Introduction
Epigenetics refers to heritable alterations in gene
expression that do not involve coding sequence modifications
(1). Epigenetic modifications may
be grouped into three primary categories: DNA methylation, histone
modifications and nucleosome positioning (2). Histone acetylation has been the most
thoroughly investigated modification. It is regulated by the
opposing activities of two enzymes, histone deacetylases (HDACs)
and histone acetyltransferases (HATs). HAT-induced histone
acetylation is associated with activation of transcription via
relaxation of the chromatin structure, whereas deacetylation by
HDACs induces a more condensed or inactive chromatin state, leading
to gene repression (3,4).
Hepatitis B virus (HBV) infection is a global public
health problem affecting >350 million individuals worldwide
(5). In China, there are ~93
million individuals who have been infected with HBV, of which 20
million are chronic hepatitis B (CHB) patients (6). CHB infection is a primary cause of
hepatic dysfunction. It is hypothesized that HBV is not directly
cytopathic and that the host immune response is responsible for the
disease. A human leucocyte antigen class I-restricted T cell
response against HBV peptides expressed on the surface of liver
cells serves an important role in the pathogenesis of liver damage
(7). In addition to the primary
damage caused by immunity, inflammatory cytokines are involved,
particularly in severe liver damage.
Epigenetic regulation of gene expression is now
regarded as a novel approach for disease treatment (8). Histone acetylation modification was
demonstrated to serve pivotal roles in numerous inflammatory
diseases, including rheumatoid arthritis (9), COPD (10) and allergic skin inflammation
(11). However, the role of
histone acetylation modification in CHB, particularly in liver
failure, remains unclear. In the present study, the association
between HDAC activity and disease severity in CHB patients was
investigated. In addition, an acute liver failure (ALF) model was
induced in mice and the RAW264.7 murine macrophage cell line was
used to evaluate the effect of acetylation regulation under
inflammatory conditions.
Materials and methods
Patients
A total of 60 patients with CHB were recruited from
the Department of Infectious Diseases, Renmin Hospital of Wuhan
University (Wuhan, China) between January and December 2013.
Informed consent was obtained from all participants in the study.
Additionally, healthy blood samples (~30) were obtained from the
Blood Bank of the Renmin Hospital of Wuhan University. The patients
were divided into two groups: CHB and CHB with liver failure
(n=30/group). CHB and CHB with liver failure were diagnosed using
guidelines of CHB and acute-on-chronic liver failure (12,13).
The present study received ethical approval from the Clinical
Research Ethics Committee of Renmin Hospital of Wuhan University
Patients with hepatitis A, C and E, autoimmune liver disease,
drug-induced hepatitis, alcoholic liver disease, and fatty liver
disease infections were excluded from the present study.
Processing of blood samples
Peripheral blood mononuclear cells (PBMCs) were
extracted from the blood samples of patients using a peripheral
blood mononuclear cell separation fluid kit (Lengton Biological
Technology, Shanghai, China) according to the manufacturer's
protocol. Blood serum and PBMCs were stored at −80°C until
required.
Processing of liver tissue
samples
Liver tissue samples were obtained from patients
with CHB and CHB with liver failure. Samples were fixed in 10%
buffered formalin for 24 h, embedded in paraffin and sliced into
5-µm thick sections. Tissue sections were dewaxed in xylene and
rehydrated in a series of dilutions of alcohol. Following this,
sections were placed in 3% H2O2 for 20 min to
block endogenous peroxidase activity and boiled in sodium citrate
buffer (pH=6.0) for 15 min. The sections were subsequently blocked
with 10% normal goat serum (Beijing Solarbio Science and Technology
Co., Ltd., Beijing, China) in a humidified chamber at 37°C for 20
min to reduce non-specific binding, following which they were
incubated with a rabbit anti-histone deacetylase 1 (HDAC1) primary
antibody (cat. no. 5356S; Cell Signaling Technology, Inc., Danvers,
MA, USA) at a 1:500 dilution at 4°C overnight. The sections were
subsequently incubated with a biotin-labelled goat anti-rabbit IgG
secondary antibody (dilution, 1:100; cat. no. BA1003; Wuhan Boster
Biological Technology, Ltd., Wuhan, China) for 1 h at 37°C.
Following this, sections were stained with 3,3-diaminobenzidine
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), counterstained
with Mayer's hematoxylin, dehydrated and mounted.
Animals
30 specific pathogen-free (SPF) male C57BL/6 mice
(weight, 16 to 30 g), were purchased from the Animal Experimental
Center, Hubei Medical University (Wuhan, China). All animals were
housed in a light-controlled room (12-h light/dark cycle) at a
temperature of 25°C and humidity of 45 to 50% with free access to
food and water. All animal experiments procedures were performed in
accordance with the institutional guidelines of the Animal Care and
Use Committee of Renmin Hospital of Wuhan University (Hubei,
China). The experimenter possessed an experimental animal
application certificate.
Model production and sample
collection
Animals were randomly divided into three groups:
Healthy (control), model and entinostat (MS275)-treated. A mouse
ALF model was induced by administration of D-Galactosamine (D-Gal;
(Sigma-Aldrich; Merck KGaA) and lipopolysaccharide (LPS;
Sigma-Aldrich; Merck KGaA). Mice in the model group (n=10 received
400 mg/kg D-Gal and 100 µg/kg LPS by intraperitoneal injection. In
addition to D-Gal and LPS, mice in the MS275-treated group (HDAC
inhibitor; HDACi; n=10) received 1 mg/kg MS275 by intraperitoneal
injection 2 h prior to ALF induction. Mice in the control group
(n=10) were injected intraperitoneally with normal saline as a
control. The time of administration of D-Gal and LPS was referred
to as the baseline (time point 0). All animals were sacrificed
following general anesthesia by intraperitoneal injection of 30
mg/kg pentobarbital sodium (Sigma-Aldrich; Merck KGaA) for orbital
blood and hepatic tissue collection at the 24 h time point.
Cell culture
The RAW264.7 murine macrophage cell line was
purchased from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China), and cultured in Dulbecco's modified Eagle's
medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 20 U/ml penicillin and 20 µg/ml
streptomycin in an incubator at 37°C with 5% CO2 under a
humidified atmosphere.
