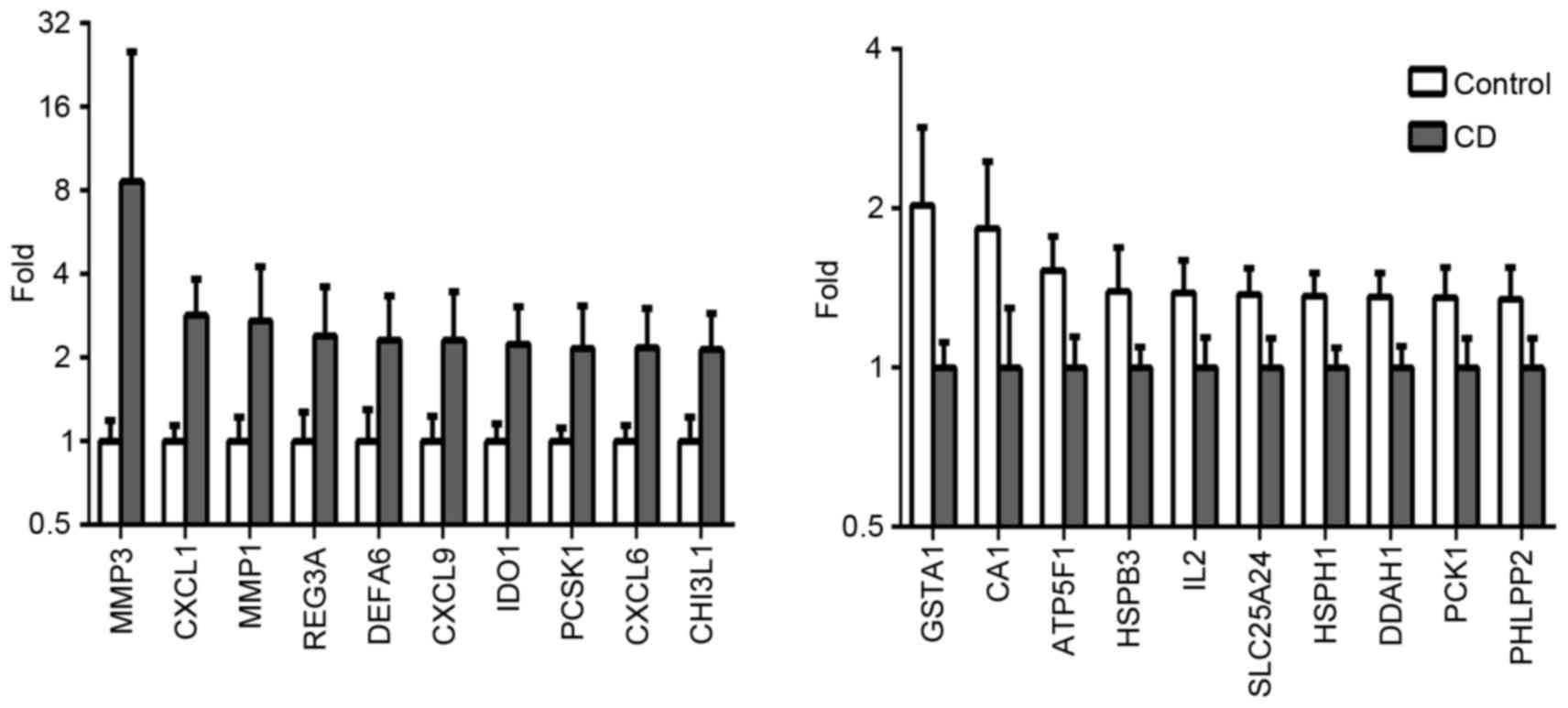
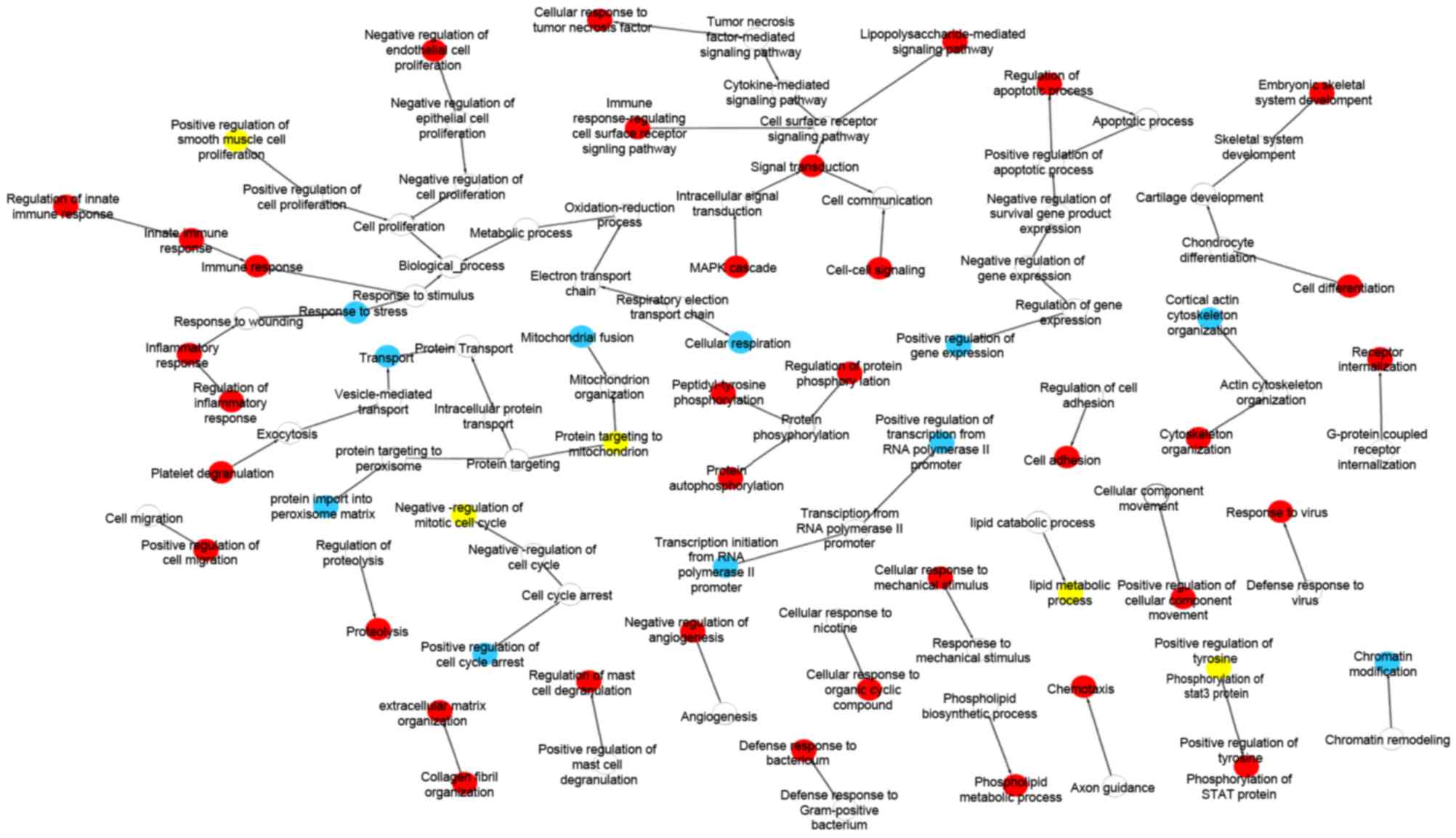
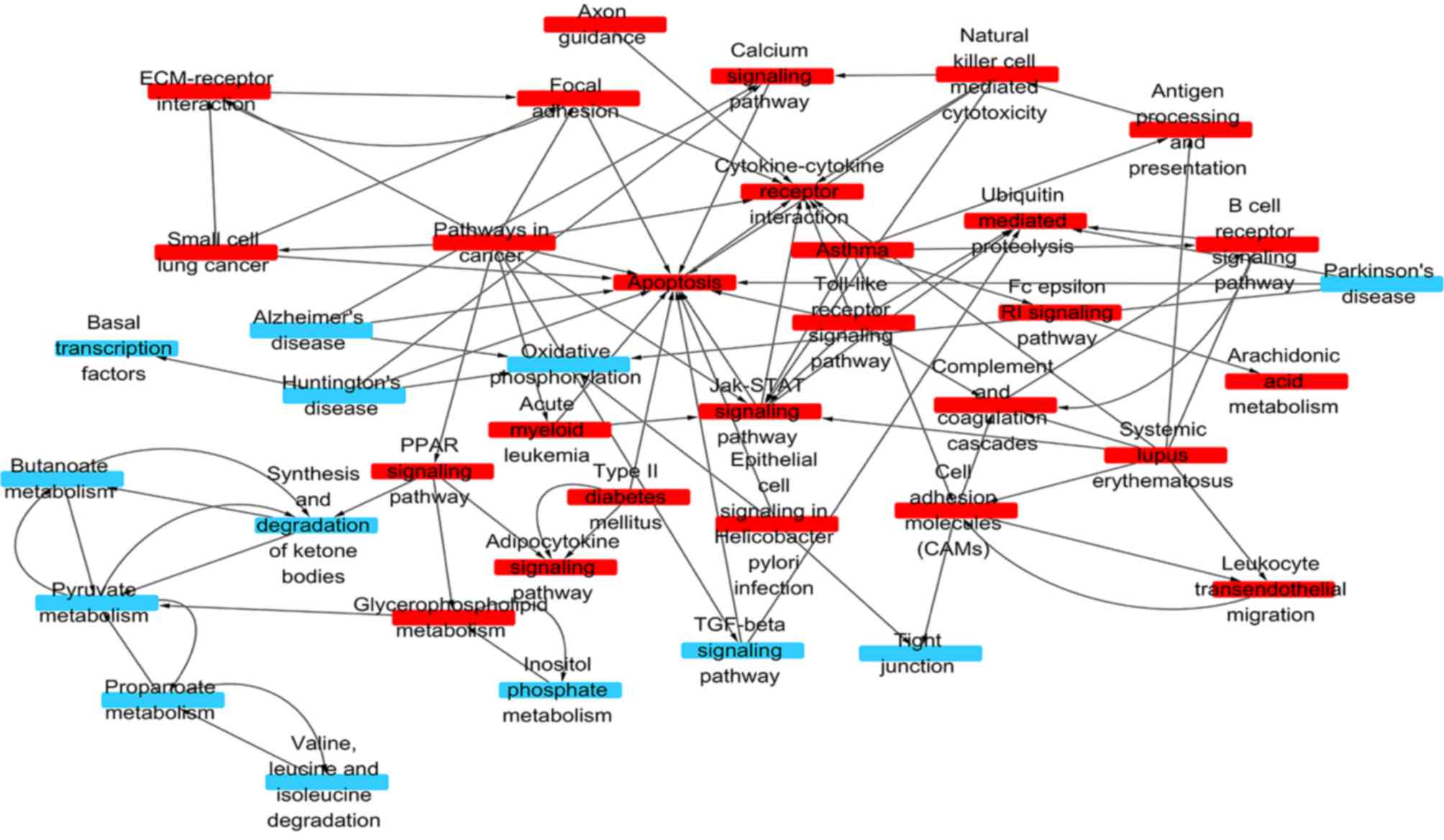
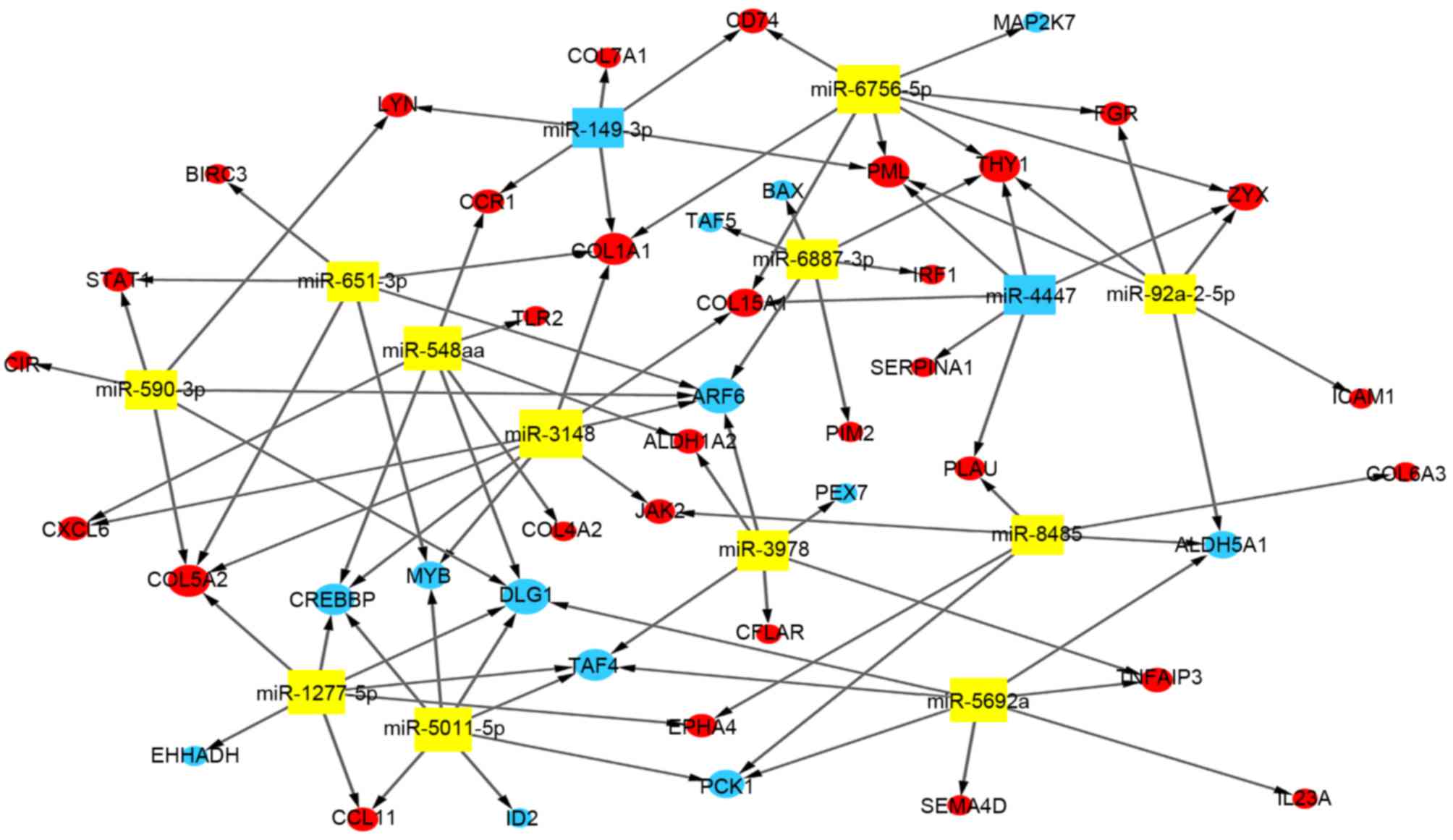
|
1
|
Molodecky NA, Soon IS, Rabi DM, Ghali WA,
Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema
HW and Kaplan GG: Increasing incidence and prevalence of the
inflammatory bowel diseases with time, based on systematic review.
Gastroenterology. 142:46–54.e42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frolkis AD, Dykeman J, Negrón ME, Debruyn
J, Jette N, Fiest KM, Frolkis T, Barkema HW, Rioux KP, Panaccione
R, et al: Risk of surgery for inflammatory bowel diseases has
