Introduction
Williams-Beuren syndrome (WBS; also termed Williams'
syndrome; Online Mendelian Inheritance in Man no. 194050) is
characterized by personality, behavioral and emotional wellbeing
irregularities, a distinctive facial appearance and congenital
cardiovascular abnormalities, particularly supravalvular aortic
stenosis (SAS). Following its first description in the 1960s
(1,2), numerous publications have reported
patients with WBS (3–5) and its estimated occurrence was ~1 in
10,000 (6). Deletion of the WBS
chromosome region (WBSCR) on the long arm of chromosome 7, spanning
1.5–1.8 million base pairs and containing 26–28 genes, was defined
as the cause of WBS (7,8). However, autism and cleft palate are
not part of the major phenotypes associated with WBS.
High-resolution single nucleotide polymorphism (SNP) arrays are
considered to be an important method for detection of
submicroscopic chromosome rearrangements; rearrangements that are
<5 Mb in size (9). The present
study provides a detailed report of a patient carrying a de
novo 1.5 Mb deletion on chromosome 7q11.23, who presented with
congenital heart disease (CHD), autism, mental retardation, growth
retardation, hypercalcemia, nephroliths and cleft palate.
Materials and methods
Clinical description
The current study was approved by the Review Board
of the Second Xiangya Hospital of Central South University
(Changsha, China). Written informed consent for the publication of
the patient images and all other details was obtained from the
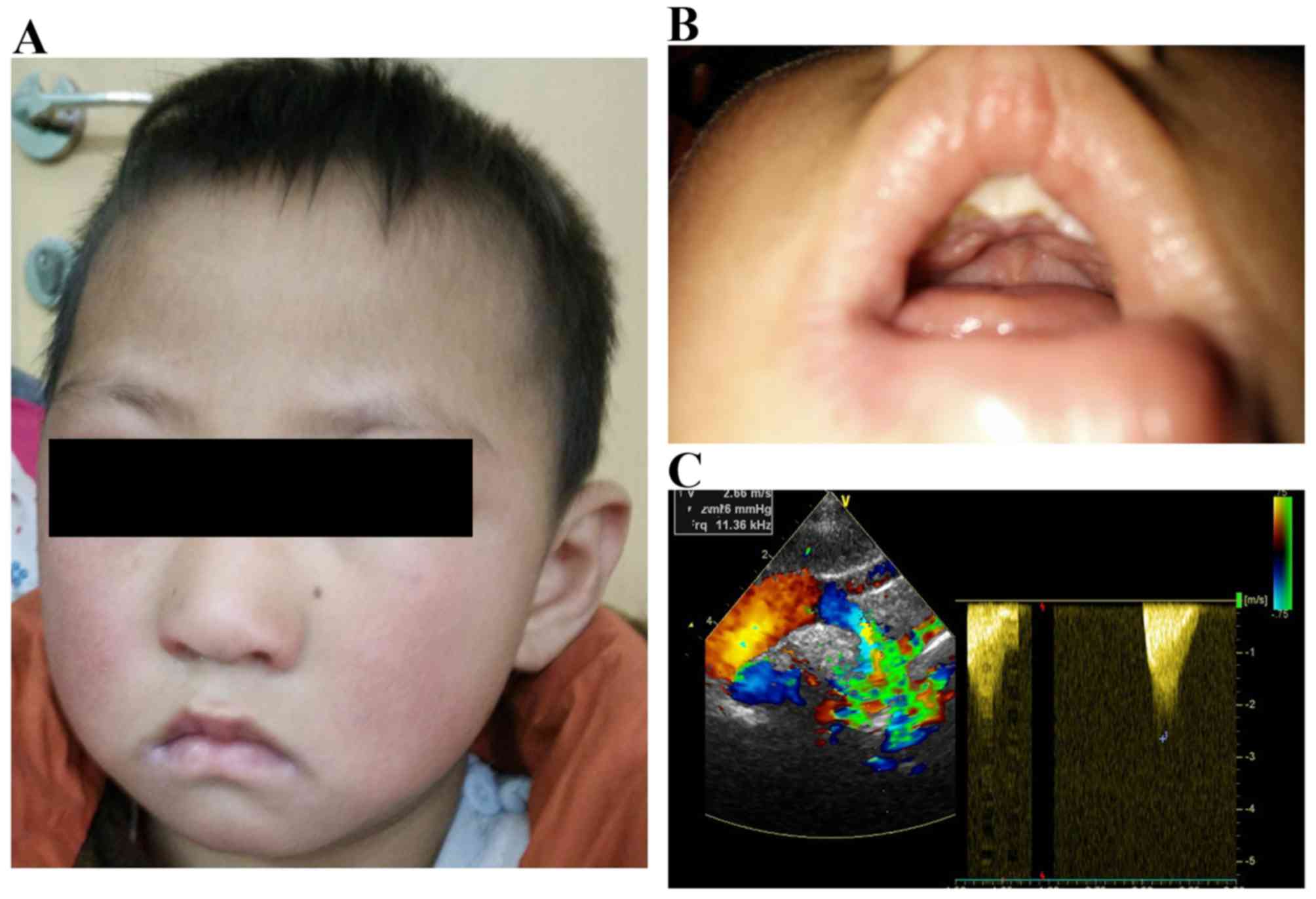
parents of the patient. The proband, a 5 year old male patient from
Hunan province (China), was born at full-term with a length of 45
cm and a birth weight of 2,650 g. He was one of two children of
non-consanguineous parents. At the birth of the patient, the father
was 24 years old and the mother was 22 years old. There was no
family history of birth defects and his brother was normal in a
pedigree analysis. The patient exhibited feeding difficulties,
cleft palate and a heart murmur following birth. Subsequently,
CHDs, SAS and patent ductus arteriosus (PDA), were identified by
transthoracic echocardiography (Fig.
1). At the age of three years, The Department of Cardiovascular