Transient transfection of small
interfering (si)RNA into RAW264.7 cells
HDAC1 (forward, 5′-GUUCUAUUCGCCCAGAUAATT-3′ and
reverse, 3′-UUAUCUGGGCGAAUAGAACTT-3′) and non-specific control
(forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) siRNA were purchased from Guangzhou
RiboBio Co., Ltd. (Guangzhou, China). Cells were seeded at a
density of 5×105 cells per well into 6-well cell culture
clusters for 4 h prior to siRNA transfection. siRNA was diluted
with Opti-Minimum Essential Medium® (Invitrogen; Thermo
Fisher Scientific, Inc.) and incubated at room temperature for 15
min with Oligofectamine™ reagent (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocol. The final concentration
in the culture was 100 nM. Following 1 h, siRNA-transfected and
control cells were treated with 1 µg/ml LPS and cultured for an
additional 48 h.
Analysis of blood samples and cell
supernatants
Blood samples were collected in medical
anticoagulant tube from patients with CHB and with CHB and liver
failure. The samples were mixed and centrifuged at 600 × g
for 30 min, at room temperature, then the supernatant were
collected and preserved in −20°C. Serum alanine aminotransferase
(ALT), aspartate aminotransferase (AST) and total bilirubin (TBil)
levels were measured using a Hitachi Automatic Analyzer (Hitachi,
Ltd., Tokyo, Japan). Tumor necrosis factor-α (TNF-α; cat. no.
BMS223HS) and interferon-γ (IFN-γ; cat. no. BMS228/BMS228TEN) serum
levels were determined using ELISA kits (eBioscience, Inc., San
Diego, CA, USA) following the manufacturer's protocol. Plasma
thromboplastin antecedent (PTA) data were taken from medical
records.
Detection of HDAC activity
The activity of HDAC was measured using an HDAC
assay kit (BioVision, Inc., Milpitas, CA, USA) according to the
manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from RAW264.7 cells and
mouse hepatic tissue, and reverse-transcribed using a PrimeScript
RT reagent kit (Takara Bio, Inc., Otsu, Japan). According to the
manufacturer's protocol, qPCR was performed with cDNA using
gene-specific primers, the SYBR®-Green Master Mix kit
(Takara Bio, Inc.) and a 7500 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primers (Table I) were developed using Primer
Express software (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Quantification cycles (Cq) were determined from the
amplification plots, and target gene expression was normalized
against the Cq of the GAPDH housekeeping gene using the
2−ΔΔCq method (14).
 | Table I.List of primer sequences used for
quantitative polymerase chain reaction. |
Table I.
List of primer sequences used for
quantitative polymerase chain reaction.
|
| Primer sequence
(5′-3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| HDAC-1 |
TGTTGCTCGCTGCTGGACTTA |
ATCTTCATCCCCACTCTCTTCG |
| HDAC-2 |
GGTCGTAGGAATGTTGCTGAT |
AAGCCAATGTCCTCAAACAGG |
| TNF-α |
CATCTTCTCAAAATTCGAGTGACAA |
TGGGAGTAGACAAGGTACAACCC |
| CSF |
TTACTTTTCCTGGGCATTGTGG |
CAGGAGGTTCAGGGCTTCTTTG |
| IL-1β |
TGCCACCTTTTGACAGTGATG |
ATGTGCTGCTGCGAGATTTG |
| Clc-2 |
ACCTGAATCGGAACCAAAT |
TGAAAGGGAATACCATAACATC |
| GAPDH |
AGGAGCGAGACCCCACTAACA |
AGGGGGGCTAAGCAGTTGGT |
Western blot analysis
Cells were washed twice using phosphate-buffered
solution (Beijing Solarbio Science and Technology Co., Ltd.). The
appropriate amount (200 µl) of Mammalian Protein Extraction Reagent
(M-PER; cat. no. 78503; Thermo Fisher Scientific, Inc.), mixed with
cOmpleteTM EDTA-free Protease Inhibitor Cocktail Tablets (cat. no.
5892791001; Roche Diagnostics, Basel, Switzerland), was added to
each well (6-well plate) and the plates were shaken gently using a
constant temperature shaker (IS-RDD3; Suzhou Jiemei Electronic Co.,
Ltd., Suzhou, China) for 15 min at 4°C. The lysates were collected
and transferred to a microcentrifuge tubes for centrifugation at
12,000 × g for 30 min at 4°C to pellet the cell debris. The
supernatant was collected and stored at −20°C. Total protein (50
µg) was subjected to 10% SDS-PAGE and subsequently transferred onto
a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA,
USA). The membrane was incubated at 4°C overnight with the
following monoclonal primary antibodies: Mouse anti-HDAC1
(dilution, 1:1,000; cat. no. 5356S), mouse anti-HDAC2 (dilution
1:1,000; cat. no. 5113P), rabbit anti-histone H3 (dilution,
1:1,000; cat. no. 9715S), mouse anti-histone H4 (cat. no. 2935),
rabbit anti-nuclear factor-κB (NF-κB) p65 (dilution, 1:1,000; cat.
no. 8242S), rabbit anti-acetyl-histone H3 (dilution, 1:1,000; cat.
no. 9649S), rabbit anti-acetyl-H4 (dilution, 1:1,000: cat. no.
2594S), rabbit anti-acetyl-NF-κB p65 (dilution, 1:1,000; cat. no.
3045S) and rabbit anti-β-actin (dilution, 1:1,000; cat. no. 4970S),
(all from Cell Signaling Technology, Inc.). This was followed by
incubation with the following secondary antibodies for 1 h at room
temperature in the dark: IRDye 800CW goat anti-mouse 926-32210
(cat. no. C50316-03, dilution, 1:10,000; LI-COR Biosciences,
Lincoln, NE, USA), IRDye 800CW goat anti-rabbit 926-32211 (cat. no.
C50602-08, dilution 1:10,000; LI-COR Biosciences). Proteins were
visualized using an ODYSSEY® infrared imaging system
(LI-COR Biosciences, USA). β-actin served as an internal control.