decreased over time: A systematic review and meta-analysis of
population-based studies. Gastroenterology. 145:996–1006. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Limbergen J, Philpott D and Griffiths
AM: Genetic profiling in inflammatory bowel disease: From
association to bedside. Gastroenterology. 141:1566–1571.e1. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen L, Zhuo D, Chen J and Yuan H:
Screening feature genes of lung carcinoma with DNA microarray
analysis. Int J Clin Exp Med. 8:12161–12171. 2015.PubMed/NCBI
|
|
5
|
Zhan C, Yan L, Wang L, Jiang W, Zhang Y,
Xi J, Jin Y, Chen L, Shi Y, Lin Z and Wang Q: Landscape of
expression profiles in esophageal carcinoma by the cancer genome
atlas data. Dis Esophagus. 29:920–928. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database issue):
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Montero-Melendez T, Llor X,
García-Planella E, Perretti M and Suárez A: Identification of novel
predictor classifiers for inflammatory bowel disease by gene
expression profiling. PLoS One. 8:e762352013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wright GW and Simon RM: A random variance
model for detection of differential gene expression in small
microarray experiments. Bioinformatics. 19:2448–2455. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng J, Wang T, Wang J, Wang Y and Chen J:
Extending gene ontology with gene association networks.
Bioinformatics. 32:1185–1194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
ELIFE. 4:2015. View Article : Google Scholar
|
|
12
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43(Database issue): D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei ZW, Xia GK, Wu Y, Chen W, Xiang Z,
Schwarz RE, Brekken RA, Awasthi N, He YL and Zhang CH: CXCL1
promotes tumor growth through VEGF pathway activation and is
associated with inferior survival in gastric cancer. Cancer Lett.
