Introduction
Hepatocellular carcinoma (HCC) is reported to be the
fifth most common cancer type and the second leading cause of
cancer-associated mortality worldwide (1,2).
Risk factors are well characterized and surgical resections well
developed; however, the long-term survival rate of patients with
HCC is poor. The five-year survival rate following resection is
limited to 30–40% (3). It is
therefore of primary concern to clarify the molecular pathogenesis
of HCC and develop novel effective treatment strategies.
RAS protein activator like 2 (RASAL2) is a protein
that contains a GTPase activating protein (GAP)-related domain,
which is characteristic of GAP proteins. A previous study
demonstrated that RASAL2 acts as an epithelial cell
transforming-2-interacting protein to mediate mesenchymal-amoeboid
transition by regulating rhodopsin activity (4). In addition, Feng et al
(5) reported that RASAL2 promotes
triple-negative breast cancer (TNBC) progression by activating
Ras-related C3 botulinum toxin substrate 1 (RAC1). RASAL2 binds to
and antagonizes RAC1-GAP protein ARHGAP24 to suppress breast cancer
invasion. Furthermore, RASAL2 has been reported to be
hypomethylated in HCC (6);
however, the potential role of RASAL2 in HCC remains to be
elucidated.
The present study aimed to research the expression
pattern and functional role of RASAL2 in HCC. The present study
demonstrated that RASAL2 was markedly upregulated in HCC tissues.
Functional assays in HuH-7 and HCC-LM3 cells demonstrated that
RASAL2 promoted proliferation and invasion in HCC. In addition,
results indicated that RASAL2 was a direct target of microRNA
(miRNA/miR)-203 in HCC. The data therefore suggested that RASAL2 is
important in HCC progression.
Materials and methods
Patients and tissue samples
The present study included 70 patients underwent
resection of HCC at the First Affiliated Hospital of Zhejiang
University School of Medicine (Hangzhou, China). The tumor and
non-tumor tissues were maintained in liquid nitrogen immediately
following resection. The inclusion criteria required were as
follows: i) Diagnosis of HCC confirmed by pathology; ii) without
anticancer treatment and distant metastases before surgery; iii)
underwent curative resection for HCC between 2010 and 2015, defined
as macroscopically complete removal of the tumor with negative
safety margin; and iv) with complete clinicopathologic and
follow-up data. The characteristics of patients are presented in
Table I. This study was approved
by the Ethical Review Committee of the First Affiliated Hospital of
Zhejiang University School of Medicine (Hangzhou, China) and
informed consent was obtained from all patients.
 | Table I.Correlations between RASAL2 levels and
clinicopathological characteristics in hepatocellular carcinoma
patients. |
Table I.
Correlations between RASAL2 levels and
clinicopathological characteristics in hepatocellular carcinoma
patients.
|
|
| RASAL2 levels |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients (n=70) | Positive | Negative | P-value |
|---|
| Age (years) |
|
|
| 1.000 |
| ≤50 | 28 | 18 | 10 |
|
|
>50 | 42 | 27 | 15 |
|
| Gender |
|
|
| 0.828 |
| Male | 55 | 35 | 20 |
|
|
Female | 15 | 10 | 5 |
|
| Tumor number |
|
|
| 0.098 |
|
Single | 30 | 16 | 14 |
|
|
Multiple | 40 | 29 | 11 |
|
| Tumor size (cm) |
|
|
| 0.015a |
| ≤5 | 26 | 12 | 14 |
|
|
>5 | 44 | 33 | 11 |
|
| Tumor node metastasis
stage |
|
|
| 0.212 |
| I,II | 35 | 20 | 15 |
|
| III | 35 | 25 | 10 |
|
| Grade |
|
|
| 0.785 |
| Well,
moderate | 49 | 31 | 18 |
|
| Poor | 21 | 14 | 7 |
|
| α-fetoprotein
(ng/ml) |
|
|
| 0.174 |
|
≤400 | 21 | 11 | 10 |
|
|
>400 | 49 | 34 | 15 |
|
| Portal vein
invasion |
|
|
| 0.237 |
|
Negative | 50 | 30 | 20 |
|
|
Positive | 20 | 15 | 5 |
|
Cell lines and culture
HuH-7 and HCC-LM3 liver cancer cell lines were
purchased from the Shanghai Institute of Cell Biology (Shanghai,
China). These two cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco,
Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing
5% CO2.