Densitometry analysis was performed using the Odyssey software
application (version, 3.0.29; LI-COR Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). One-way
analysis of the variance was performed, with multiple comparisons
between groups compared using the Student-Newman-Kuels method for
post hoc tests. Data are presented as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Increased inflammatory cytokine levels
in patients with CHB and CHB with liver failure
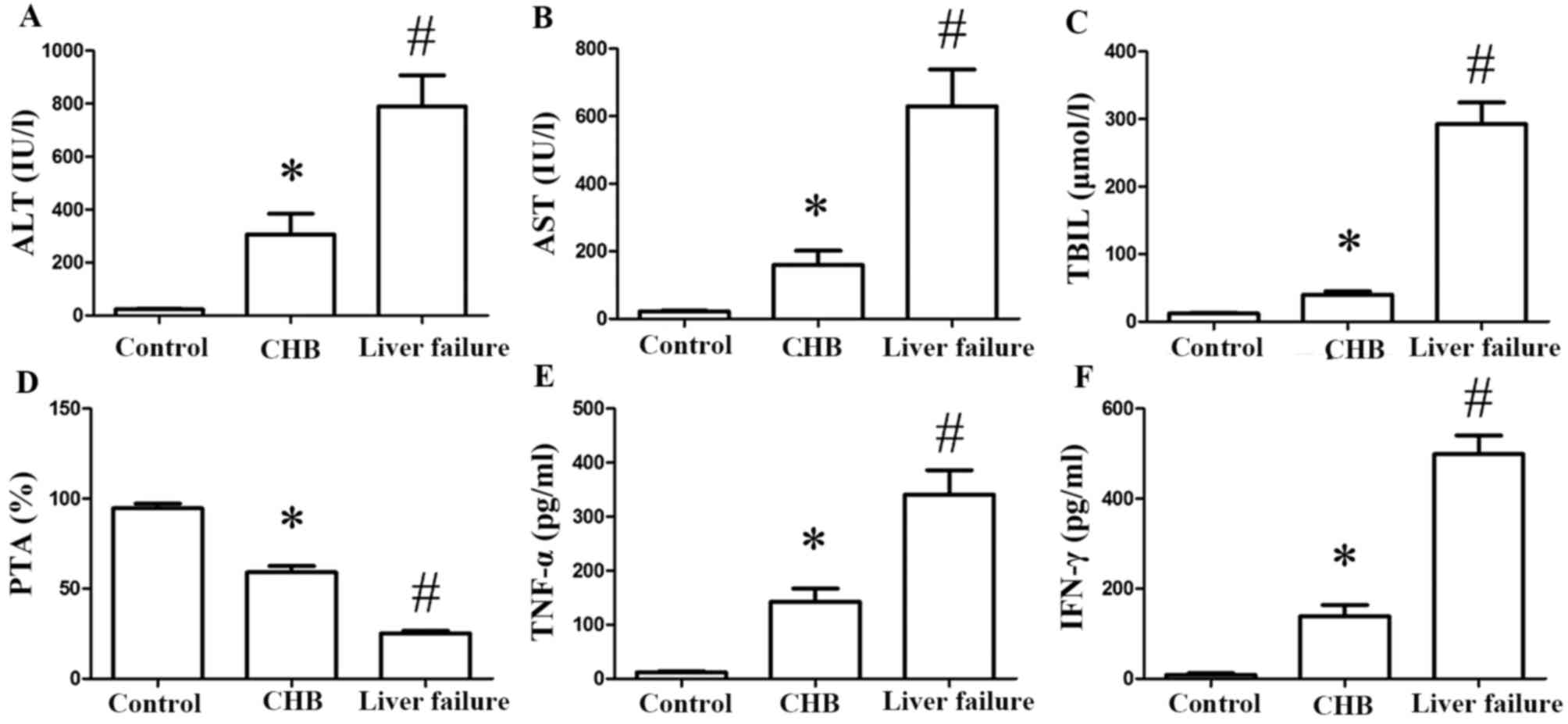
Serum levels of ALT (Fig. 1A), AST (Fig. 1B) and TBil (Fig. 1C) increased significantly in the
CHB and CHB with liver failure groups compared with the control
group (P<0.05). In addition, serum levels of ALT, AST and TBil
were markedly increased in the CHB with liver failure group
compared with the CHB group (P<0.05). PTA levels decreased in
the CHB and CHB with liver failure groups compared with the control
group (P<0.05), and levels in the CHB and liver failure group
were markedly reduced compared with the CHB group (P<0.05;
Fig. 1D). As measured by ELISA,
serum levels of TNF-α (Fig. 1E)
and IFN-γ (Fig. 1F) were markedly
increased in CHB patients compared with the control group
(P<0.05). Compared with the CHB group, TNF-α and IFN-γ levels in
the CHB with liver failure group increased significantly
(P<0.05).
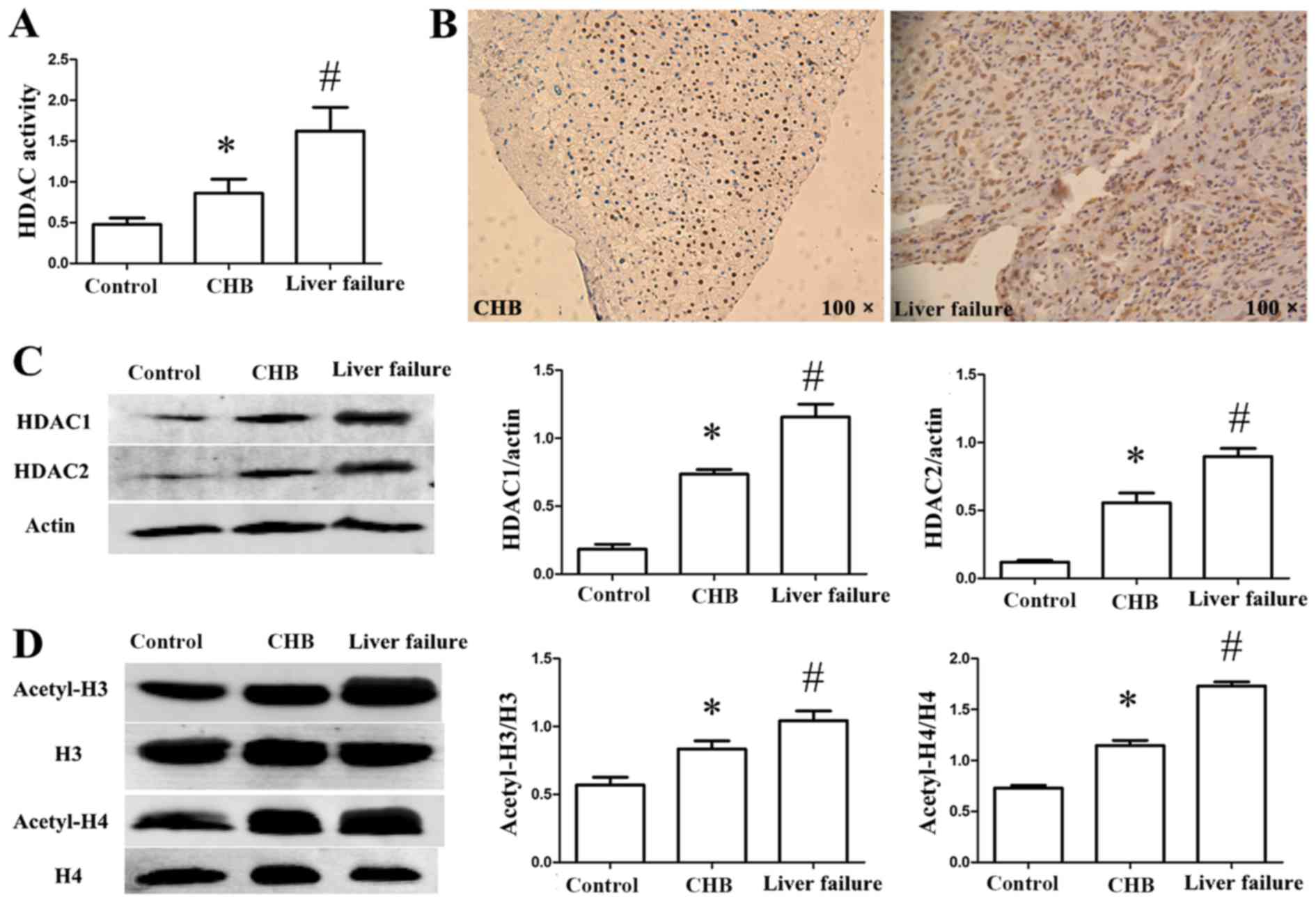
Increased HDAC activity and HDAC
expression levels in patients
HDAC activity was detected in the sera of the
patients. As presented in Fig. 2A,
HDAC activity increased markedly in the CHB and CHB with liver
failure groups compared with the control group (P<0.05). HDAC
activity in the CHB with liver failure group was markedly increased