359:335–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peter MR, Jerkic M, Sotov V, Douda DN,
Ardelean DS, Ghamami N, Lakschevitz F, Khan MA, Robertson SJ,
Glogauer M, et al: Impaired resolution of inflammation in the
Endoglin heterozygous mouse model of chronic colitis. Mediators
Inflamm. 2014:7671852014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baugh MD, Perry MJ, Hollander AP, Davies
DR, Cross SS, Lobo AJ, Taylor CJ and Evans GS: Matrix
metalloproteinase levels are elevated in inflammatory bowel
disease. Gastroenterology. 117:814–822. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kusugami K, Nobata K, Tsuzuki T, Ando T
and Ina K: Mucosal expression of matrix metalloproteinases and
their tissue inhibitors in ulcerative colitis patients. J
Gastroenterol. 38:412–413. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
De Bruyn M, Machiels K, Vandooren J,
Lemmens B, Van Lommel L, Breynaert C, van der Goten J, Staelens D,
Billiet T, De Hertogh G, et al: Infliximab restores the
dysfunctional matrix remodeling protein and growth factor gene
expression in patients with inflammatory bowel disease. Inflamm
Bowel Dis. 20:339–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Biancheri P, Brezski RJ, Di Sabatino A,
Greenplate AR, Soring KL, Corazza GR, Kok KB, Rovedatti L,
Vossenkämper A, Ahmad N, et al: Proteolytic cleavage and loss of
function of biologic agents that neutralize tumor necrosis factor
in the mucosa of patients with inflammatory bowel disease.
Gastroenterology. 149:1564–1574.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Russo I, Luciani A, De Cicco P, Troncone E
and Ciacci C: Butyrate attenuates lipopolysaccharide-induced
inflammation in intestinal cells and Crohn's mucosa through
modulation of antioxidant defense machinery. PLoS One.
7:e328412012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Loddo I and Romano C: Inflammatory bowel
disease: Genetics, epigenetics, and pathogenesis. Front Immunol.
6:5512015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirano T, Ishihara K and Hibi M: Roles of
STAT3 in mediating the cell growth, differentiation and survival
signals relayed through the IL-6 family of cytokine receptors.
Oncogene. 19:2548–2556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aaronson DS and Horvath CM: A road map for
those who don't know JAK-STAT. Science. 296:1653–1655. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Coskun M, Salem M, Pedersen J and Nielsen
OH: Involvement of JAK/STAT signaling in the pathogenesis of
inflammatory bowel disease. Pharmacol Res. 76:1–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Warnecke-Eberz U, Chon SH, Hölscher AH,
Drebber U and Bollschweiler E: Exosomal onco-miRs from serum of
patients with adenocarcinoma of the esophagus: Comparison of miRNA
profiles of exosomes and matching tumor. Tumour Biol. 36:4643–4653.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo G, Chao YL, Tang B, Li BS, Xiao YF,
Xie R, Wang SM, Wu YY, Dong H, Liu XD and Yang SM: miR-149
represses metastasis of hepatocellular carcinoma by targeting
actin-regulatory proteins PPM1F. Oncotarget. 6:37808–37823.
2015.PubMed/NCBI
|
|
26
|
Wang F, Ma YL, Zhang P, Shen TY, Shi CZ,
Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y and Qin HL: SP1
mediates the link between methylation of the tumour suppressor
miR-149 and outcome in colorectal cancer. J Pathol. 229:12–24.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G,
Yang J, Xia L, Wang R, Cai X, Hu H, et al: MicroRNA-149 inhibits
proliferation and cell cycle progression through the targeting of
ZBTB2 in human gastric cancer. PLoS One. 7:e416932012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clay CC, Maniar-Hew K, Gerriets JE, Wang
TT, Postlethwait EM, Evans MJ, Fontaine JH and Miller LA: Early
life ozone exposure results in dysregulated innate immune function
and altered microRNA expression in airway epithelium. PLoS One.
9:e904012014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carey R, Jurickova I, Ballard E, Bonkowski
E, Han X, Xu H and Denson LA: Activation of an IL-6:STAT3-dependent
transcriptome in pediatric-onset inflammatory bowel disease.
Inflamm Bowel Dis. 14:446–457. 2008. View Article : Google Scholar : PubMed/NCBI
|