RNA oligoribonucleotides and
transfection
Small interfering RNA (si)-RASAL2 (SI04200945),
AllStars negative control (NC) siRNA (SI03650318) and miR-203
(MSY0000264) were purchased from Qiagen GmbH (Hilden, Germany). The
transfection was performed using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol once cells reached 30–50% confluence.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blotting
Total RNA from the HCC cell lines was isolated using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. Subsequently, cDNA was
synthesized and qPCR reactions were performed using the ABI7500
Fast system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Briefly, reverse transcription was performed as follows the total
20 µl reaction compound consisted of 10 µl 2xmiRNA Reaction Buffer
mix, 0.1% 2 µl BSA, 2 µl miRNA PrimeScript RT Enzyme Mix, 1 µl RNA
and 5 µl RNase Free dH2O. All regents used were
purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
Next, the reaction compound was then incubated at 37°C for 1 h and
then 85°C for 5 min. Subsequently 80 ul RNase Free dH2O
was added to the compound and it was stored at −80°C until use.
qPCR was performed using a total 20 µl reaction volume consisting
of 2 µl SYBR Premix Ex TaqTM II (2x), 0.8 µl PCR Forward Primer (10
µM), 0.8 µl Uni-miR qPCR Primer (10 µM), 0.4 µl ROX Reference Dye
II (50x), 2 µl synthesized cDNA and 6 µl dH2O. All the
regents used were purchased from Takara Biotechnology Co., Ltd. The
thermocycling conditions were as follows: 40 cycles of denaturation
for 15 sec at 95°C and extension for 60 sec at 60°C. GAPDH and U6
served as internal controls. The results were quantified by
2−∆∆Cq method (7). The
sequences of primers used in the present study were as follows:
RASAL2, forward CCA AAT GTC AGT GGA AGC CTC TC, reverse CTG TGT TGT
CCT GGC TTG GAG A; and GAPDH, forward GTC TCC TCT GAC TTC AAC AGC G
and reverse, ACC ACC CTG TTG CTG TAG CCA A. The primers for miR-203
(MS00003766) and U6 (MS00033740) were obtained from Qiagen
GmbH.
For western blotting, cells were lysed in
radioimmunoprecipitation buffer (Beyotime Institute of
Biotechnology, Haimen, China) accompanied by phenylmethane sulfonyl
fluoride (Beyotime Institute of Biotechnology). The concentration
of each protein was quantified by Pierce BCA Protein Assay kit
(23225, Thermo Fisher Scientific, Inc.). For each sample, 30 µg
protein was separated by 10% SDS-PAGE. The proteins were then
transferred to polyvinylidene membranes, which were blocked with 5%
non-fat milk at room temperature for 1 h. The washing regent used
for the western blotting analysis was PBST. The primary antibodies
used in the study were as follows: RASAL2 (1:300; ab121578; Abcam,
Cambridge, MA, USA), phosphorylated-AKT (p-AKT; 1:3,000; ab81283;
Abcam, Cambridge, MA, USA), β-actin (1:4,000; A5441; Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany), Anti-rabbit IgG, horseradish
peroxidase (HRP)-conjugated antibody (7074, Cell Signaling
Technology) and anti-mouse IgG, HRP-conjugated antibody (7076, Cell
Signaling Technology). An enhanced chemiluminescence kit (NCI4106,
Huiying Biotec, Shanghai, China) was used to visualize the
membranes.
Immunohistochemistry (IHC)
Immunostaining was performed on paraffin-embedded
specimens as previously described (8). The antibody used for IHC was RASAL2
(1:400; ab121578; Abcam, Cambridge, MA, USA). For evaluation of the
staining score, semi-quantitative estimation was applied as
previously described (9).
Intensity was graded as 0, no staining; i) weak staining; ii)
moderate staining; or iii) strong staining. The abundance of
positive cells was graded from 0 to 4 (0, <5% positive cells; i)
5–25% positive cells; ii) 26–50% positive cells; iii) 51–75%
positive cells; iv) >75% positive cells). The final score was
obtained by multiplying the two values and all the cases were
grouped as RASAL2-negative (scores between 1 and 6) and
RASAL2-positive (scores between 7 and 12).