Surgery (The Second Xiangya Hospital) suggested that his cardiac
defects required treatment for the mild symptoms (PDA, 2 mm; peak
SAS gradients, 30 mmHg). At that time, the patient had moderate
growth retardation, weighed 11 kg and had a height of 85 cm. The
patient was later also diagnosed with hypercalcemia, nephroliths
and impaired visuospatial abilities following examination. The
patient was further diagnosed with autism and severe expressive
language impairment by the Mental Health Institute of The Second
Xiangya Hospital. The current study used the Autism Diagnostic
Interview-Revised (ADI-R) (10,11)
to perform diagnostic and behavioral assessments. The patient met
the ADI-R criteria for autism in all three domains. The symptoms of
the patient included severe deficits in the comprehension of simple
language, reciprocal conversation, articulation and socializing
with peers. The patient also met the Autism Diagnostic Observation
Schedule criteria (10,12) for autism in the restricted
behavior, social and communication domain. The patient also
presented with excessive non-social anxiety and was easily angered.
The cognitive profile of the patient on the Wechsler Preschool and
Primary Scale of Intelligence-3rd edition (13,14)
highlighted a low IQ range compared with his peers.
Cytogenetic analysis
Peripheral blood (5 ml) from the patient and parents
were collected and chromosome analysis by conventional G-banded
techniques (550 bands resolution) was performed as previously
described (15,16). According to standard cytogenetic
protocol (16), all samples were
subjected to lymphocyte culture.
DNA extraction
The genomic DNA was extracted from peripheral blood
of the patient and his parents. Genomic DNA was prepared using a
DNeasy Blood & Tissue kit (Qiagen, Inc., Valencia, CA, USA) on
the QIA cube automated DNA extraction robot (Qiagen, GmbH, Hilden,
Germany).
SNP-array analysis
Genomic DNA samples, at a final concentration of 50
ng/ml, of the patient and his parents were used in the present
study. The Illumina BeadScan genotyping system (Beadstation Scanner
500; Illumina, Inc., San Diego, CA, USA) and the HumanOmni1-Quad
Chip (Illumina, Inc.) were employed to obtain signal intensities of
SNP probes.