compared with the CHB group (P<0.05). The expression of HDAC1 in
the liver tissue was detected by immunohistochemistry. HDAC1 was
primarily located in the nucleus, and increased positive staining
was observed in the CHB with liver failure group compared with the
CHB group (Fig. 2B). To observe
alterations in HDAC expression, the protein expression levels of
HDAC1 and HDAC2 in PBMCs were measured by western blot analysis. As
presented in Fig. 2C, the
expression levels of HDAC1 and HDAC2 increased significantly in the
CHB and CHB with liver failure groups compared with the control
group (P<0.05), and were increased in the CHB with liver failure
group compared with the CHB group (P<0.05).
Acetylation levels of H3/H4 are
increased in PBMCs in patients
To investigate histone acetylation alterations in
patients with CHB, the acetylation levels of H3 and H4 in PBMCs
were assessed. As presented in Fig.
2D, the acetylation levels of H3 and H4 were markedly increased
in the CHB and CHB with liver failure groups compared with the
control group (P<0.05). In addition, the acetylation levels of
H3 and H4 were markedly increased in the CHB with liver failure
group compared with the CHB group (P<0.05).
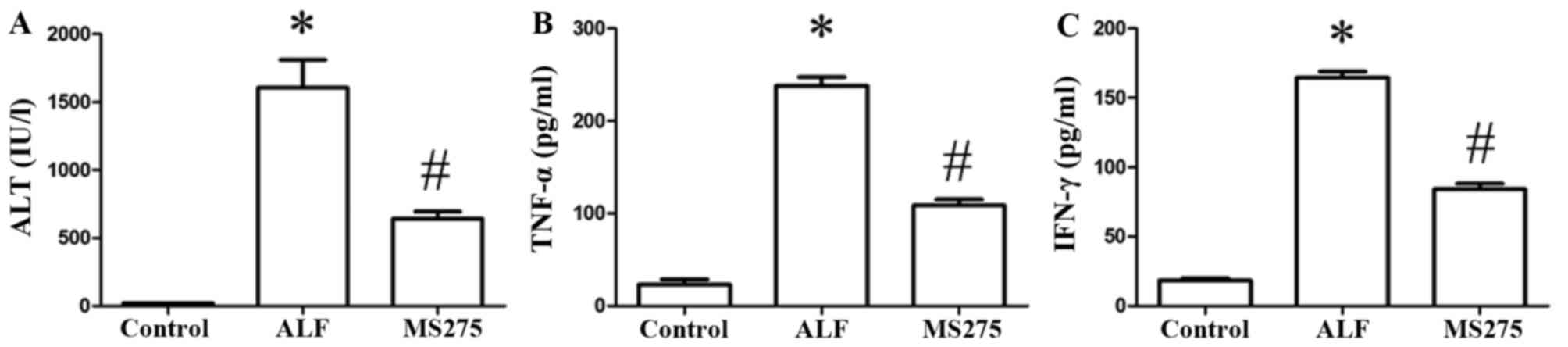
MS275 alleviates liver injury and the
production of inflammatory cytokines in ALF mice
Serum levels of ALT increased significantly in the
ALF group compared with the control group (P<0.05) and decreased
markedly in the MS275-treated group compared with the ALF group
(P<0.05; Fig. 3A). Serum levels
of TNF-α (Fig. 3B) and IFN-γ
(Fig. 3C) were markedly increased
in the ALF group compared with control group (P<0.05), and were
significantly decreased in the MS275-treated group compared with
the ALF group (P<0.05).