Cell proliferation and transwell
assay
The proliferation of the HuH-7 and HCC-LM3 cells was
evaluated using the Cell Counting Kit (CCK)-8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan). The transfected cells were
plated into 96-well plates at a density of 5×103
cells/well. Following incubation with the CCK reagent for 2 h, the
optical density of each well was measured with a Thermomax
microplate reader (Sunnyvale, CA, USA) at a wavelength of 450 nm.
The assays were conducted at three different time points (24, 48
and 72 h) and the experiments were conducted in triplicate.
For the transwell assay, 5×104
transfected cells in serum-free DMEM were seeded into the upper
chambers of inserts (Corning Incorporated-Life Sciences,
Tewkesbury, MA, USA), which were coated with Matrigel (BD
Biosciences, San Jose, CA, USA). DMEM supplemented with 10% fetal
bovine serum was added to the lower chambers. Following incubation
for 48 h, the lower surface was stained with crystal violet. The
cells were then counted under an inverted microscope with ×100
magnification.
Luciferase activity assay
A luciferase reporter assay was conducted using the
Dual-Luciferase Reporter Assay system (E2920; Promega Corporation,
Madison, WI, USA) according to the manufacturer's protocol. HuH-7
cells were seeded in 24-well plates and were co-transfected with
miR-203 mimics or NC, and wild-type or mutant-type RASAL2
3′untranslated region (UTR) plasmid, which was synthesized and
mutated by Genechem Co., Ltd. (Shanghai, China). All the binding