Results
The karyotypes of the patient and his parents were
normal. The current study employed a high-resolution SNP array
system to analyze the whole genome for copy number variations
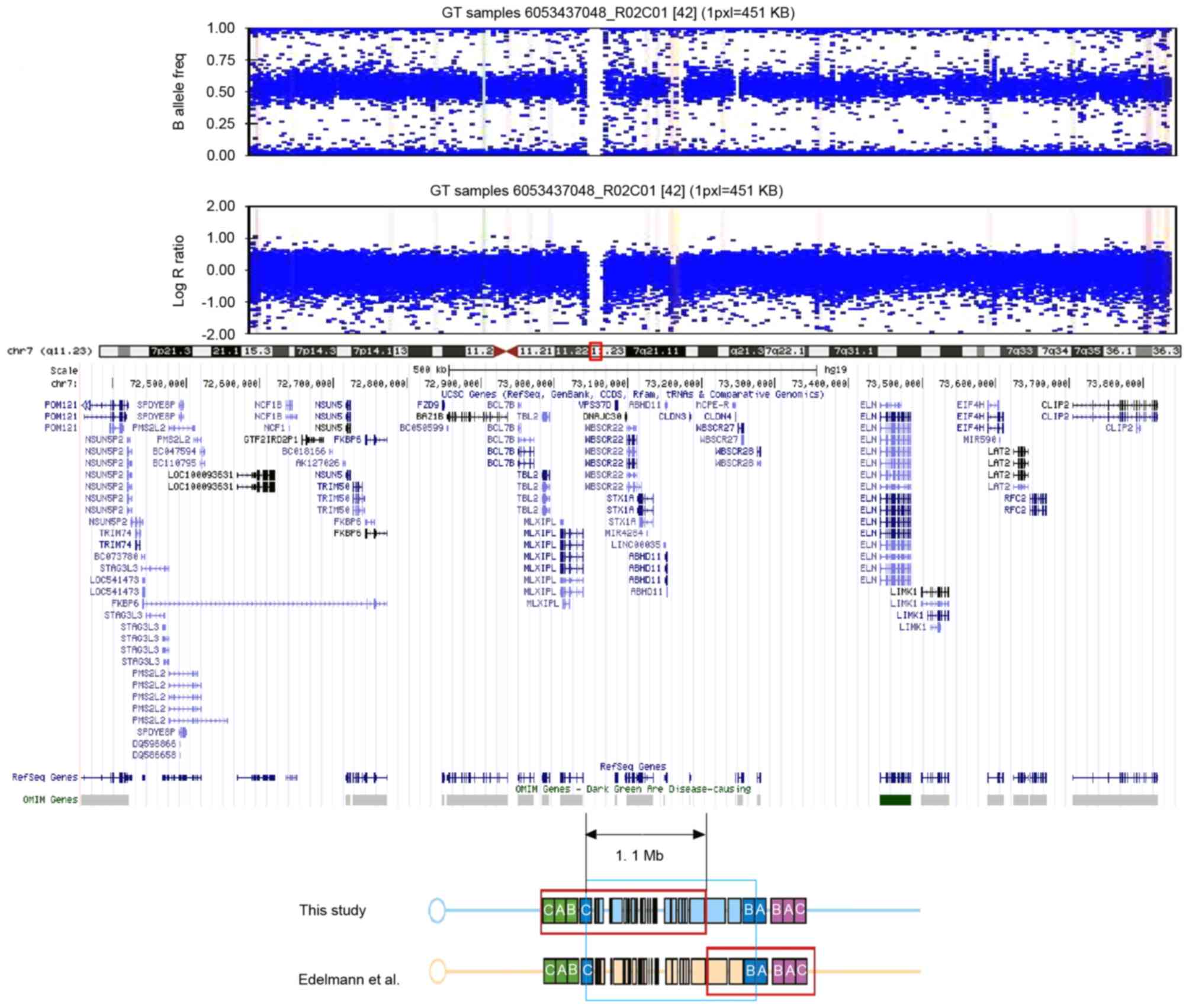
(CNVs). A total of 309 CNVs were discovered. A de novo 1.5
Mb deletion of chromosome 7q11.23 (Chr7: 72,357,322–73,856,472) was
identified following the exclusion of CNVs presented in the
Database of Genomic Variants (17,18).
