
|