sites were mutated. The firefly and Renilla luciferase activities
were measured by a dual-luciferase reporter assay (Promega
Corporation) following transfection for 48 h.
Statistical analysis
The correlations between RASAL2 levels and
clinicopathological data were analyzed using a χ2 test.
The comparisons between groups were analyzed using paired Student's
t-test. The data is presented as the mean ± standard deviation.
Statistical analyses were performed with SPSS software version 17.0
(SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5 (Graphpad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
RASAL2 is upregulated in HCC
tissues
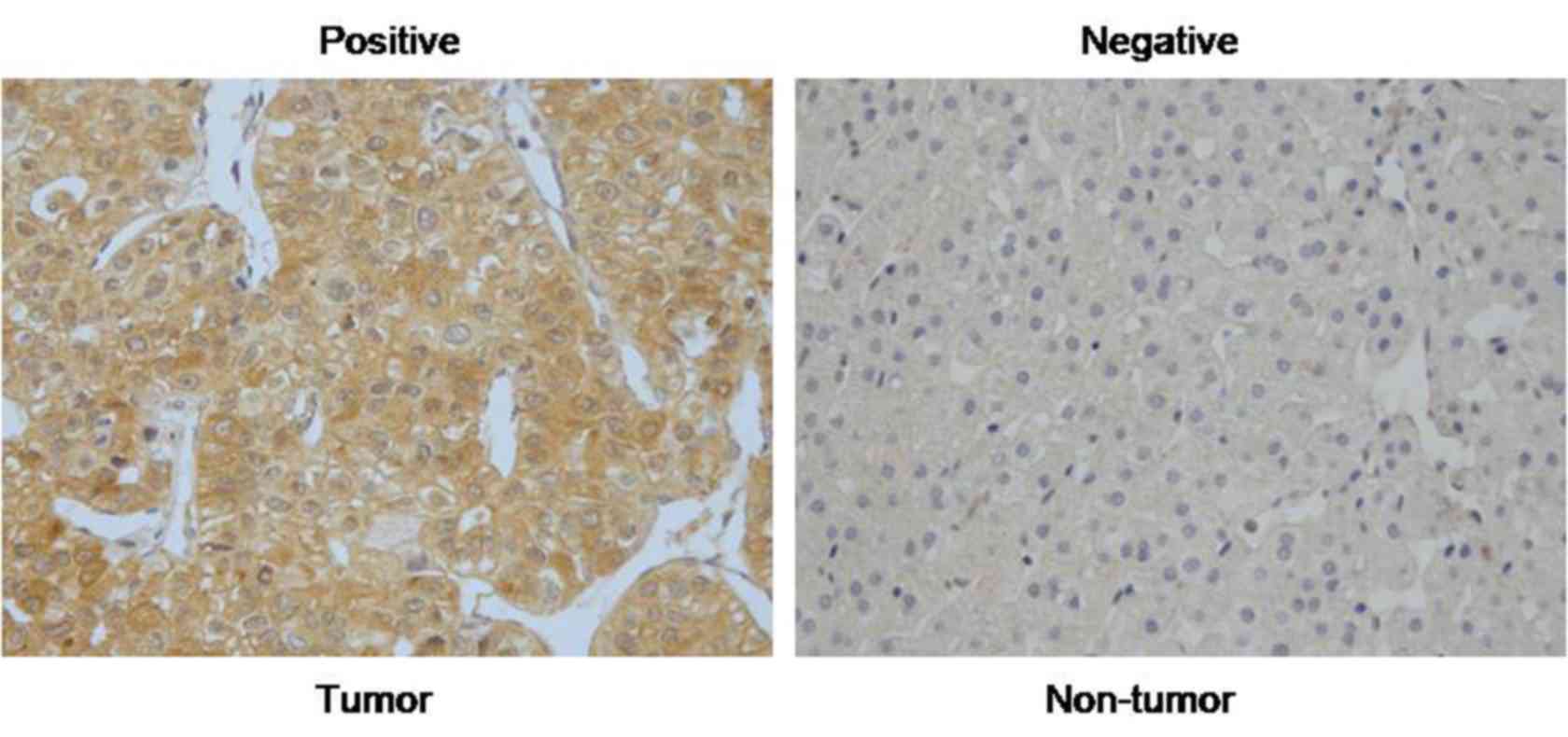
In order to explore the expression pattern of RASAL2
in HCC, the present study performed IHC on 70 pairs of HCC tissues.
As presented in Fig. 1, the
immunoreactivity of RASAL2 was primarily located in the cytoplasm.
The results indicated that the levels of RASAL2 were markedly
increased in HCC compared with in normal liver tissues. Of all the
tissues, 45 of 70 (64.3%) HCC tissues were positive for RASAL2,
whereas only 15 of 70 (21.4%) normal liver tissues were
positive.
Subsequently, the present study investigated whether
the levels of RASAL2 were correlated with clinicopathological
parameters in these 70 HCC tissues by applying χ2 test
(Table I). The results revealed
that the positive RASAL2 expression group exhibited a larger tumor
size (P=0.015). However, no further correlations between RASAL2
levels and other clinicopathological characteristics were observed,