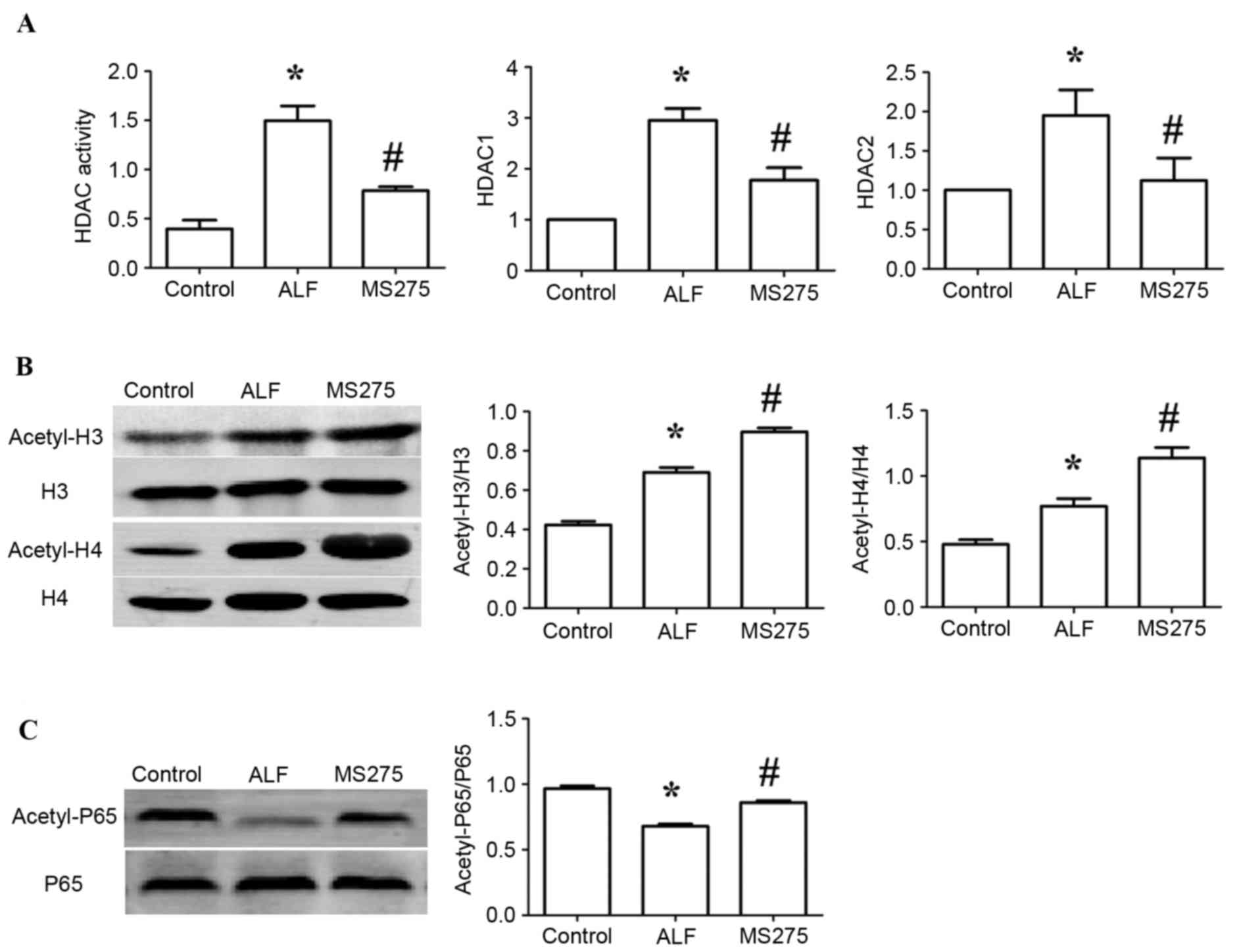
MS275 inhibits HDAC activity and HDAC
expression levels in ALF mice
The present study assessed HDAC activity in the sera
of ALF mice. HDAC activity increased significantly in the ALF group
compared with the control group (P<0.05), and HDAC activity in
the MS275-treated group was markedly reduced compared with the ALF
group (P<0.05). To observe the alterations in HDAC expression,
mRNA expression levels of HDAC1 and HDAC2 in liver tissue were
measured by RT-qPCR. The expression levels of HDAC1 and HDAC2
increased significantly in the ALF group compared with the control
group (P<0.05), and were decreased in the MS275-treated group
compared with the ALF group (P<0.05; Fig. 4A).
MS275 promotes the acetylation of
H3/H4 in the ALF mice
To investigate alterations in histone acetylation,
the acetylation levels of H3 and H4 in liver tissue were detected
in ALF mice. As presented in Fig.
4B, the acetylation levels of H3 and H4 were markedly increased
in the ALF group compared with the control group (P<0.05), and
were enhanced in the MS275-treated group compared with the ALF
group (P<0.05).
MS275 promotes the acetylation of
NF-κB p65 in the ALF mice
To investigate alterations in non-histone
acetylation, the acetylation levels of NF-κB p65 in liver tissue
were detected in ALF mice. As presented in Fig. 4C, the acetylation levels of p65
were decreased in the ALF group compared with the control group
(P<0.05), and were enhanced in the MS275-treated group compared
with the ALF group (P<0.05).