This chromosome region contains ~17 notable genes, including
elastin (ELN) and the general transcription factor IIi (GTF2I)
family (Fig. 2). The parents did
not carry this deletion. The 1.1 Mb region that overlapped with the
WBSCR interval is indicated in Fig.
2. This region contains certain genes typically associated with
Williams-Beuren syndrome, including ELN, GTF2I and bromodomain
adjacent to zinc finger domain 1B (BAZ1B).
Discussion
The patient in the current study presented with
atypical phenotypes of CHD, autism, hypercalcemia and cleft palate,
which are not usually linked to a diagnosis of WBS. However,
high-resolution SNP arrays have proved an effective method of
diagnosing syndromes when atypical phenotypes were observed. The
deleted region of chromosome 7q11.23 in the patient, reported in
the current study, spans 1.5 Mb and includes the genes ELN and the
GTF2I family, amongst others. To the best of our knowledge, this is
the first patient with WBS to also present with autism and cleft
palate reported to date. Most notably, the patient presented with
mild SAS and various other phenotypes, including autism and cleft
palate, due to the different deleted region compared with the
typically reported WBSCR.
Numerous previous publications have reported the
co-existence of WBS and autistic disorders with impairments in
socialization and communication. Reiss et al (19) initially identified two patients
that presented with autism alongside WBS in 1985. Later, Gillberg
and Rasmussen (20) (n=4), Gosch
and Pankau (21) (n=2), Herguner
and Mukaddes (22) (n=1), Leyfer
et al (23) (n=9), Edelmann
et al (24) (n=1), Lincoln
et al (25) (n=1),
Klein-Tasman et al (26)
(n=3) and Tordjman et al (27,28)
(n=9) reported the dual presence of both disorders. Most patients
were diagnosed by clinical manifestation or fluorescence in
situ hybridization, only Edelmann et al (24) identified the exact deletion in the
chromosome by array-based comparative genomic hybridisation
(Fig. 2). Sanders et al
(29) reported an association
between autism and de novo duplications of 7q11.23, (where
the reciprocal deletion causes WBS) characterized by a highly
social personality. The patient in the current study presented with
certain typical phenotypic characteristics of WBS, including SAS,
hypercalcemia, and growth and intelligence retardation. It has been
previously suggested that deletion of ELN contributes to SAS
(30), and the BAZ1B gene was
suggested to contribute to hypercalcemia (31). The GTF2I family, including GTF2I
repeat domain containing 1, was suggested to contribute to the
specific cognitive deficit exhibited in affected individuals
(32–34). However, autism and fear of
strangers were not considered to be phenotypes of WBS. Sanders
et al (29) reported that
duplications of the 7q11.23 WBSCR are strongly associated with
autism. As increasing number of patients with WBS are also
presenting with autism, and the results of the current study
indicate that autistic disorder should be considered as part of the
phenotype of WBS. Cleft palate is one of most common congenital
craniofacial deformities, but is rarely reported in WBS. Only five
cases were reported in a previously published article (35–38),
two of them were monozygotic twins and the others were sporadic
cases.
In conclusion, to the best of our knowledge, the
present study is the first to describe a case of WBS where SAS,
autism and cleft palate are also present, and an atypical deletion
of 7q11.23 was identified. High-resolution SNP arrays were
demonstrated to be an effective method for providing a specific
diagnosis of WBS despite the presence of atypical phenotypes.
Acknowledgements
This study was supported by Hunan Provincial Natural
Science Foundation of China (grant no. 2015JJ4085) and the
Scientific Research Foundation for the Returned Overseas Chinese
Scholars, State Education Ministry (2014; grant no. 1685).
References
|
1
|
Williams JC, Barratt-Boyes BG and Lowe JB:
Supravalvular aortic stenosis. Circulation. 24:1311–1318. 1961.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beuren AJ, Apitz J and Harmjanz D:
Supravalvular aortic stenosis in association with mental
retardation and a certain facial appearance. Circulation.
26:1235–1240. 1962. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wuang YP and Tsai HY: Sensorimotor and
visual perceptual functioning in school-aged children with Williams
syndrome. J Intellect Disabil Res. Nov 30–2016.(Epub ahead of
print). PubMed/NCBI
|
|
4
|
Sammour ZM, de Bessa J Jr, Hisano M,
Bruschini H, Kim CA, Srougi M and Gomes CM: Lower urinary tract
symptoms in children and adolescents with Williams-Beuren syndrome.