including, gender, age, tumor number, grade and α-fetoprotein.
RASAL2 knockdown inhibits cell
proliferation and metastasis in HCC cells
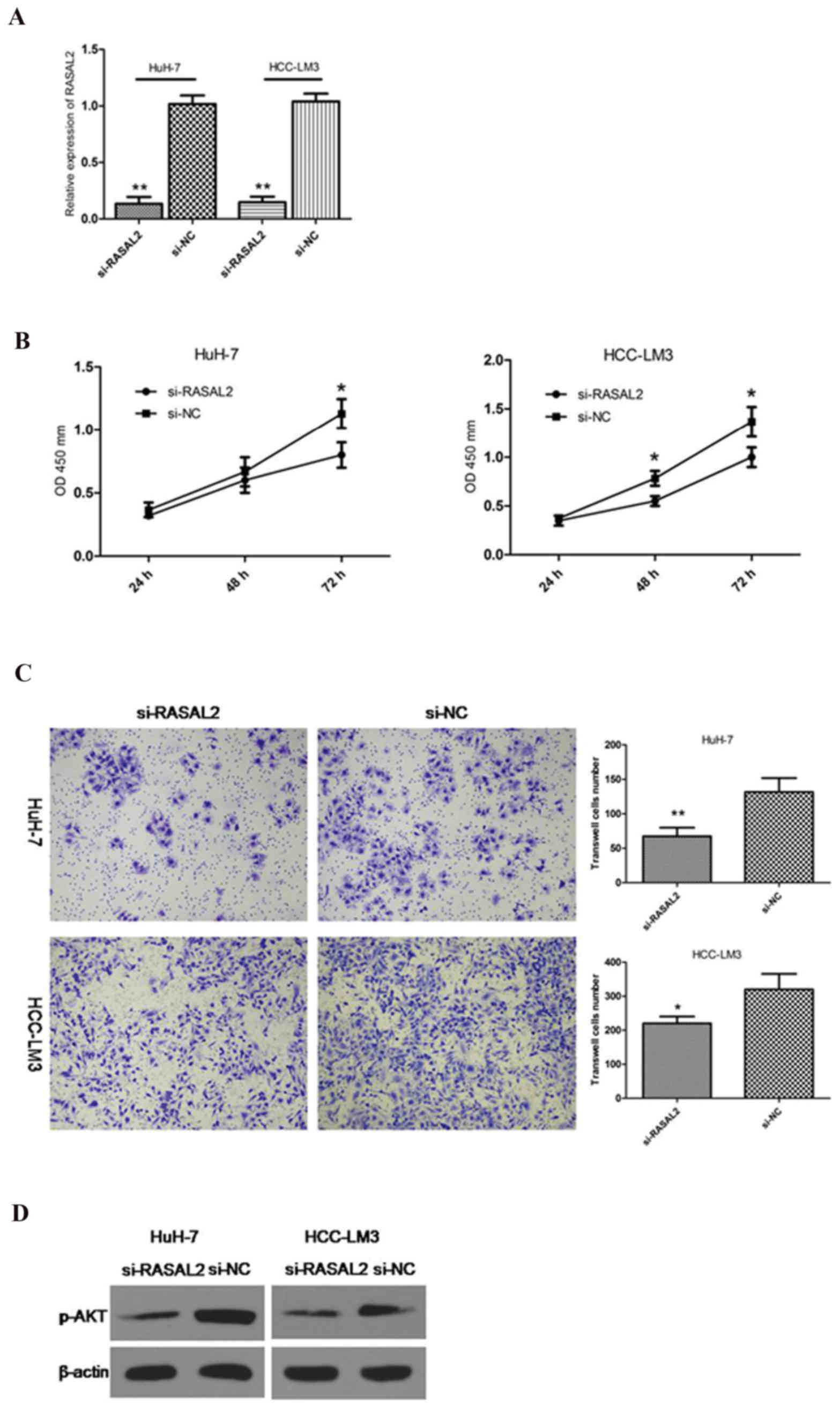
Chemically synthesized siRNA was used to knockdown
RASAL2 in HuH-7 and HCC-LM3 cells. RT-qPCR was applied to detect
the knockdown efficiency of si-RASAL2 (Fig. 2A). In order to evaluate the
proliferation rate, a CCK-8 assay was conducted. The results
revealed that the growth rate decreased following inhibition of
RASAL2 in Huh7 and HCC-LM3 cells (Fig.
2B). To further determine the significance of RASAL2 in HCC, a
transwell assay was applied to investigate the effects of RASAL2 on
HCC cell invasion. It was observed that RASAL2 knockdown markedly
reduced the invasion of HCC cells (Fig. 2C). The phosphoinositide 3 kinase
(PI3K)/AKT pathway has been reported to be important in HCC
proliferation and metastasis (10,11).
The present study hypothesized that RASAL2 may regulate the
PI3K/AKT pathway in HCC cells. To verify this, the present study
investigated p-AKT levels, since p-AKT is the key protein of the
AKT pathway. It was observed that RASAL2 suppression markedly
reduced the expression levels of p-AKT compared with the control
group in the two cell lines (Fig.
2D). These results indicated that RASAL2 promotes proliferation
and invasion in HCC.
RASAL2 is a direct target of miR-203
in HCC
miRNAs have recently been demonstrated to act as
critical molecules that regulate proliferation and metastatic
progression by binding target genes in HCC (12–14).