siRNA inhibits HDAC1 expression and
HDAC activity in RAW264.7 cells
To investigate the function of HDAC in inflammatory
responses, siRNA was transfected into RAW264.7 cells to silence
HDAC1. siRNA transfection significantly inhibited HDAC1 expression
and decreased HDAC activity in LPS-treated cells compared with
control cells (Fig. 5A).
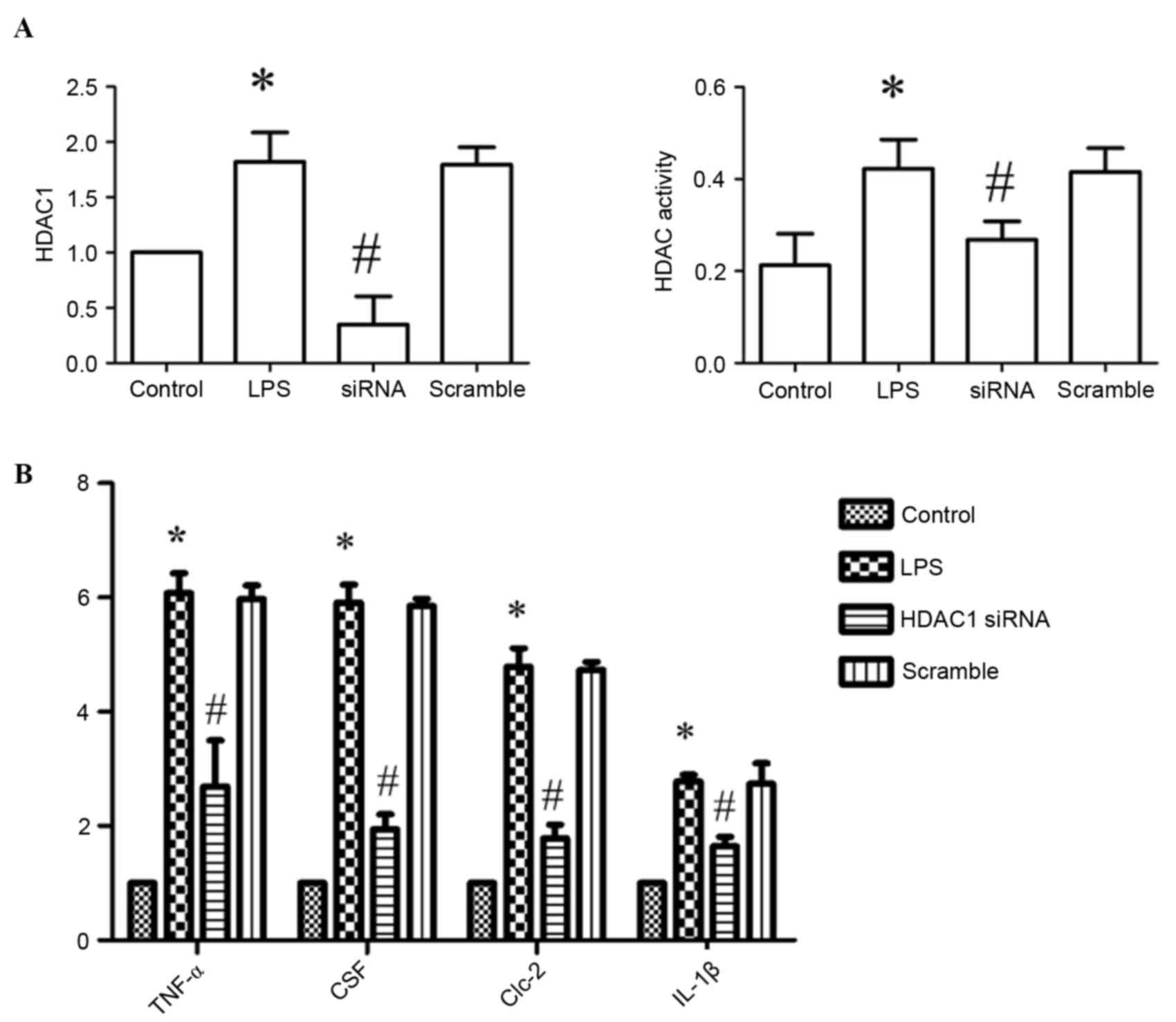 | Figure 5.Effect of silencing HDAC1 on HDAC
activity and cytokine production. (A) HDAC1 expression levels and
HDAC activity in RAW264.7 cells. (B) Expression levels of TNF-α,
CSF, Clc-2 and IL-1β, as assessed by reverse
transcription-quantitative polymerase chain reaction. Data are
presented as the mean ± standard deviation. *P<0.05 vs. control
group; #P<0.05 vs. LPS-treated group. LPS,
lipopolysaccharide; HDAC, histone deacetylase; siRNA, small
interfering RNA; TNF-α, tumor necrosis factor-α; CSF, cerebrospinal
fluid protein; Clc-2, chloride channel-2; IL-1β,
interleukin-1β. |
siRNA inhibits the production of
cytokines in RAW264.7 cells
To evaluate the effect of HDAC1-silencing on
cytokine production in RAW264.7 cells, the mRNA expression levels
of TNF-α, cerebral spinal fluid (CSF) protein, chloride channel
(Clc)-2 and interleukin (IL)-1β were measured by RT-qPCR.
Expression levels of TNF-α, CSF, Clc-2 and IL-1β decreased
significantly in siRNA-transfected cells compared with LPS-treated
cells (P<0.05; Fig. 5B).
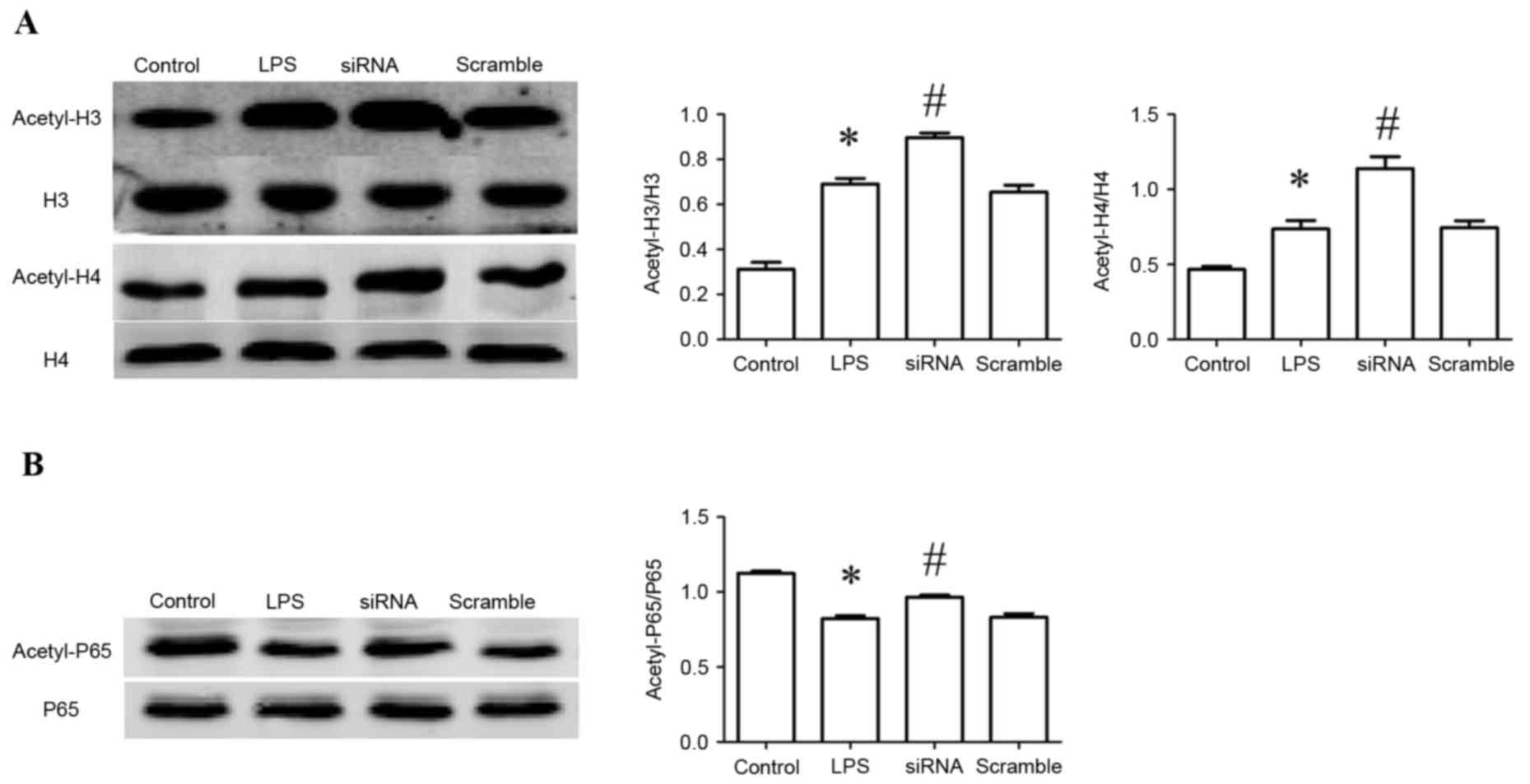
siRNA promotes acetylation of H3/H4 in
RAW264.7 cells
Acetylation of H3/H4 in RAW26.7 cells was assessed.