J Pediatr Urol. Nov 2–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirai M, Muramatsu Y, Mizuno S, Kurahashi
N, Kurahashi H and Nakamura M: Typical visual search performance
and atypical gaze behaviors in response to faces in Williams
syndrome. J Neurodev Disord. 8:382016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Strømme P, Bjørnstad PG and Ramstad K:
Prevalence estimation of Williams syndrome. J Child Neurol.
17:269–271. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abbas E, Cox DM, Smith T and Butler MG:
The 7q11.23 microduplication syndrome: A clinical report with
review of literature. J Pediatr Genet. 5:129–140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barak B and Feng G: Neurobiology of social
behavior abnormalities in autism and Williams syndrome. Nat
Neurosci. 19:647–655. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinto D, Darvishi K, Shi X, Rajan D,
Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald
JR, Mills R, et al: Comprehensive assessment of array-based
platforms and calling algorithms for detection of copy number
variants. Nat Biotechnol. 29:512–520. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Falkmer T, Anderson K, Falkmer M and
Horlin C: Diagnostic procedures in autism spectrum disorders: A
systematic literature review. Eur Child Adolesc Psychiatry.
22:329–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Bildt A, Sytema S, Zander E, Bölte S,
Sturm H, Yirmiya N, Yaari M, Charman T, Salomone E, LeCouteur A, et
al: Autism diagnostic interview-revised (adi-r) algorithms for
toddlers and young preschoolers: Application in a non-us sample of
1,104 children. J Autism Dev Disord. 45:2076–2091. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zander E, Willfors C, Berggren S,
Choque-Olsson N, Coco C, Elmund A, Moretti ÅH, Holm A, Jifält I,
Kosieradzki R, et al: The objectivity of the autism diagnostic
observation schedule (ADOS) in naturalistic clinical settings. Eur
Child Adolesc Psychiatry. 25:769–780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu TY, Chen KL, Chou W, Yang SH, Kung SC,
Lee YC and Tung LC: Intelligence quotient discrepancy indicates
levels of motor competence in preschool children at risk for
developmental delays. Neuropsychiatr Dis Treat. 12:501–510.
2016.PubMed/NCBI
|
|
14
|
Trumpff C, De Schepper J, Vanderfaeillie
J, Vercruysse N, Van Oyen H, Moreno-Reyes R, Tafforeau J, Vanderpas
J and Vandevijvere S: Thyroid-stimulating hormone (TSH)
concentration at birth in belgian neonates and cognitive
development at preschool age. Nutrients. 7:9018–9032. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goud TM, Al Salmani KK, Al Harasi SM, Al
Musalhi M, Wasifuddin SM and Rajab A: Importance of FISH combined
with morphology, immunophenotype and cytogenetic analysis of
childhood/adult acute lymphoblastic leukemia in Omani patients.
Asian Pac J Cancer Prev. 16:7343–7350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peterson JF, Aggarwal N, Smith CA, Gollin
SM, Surti U, Rajkovic A, Swerdlow SH and Yatsenko SA: Integration
of microarray analysis into the clinical diagnosis of hematological
malignancies: How much can we improve cytogenetic testing.
Oncotarget. 6:18845–18862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun G, Tan Z, Fan L, Wang J, Yang Y and
Zhang W: 1q21.1 microduplication in a patient with mental
impairment and congenital heart defect. Mol Med Rep. 12:5655–5658.
2015.PubMed/NCBI
|
|
18
|
MacDonald JR, Ziman R, Yuen RK, Feuk L and
Scherer SW: The database of genomic variants: A curated collection
of structural variation in the human genome. Nucleic Acids Res.