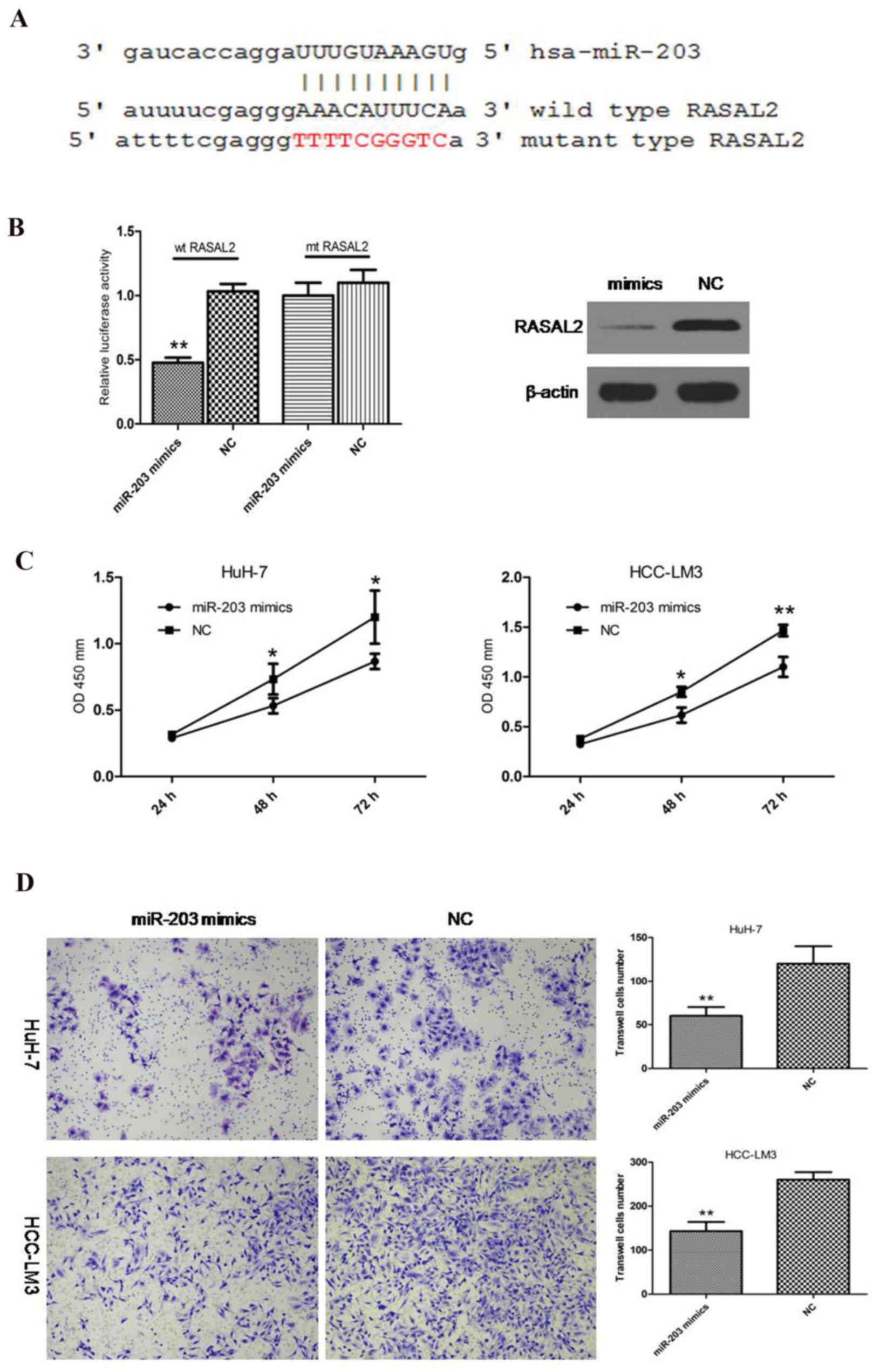
The present study hypothesized that RASAL2 may be targeted by a
specific miRNA in HCC. Following application of TargetScan
(http://www.targetscan.org) and miRanda
(http://www.microrna.org) computational tools,
miR-203 was investigated (Fig.
3A). This specific miRNA was identified by the two
bioinformatics tools. Previous studies have reported that miR-203
is downregulated in HCC, and inhibits proliferation and metastasis
in HCC (15,16). The miR-203 mimics were used to
enhance the level of miR-203 in HCC cells (Fig. 3A). A dual-luciferase reporter assay
was conducted in HuH-7 cells and the results revealed that miR-203
mimics significantly inhibited the luciferase activity of wild-type
RASAL2 3′UTR rather than the mutant type (Fig. 3B). In addition, western blotting
indicated that the protein levels of RASAL2 were decreased
following miR-203 overexpression (Fig.
3B). Furthermore, similar to si-RASAL2, miR-203 mimics exerted
anti-proliferative and anti-metastatic effects in HCC cells
(Fig. 3C and D). These data
suggested that RASAL2 may be a direct target of miR-203 in HCC.