Compared with the control group, the acetylation levels of H3/H4 in
LPS-treated cells were increased significantly (P<0.05), and
enhanced in siRNA-transfected cells compared to LPS-treated cells
(P<0.05; Fig. 6A).
siRNA promotes acetylation of NF-κB
p65 in RAW264.7 cells
In addition to histone acetylation, acetylation of
NF-κB p65 in liver tissue was examined. The acetylation levels of
NF-κB p65 decreased in LPS-treated cells compared with the control
group (P<0.05), and were enhanced in the MS275-treated group
compared with the LPS-treated group (P<0.05; Fig. 6B).
Discussion
Epigenetic mechanisms have been identified as a
primary determination of gene expression and regulate complex
physiological and pathological processes. In addition to
methylation, histone acetylation is considered a key component of
epigenetic regulation. The nucleosome is composed of an octamer of
four core histones, an H3/H4 tetramer and two H2A/H2B dimers,
surrounded by 146 bp of DNA (15).
This architecture of chromatin is strongly influenced by histone
acetylation. Histone acetylation is controlled by HATs and HDACs.
To date, 18 members of the HDAC family have been identified
(16). The class I (HDAC1, 2, 3
and 8) and class II (HDAC4, 5, 6, 7, 9, 10 and 11) isoforms are
zinc-dependent, whereas class III HDACs (Sirtuins1, 2 and 7) are
nicotinamide adenine dinucleotide-dependent.
The present study demonstrated that HDAC activity,
and HDAC1 and HDAC2 expression levels, increased significantly in
patients with CHB, particularly in those with liver failure. These
results indicated that the aberrant status of HDAC activity and
expression levels may be associated with the pathogenesis of
CHB.
Additionally, the acetylation levels of H3 and H4
were assessed; acetylation levels were markedly increased in CHB
patients, particularly in those with liver failure. These results
indicated that the acetylation of histone is associated with the
disease progression of CHB.
Histone acetylation has previously been demonstrated
to be associated with activation of transcription, whereas
deacetylation is associated with gene repression (3,4).
Previous studies have additionally revealed that the acetylation of
histones is closely associated with activation of gene
transcription (17–20), which may promote inflammatory
responses by enhancing the expression levels of proinflammatory
genes (21). The present study
demonstrated increased acetylation levels of H3 and H4, and serum
HDAC expression levels and activity, in PBMCs of patients with CHB
and liver failure. This indicated that increased acetylation levels
of histones and HDAC activity are associated with the progression
of CHB. HDACis, including valproic acid, suberolyanilide hydroxamic
acid and peroxiredoxin, may suppress the expression levels of
proinflammatory cytokines and increase the survival rate of mice in
a septic shock model (22–24). The results of the present study
suggested that inhibiting the activity of HDACs may be beneficial
for the treatment of inflammation. However, histone acetylation
levels and HDACis require further investigation. Acetylation of
histones is associated with activation of transcription via
relaxation of the chromatin structure, whereas deacetylation
induces a condensed or inactive chromatin state, leading to gene
repression. This may explain the increased histone acetylation
levels observed in CHB patients. HDAC expression levels and
activity were expected to increase due to the deacetylation of
histones and the subsequent activation of transcription. However,
the effect of transcription regulation was interfered by the
acetylation of non-histones and/or other target genes of MS275
(only HDAC1 and HDAC2 were detected in this study). Histone
acetylation levels increased following MS275 treatment, indicating
that endogenous HDAC expression levels and activity affects
deacetylation of histones.
To investigate the roles of acetylation in
inflammatory responses, the present study examined the effect of
MS275 treatment on ALF mice, and silencing of HDAC1 in LPS-treated
RAW264.7 cells. MS275 is a class I-specific HDAC inhibitor, which
has previously been used as an antitumor drug (25). In the present study, MS275 was
demonstrated to protect liver tissue and inhibit the production of
pro-inflammatory cytokines in ALF mice. In addition, the expression
levels of HDAC1 and HDAC2, and HDAC activity, were decreased by
MS275 treatment. However, the acetylation levels of H3 and H4 were
enhanced by MS275 administration. These effects were additionally
observed in LPS-treated RAW264.7 cells.
These results indicated that HDAC1 serves a role in
the inflammatory response and that MS275 may represent a potential
therapeutic agent for the treatment of inflammation. Acetylation of
histones may not be involved in this anti-inflammatory effect, as
acetylation of H3 and H4 were promoted by MS275 and HDAC1 siRNA,
potentially activating gene transcription. Therefore, alterations
in HDAC activity and expression levels may contribute more to
inflammation, compared with histone acetylation.
In addition to histones, non-histones are
hypothesized to be modified by HDACs and HATs. A previous study
reported that >1,750 proteins are acetylated at their lysine
residuals (26). Furthermore,
>60 transcription factors were identified to be acetylated,
including signal transducer and activator of transcription
proteins, NF-κB, p53 and forkhead box O. The acetylation of these
proteins regulates multiple processes, including gene expression
and protein activity (27). To
evaluate whether the acetylation of non-histones is involved in the
anti-inflammatory effect of MS275 treatment and HDAC1 silencing,
the acetylation levels of NF-κB p65 were detected in ALF mice and
LPS-treated RAW264.7 cells. The results demonstrated that the
acetylation levels of NF-κB p65 decreased in LPS-treated cells and
ALF mice, and were promoted by MS275 treatment and HDAC1 silencing.
This indicated that the acetylation of non-histones may be
associated with the anti-inflammatory effects of MS275 treatment
and HDAC1 silencing.
It has previously been reported that acetylation of
histones affects HBV replication (28,29).
In patients with CHB, the HBV infection is the original cause of
liver injury. The immune system eliminates the virus by initiating
an inflammatory response. In the present study, the association
between acetylation of histones and HBV DNA load was not evaluated;
acetylation modification of histones/non-histones in the
inflammatory process was examined in vitro, in vivo
and in CHB patients. Therefore, inhibition of histone/non-histone
acetylation may have an anti-inflammatory effect.