42:D986–D992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reiss AL, Feinstein C, Rosenbaum KN,
Borengasser-Caruso MA and Goldsmith BM: Autism associated with
Williams syndrome. J Pediatr. 106:247–249. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gillberg C and Rasmussen P: Brief report:
Four case histories and a literature review of Williams syndrome
and autistic behavior. J Autism Dev Disord. 24:381–393. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gosch A and Pankau R: Personality
characteristics and behaviour problems in individuals of different
ages with Williams syndrome. Dev Med Child Neurol. 39:527–533.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herguner S and Mukaddes NM: Autism and
Williams syndrome: A case report. World J Biol Psychiatry.
7:186–188. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leyfer OT, Woodruff-Borden J, Klein-Tasman
BP, Fricke JS and Mervis CB: Prevalence of psychiatric disorders in
4 to 16-year-olds with Williams syndrome. Am J Med Genet B
Neuropsychiatr Genet. 141B:615–622. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edelmann L, Prosnitz A, Pardo S, Bhatt J,
Cohen N, Lauriat T, Ouchanov L, González PJ, Manghi ER, Bondy P, et
al: An atypical deletion of the Williams-Beuren syndrome interval
implicates genes associated with defective visuospatial processing
and autism. J Med Genet. 44:136–143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lincoln AJ, Searcy YM, Jones W and Lord C:
Social interaction behaviors discriminate young children with
autism and Williams syndrome. J Am Acad Child Adolesc Psychiatry.
46:323–331. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klein-Tasman BP, Mervis CB, Lord C and
Phillips KD: Socio-communicative deficits in young children with
Williams syndrome: Performance on the autism diagnostic observation
Schedule. Child Neuropsychol. 13:444–467. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tordjman S, Anderson GM, Botbol M, Toutain
A, Sarda P, Carlier M, Saugier-Veber P, Baumann C, Cohen D,
Lagneaux C, et al: Autistic disorder in patients with
Williams-Beuren syndrome: A reconsideration of the Williams-Beuren
syndrome phenotype. PLoS One. 7:e307782012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tordjman S, Anderson GM, Cohen D,
Kermarrec S, Carlier M, Touitou Y, Saugier-Veber P, Lagneaux C,
Chevreuil C and Verloes A: Presence of autism, hyperserotonemia,
and severe expressive language impairment in Williams-Beuren
syndrome. Mol Autism. 4:292013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sanders SJ, Ercan-Sencicek AG, Hus V, Luo
R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR,
Thomson SA, et al: Multiple recurrent de novo CNVs, including
duplications of the 7q11.23 Williams syndrome region, are strongly
associated with autism. Neuron. 70:863–885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Curran ME, Atkinson DL, Ewart AK, Morris
CA, Leppert MF and Keating MT: The elastin gene is disrupted by a
translocation associated with supravalvular aortic stenosis. Cell.
73:159–168. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pober BR: Williams-Beuren syndrome. N Engl
J Med. 362:239–252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gagliardi C, Bonaglia MC, Selicorni A,
Borgatti R and Giorda R: Unusual cognitive and behavioural profile
in a Williams syndrome patient with atypical 7q11.23 deletion. J
Med Genet. 40:526–530. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hirota H, Matsuoka R, Chen XN, Salandanan
LS, Lincoln A, Rose FE, Sunahara M, Osawa M, Bellugi U and
Korenberg JR: Williams syndrome deficits in visual spatial
processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23.
Genet Med. 5:311–321. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Morris CA, Mervis CB, Hobart HH, Gregg RG,
Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA,
et al: GTF2I hemizygosity implicated in mental retardation in
Williams syndrome: Genotype-phenotype analysis of five families
with deletions in the Williams syndrome region. Am J Med Genet A.
123A:45–59. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pankau R, Gosch A, Simeoni E and Wessel A:
Williams-Beuren syndrome in monozygotic twins with variable
expression. Am J Med Genet. 47:475–477. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Blanco-Dávila F and Olveda-Rodriguez JA:
Cleft palate in a patient with Williams' syndrome. J Craniofac
Surg. 12:145–147. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vincent C, Mercier JM and David A: Cleft
palate and Williams syndrome. Rev Stomatol Chir Maxillofac.
109:44–47. 2008.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Domenico S, Orlando C, Graziana FF, Papi P
and Giulia A: Cleft palate in Williams syndrome. Ann Maxillofac
Surg. 3:84–86. 2013. View Article : Google Scholar : PubMed/NCBI
|