Discussion
HCC is one of the leading causes of
malignancy-associated mortality in humans worldwide. The
development of surgical resection, drug targets and other
therapeutic strategies is well established; however, the prognosis
of HCC remains poor. The identification of molecular markers in HCC
has been the primary target of research (17–19);
therefore, the present study aimed to identify a novel therapeutic
biomarker for HCC.
The present study demonstrated that the expression
levels of RASAL2 were markedly overexpressed in HCC tissues
compared with in normal liver tissues. In a previous study,
Stefanska et al (6)
reported that the promoters of RASAL2 are hypomethylated, which
results in the increased expression of RASAL2 in HCC tissues and
cell lines. The data from the present study suggested that RASAL2
acts as an oncogene in HCC development. In addition, the
correlation between RASAL2 and clinicopathological features in HCC
was investigated, and it was observed that high RASAL2 expression
was closely correlated with tumor size. Subsequently, RASAL2
expression was suppressed in HuH-7 and HCC-LM3 cells based on the
observations from the HCC tissues, in order to elucidate its
functional role in HCC. The results demonstrated that knockdown of
RASAL2 decreased the growth rate and metastatic ability of HCC
cells. It has previously been reported that RASAL2 functions as a
tumor oncogene in TNBC (5), and
this is consistent with the results of the present study. The
overexpression of RASAL2 in TNBC drives mesenchymal invasion and is
correlated with poor outcomes. This further confirms the oncogenic
role of RASAL2 in human cancer.
The present study aimed to identify pathways
mediating proliferation and metastasis in HCC. The PI3K/AKT
pathway, which regulates growth and invasion during cancer
development, including HCC, was studied (20,21).
Following suppression of RASAL2, the phosphorylation levels of AKT
were reduced in the two HCC cell lines. This result suggested that
the over-expression of RASAL2 in HCC may activate the AKT pathway
resulting in increases in proliferation and invasion.
miRNAs are small non-coding RNAs that regulate gene
expression by inhibiting translation and/or stability of mRNA, and
thereby contribute to a wide range of physiological and
pathological processes (22–24).
In human cancer, numerous miRNAs act as potential tumor suppressor
genes and low levels of these miRNAs may contribute to
overexpression of oncogenic genes (25,26).
RASAL2 was revealed to serve an oncogene role in HCC; therefore,
the present study hypothesized that high levels of RASAL2 may be
attributed to the downregulation of a specific miRNA. miR-203 has
been reported to be frequently downregulated in various cancer
types, including HCC (27–29). A previous study revealed that
RASAL2 is a direct target of miR-203 in breast cancer. The present
study, to the best of our knowledge, is the first to confirm that
miR-203 directly targets RASAL2 in HCC, according to the following
evidence: i) Dual-luciferase reporter assay revealed a decrease in
luciferase activity in cells co-transfected with miR-203 mimics and
wild-type RASAL2 3′UTR, whereas no change was observed in cells
transfected with the mutant RASAL2 3′UTR; ii) the protein levels of
RASAL2 were decreased following transfection with miR-203 mimics;
iii) the effects of miR-203 mimics on proliferation and metastasis
were similar to those exhibited following inhibition of RASAL2.
These data therefore suggested that miR-203 may directly target
RASAL2 in HCC.
In conclusion, the results of the present study
indicated that RASAL2 is significantly upregulated in HCC tissues.
In addition, it was confirmed that RASAL2 promotes proliferation
and metastasis by regulating the AKT pathway. Furthermore, it was
demonstrated that RASAL2 is the direct target of the tumor
suppressor gene miR-203. These data are expected to contribute to
mechanistic HCC development and RASAL2 may be considered a
potential novel molecular target for the development of future
therapeutic technologies.