In conclusion, the present study demonstrated
aberrant histone acetylation, and HDAC activity and expression
levels, in patients with CHB; these were associated with the
severity of the disease. Additionally, MS275 treatment and HDAC1
silencing had an anti-inflammatory effect by decreasing the
expression levels of pro-inflammatory cytokines. Alterations in
HDAC activity and expression levels demonstrated a greater effect
on inflammation compared with histone acetylation; therefore, the
underlying mechanisms may be associated with the acetylation of
non-histones. These results provide a potential novel therapeutic
strategy for the treatment of CHB.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81371789).
Glossary
Abbreviations
Abbreviations:
|
CHB
|
chronic hepatitis B
|
|
HDACs
|
histone deacetylases
|
|
HATs
|
histone acetyltransferases
|
|
HDACi
|
histone deacetylase inhibitor
|
|
NF-κB
|
nuclear factor-κB
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
SPF
|
specific pathogen-free
|
References
|
1
|
Lu Q, Qiu X, Hu N, Wen H, Su Y and
Richardson BC: Epigenetics, disease, and therapeutic interventions.
Ageing Res Rev. 5:449–467. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forsberg EC and Bresnick EH: Histone
acetylation beyond promoters: Long-range acetylation patterns in
the chromatin world. Bioessays. 23:820–830. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wade PA: Transcriptional control at
regulatory checkpoints by histone deacetylases: Molecular
connections between cancer and chromatin. Hum Mol Genet.
10:693–698. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Merican I, Guan R, Amarapuka D, Alexander
MJ, Chutaputti A, Chien RN, Hasnian SS, Leung N, Lesmana L, Phiet
PH, et al: Chronic hepatitis B virus infection in Asian countries.
J Gastroenterol Hepatol. 15:1356–1361. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
The guideline of prevention and treatment
for chronic hepatitis B (2010 version). Zhonghua Gan Zang Bing Za
Zhi. 19:13–24. 2011.(In Chinese). PubMed/NCBI
|
|
7
|
Yim HJ and Lok AS: Natural history of
chronic hepatitis B virus infection: What we knew in 1981 and what
we know in 2005. Hepatology. 43:(2 Suppl 1). S173–S181. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cantley MD and Haynes DR: Epigenetic
regulation of inflammation: Progressing from broad acting histone
deacetylase (HDAC) inhibitors to targeting specific HDACs.
Inflammopharmacology. 21:301–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gillespie J, Savic S, Wong C, Hempshall A,
Inman M, Emery P, Grigg R and McDermott MF: Histone deacetylases
are dysregulated in rheumatoid arthritis and a novel histone
deacetylase 3-selective inhibitor reduces interleukin-6 production
by peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Rheum. 64:418–422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barnes PJ: Role of HDAC2 in the
pathophysiology of COPD. Annu Rev Physiol. 71:451–464. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim Y, Kim K, Park D, Lee E, Lee H, Lee
YS, Choe J and Jeoung D: Histone deacetylase 3 mediates allergic
skin inflammation by regulating expression of MCP1 protein. J Biol
Chem. 287:25844–25859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarin SK, Kumar A, Almeida JA, Chawla YK,
Fan ST, Garg H, de Silva HJ, Hamid SS, Jalan R, Komolmit P, et al:
Acute-on-chronic liver failure: Consensus recommendations of the
Asian pacific association for the study of the liver (APASL).
Hepatol Int. 3:269–282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liaw YF, Kao JH, Piratvisuth T, Chan HL,
Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH, et al:
Asian-Pacific consensus statement on the management of chronic
hepatitis B: A 2012 update. Hepatol Int. 6:531–561. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J.
370:737–749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chung S, Sundar IK, Yao H, Ho YS and
Rahman I: Glutaredoxin 1 regulates cigarette smoke-mediated lung
inflammation through differential modulation of I{kappa}B kinases
in mice: Impact on histone acetylation. Am J Physiol Lung Cell Mol
Physiol. 299:L192–L203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Natsume-Kitatani Y, Shiga M and Mamitsuka
H: Genome-wide integration on transcription factors, histone
acetylation and gene expression reveals genes co-regulated by
histone modification patterns. Plos One. 6:e222812011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chung S, Sundar IK, Hwang JW, Yull FE,
Blackwell TS, Kinnula VL, Bulger M, Yao H and Rahman I: NF-κB
inducing kinase, NIK mediates cigarette smoke/TNFα-induced histone
acetylation and inflammation through differential activation of
IKKs. PLoS One. 6:e234882011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balasubramani A, Winstead CJ, Turner H,
Janowski KM, Harbour SN, Shibata Y, Crawford GE, Hatton RD and
Weaver CT: Deletion of a conserved cis-element in the Ifng locus
highlights the role of acute histone acetylation in modulating
inducible gene transcription. PLoS Genet. 10:e10039692014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khan N, Jeffers M, Kumar S, Hackett C,
Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, et al:
Determination of the class and isoform selectivity of
small-molecule histone deacetylase inhibitors. Biochem J.
409:581–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao W, Bao C, Padalko E and Lowenstein CJ:
Acetylation of mitogen-activated protein kinase phosphatase-1
inhibits Toll-like receptor signaling. J Exp Med. 205:1491–1503.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Liu B, Zhao H, Sailhamer EA,
Fukudome EY, Zhang X, Kheirbek T, Finkelstein RA, Velmahos GC,
deMoya M, et al: Protective effect of suberoylanilide hydroxamic
acid against LPS-induced septic shock in rodents. Shock.
32:517–523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Wan J, Jiang R, Wang W, Deng H,
Shen Y, Zheng W and Wang Y: Protective effects of trichostatin A on
liver injury in septic mice. Hepatol Res. 39:931–938. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Flis S, Gnyszka A and Spławiński J: HDAC
inhibitors, MS275 and SBHA, enhances cytotoxicity induced by
oxaliplatin in the colorectal cancer cell lines. Biochem Biophys
Res Commun. 387:336–341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Spange S, Wagner T, Heinzel T and Krämer
OH: Acetylation of non-histone proteins modulates cellular
signalling at multiple levels. Int J Biochem Cell Biol. 41:185–198.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang DY, Zou LP, Liu XJ, Zhu HG and Zhu R:
Hepatitis B virus X protein induces the histone H3 lysine 9
trimethylation on the promoter of p16 gene in hepatocarcinogenesis.
Exp Mol Pathol. 99:399–408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tropberger P, Mercier A, Robinson M, Zhong
W, Ganem DE and Holdorf M: Mapping of histone modifications in
episomal HBV cccDNA uncovers an unusual chromatin organization
amenable to epigenetic manipulation. Proc Natl Acad Sci USA.
112:E5715–E5724. 2015. View Article : Google Scholar : PubMed/NCBI
|