References
|
1
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Altekruse SF, McGlynn KA and Reichman ME:
Hepatocellular carcinoma incidence, mortality, and survival trends
in the United States from 1975 to 2005. J Clin Oncol. 27:1485–1491.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weeks A, Okolowsky N, Golbourn B, Ivanchuk
S, Smith C and Rutka JT: ECT2 and RASAL2 mediate
mesenchymal-amoeboid transition in human astrocytoma cells. Am J
Pathol. 181:662–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng M, Bao Y, Li Z, Li J, Gong M, Lam S,
Wang J, Marzese DM, Donovan N, Tan EY, et al: RASAL2 activates RAC1
to promote triple-negative breast cancer progression. J Clin
Invest. 124:5291–5304. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stefanska B, Cheishvili D, Suderman M,
Arakelian A, Huang J, Hallett M, Han ZG, Al-Mahtab M, Akbar SM,
Khan WA, et al: Genome-wide study of hypomethylated and induced
genes in patients with liver cancer unravels novel anticancer
targets. Clin Cancer Res. 20:3118–3132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R,
Zhang W, Lv Z, Zheng S and Zhou L: NDRG1 as a biomarker for
metastasis, recurrence and of poor prognosis in hepatocellular
carcinoma. Cancer Lett. 310:35–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Noske A, Denkert C, Schober H, Sers C,
Zhumabayeva B, Weichert W, Dietel M and Wiechen K: Loss of Gelsolin
expression in human ovarian carcinomas. Eur J Cancer. 41:461–469.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bleeker FE, Felicioni L, Buttitta F, Lamba
S, Cardone L, Rodolfo M, Scarpa A, Leenstra S, Frattini M,
Barbareschi M, et al: AKT1 (E17K) in human solid tumours. Oncogene.
27:5648–5650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sahin F, Kannangai R, Adegbola O, Wang J,
Su G and Torbenson M: mTOR and P70 S6 kinase expression in primary
liver neoplasms. Clin Cancer Res. 10:8421–8425. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohta K, Hoshino H, Wang J, Ono S, Iida Y,
Hata K, Huang SK, Colquhoun S and Hoon DS: MicroRNA-93 activates
c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by
directly inhibiting PTEN and CDKN1A. Oncotarget. 6:3211–3224. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z, Zhang Y, Sun XX, Ma X and Chen
ZN: microRNA-146a inhibits cancer metastasis by downregulating VEGF
through dual pathways in hepatocellular carcinoma. Mol Cancer.
14:52015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yunqiao L, Vanke H, Jun X and Tangmeng G:
MicroRNA-206, down-regulated in hepatocellular carcinoma,
suppresses cell proliferation and promotes apoptosis.
Hepatogastroenterology. 61:1302–1307. 2014.
|
|
15
|
Wei W, Wanjun L, Hui S, Dongyue C, Xinjun
Y and Jisheng Z: miR-203 inhibits proliferation of HCC cells by
targeting survivin. Cell Biochem Funct. 31:82–85. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wan D, Shen S, Fu S, Preston B, Brandon C,
He S, Shen C, Wu J, Wang S, Xie W, et al: miR-203 suppresses the
proliferation and metastasis of hepatocellular carcinoma by
targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC.
Anticancer Agents Med Chem. 16:414–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Minguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mann CD, Neal CP, Garcea G, Manson MM,
Dennison AR and Berry DP: Prognostic molecular markers in
hepatocellular carcinoma: A systematic review. Eur J Cancer.
43:979–992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang N, Yao M, Xu J, Quan Y, Zhang K, Yang
R and Gao WQ: Autocrine activation of CHRM3 promotes prostate
cancer growth and castration resistance via CaM/CaMKK-mediated
phosphorylation of Akt. Clin Cancer Res. 21:4676–4685. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang B, Qi G, Tang F, Yuan S, Wang Z,
Liang X, Li B, Yu S, Liu J, Huang Q, et al: JARID1B promotes
metastasis and epithelial-mesenchymal transition via PTEN/AKT
signaling in hepatocellular carcinoma cells. Oncotarget.
6:12723–12739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Di Leva G and Croce CM: miRNA profiling of
cancer. Curr Opin Genet Dev. 23:3–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao H, Bai Y, Qiu S, Zheng L, Huang L,
Liu T, Wang X, Liu Y, Xu N, Yan X and Guo H: miR-203 downregulation
is responsible for chemoresistance in human glioblastoma by
promoting epithelial-mesenchymal transition via SNAI2. Oncotarget.
6:8914–8928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu M, Gu M, Zhang K, Zhou J, Wang Z and Da
J: miR-203 inhibition of renal cancer cell proliferation, migration
and invasion by targeting of FGF2. Diagn Pathol. 10:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Y, Ren F, Rong M, Luo Y, Dang Y and
Chen G: Association between underexpression of microrna-203 and
clinicopathological significance in hepatocellular carcinoma
tissues. Cancer Cell Int. 15:622015. View Article : Google Scholar : PubMed/NCBI
|