Introduction
Gastric cancer is one of the most common malignant
tumors worldwide, causing ~738,000 deaths in 2008 (1). Major contributory factors to gastric
cancer include Helicobacter pylori infection, diet,
alcoholic consumption and smoking (2,3).
Receptor tyrosine kinase (RTK) pathways have key
roles in the progression of various tumors (4–6),
with aberrant epidermal growth factor receptor (EGFR) signaling
demonstrated to be particularly common (7); EGF family proteins have been revealed
to be significantly overexpressed in >60% of tumors (8,9). The
main EGF family members include EGFR [also termed human epidermal
growth factor receptor 1 (HER1)], HER2 (also termed ErbB2), HER3
(also termed ErbB3) and HER4 (also termed ErbB4). EGFR and HER2 are
often significantly upregulated in gastric cancer, and are
considered to be well-established oncogenes (10). Mucin 13 (MUC13) has also been
demonstrated to be aberrantly upregulated in various tumors
(11–13). Exogenous expression of MUC13
contributes to abnormal cell proliferation, motility and tumor
growth (13), and overexpression
of MUC13 results in the activation of HER2, extracellular
signal-regulated kinase (ERK) and Akt serine/threonine kinase
(Akt), and the reduction of p53 expression (12). However, few studies have, thus far,
investigated the expression of MUC13 in gastric cancer.
MicroRNAs (miRNAs) are small non-coding RNAs that
widely control gene expression at the post-transcriptional level
(14–16). Due to the oncogenic or tumor
suppressive roles of miRNAs, abnormal expression can lead to the
initiation, formation and progression of tumors. For example,
numerous miRNAs are differentially expressed in gastric cancer,
including miRNA-199a-3p (miR-199a-3p), miR-429 and miR-34a
(14–16). The present study aimed to
investigate miRNAs that regulate the expression of MUC13 in gastric
cancer.
Materials and methods
Patient selection and biopsy
collection
In the present study, biopsies were taken from both
tumor tissue and the adjacent normal tissue of 40 patients
receiving adenocarcinoma surgery of the stomach or esophageal
junction. Samples were collected from July 2012 to November 2014
and written consent was provided by all individuals. The collection
of biopsies was approved by the Ethics Committee of the First
Hospital of Jilin University (Changchun, China) in accordance with
the Declaration of Helsinki. All biopsy samples were reviewed by an
experienced pathologist to validate the diagnosis.
Immunohistochemistry
Paraffin-embedded tissues fixed in 4% buffered
paraformaldehyde were cut into 5 µm sections and washed three times
(5 min per wash) in phosphate-buffered saline (PBS), then incubated
in 3% H2O2 for 30 min at room temperature.
Sections were blocked by incubation with 10% goat serum in PBS
(Origene Technologies, Inc., Rockville, MD, USA) for 30 min at
37°C, then incubated with the MUC13 primary antibody (1:80; catalog
no. ab124654; Abcam, Cambridge, UK) for 24 h at 4°C. Sections were
washed with PBS, then incubated with secondary antibody
(biotin-labelled goat anti-mouse immunoglobulin G; 1:200; catalog
no. SP-9000D; Origene Technologies, Inc.) for 1 h at 4°C. Following
washing with PBS, sections were incubated with horseradish
peroxidase conjugated streptavidin (1:200) for 1 h at room
temperature, then with diaminobenzidine/H2O2
for 15 min at room temperature. Following dehydration in gradient
alcohol, and transparentizing in xylene, sections were mounted with
glycerol and observed under a microscope. In control sections, the
primary antibody was replaced with 1% calf serum (Origene
Technologies, Inc.).
Cell culture
Gastric cancer cell line, MKN28 was purchased from
American Type Culture Collection (ATCC, Manassas, VA, USA) and
cultured in RPMI-1640 (GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), streptomycin (100
mg/ml) and penicillin (100 U/ml) at 37°C in a humidified atmosphere
containing 5% CO2. Recent reports have suggested that
the MKN28 gastric carcinoma cell line used in this study is
contaminated with another gastric carcinoma cell line, MKN74
(17).
RNA extraction
Total RNA was extracted from gastric tissues or
MKN28 cells using TRIzol reagent according to the manufacturers'
instructions (Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was reverse transcribed using Takara
MicroRNA Reverse Transcription Kit (Takara Bio, Inc., Otsu, Japan)
with specific primers for miR-132-3p (GTC GT ATC CAG TGC AGG GTC
CGA GGT ATT CGC ACT GGA TAC GAC CGA CC) and U6 (GTC GTAT CCA GTG
CAG GGT CCG AGG TAT TCG CAC TGG ATA CGA CAA ATA T). Subsequently,
the PCR amplification was performed. 1 mg of cDNA was used for qPCR
using SYBR green Master mix (Roche Diagnostics, Basel, Switzerland)
on a Roche Lightcycler 480 (Roche Diagnostics) at 95°C for 10 min
followed by 50 cycles of 95°C for 10 sec, 55°C for 10 sec, 72°C for
5 sec; 99°C for 1 sec; 59°C for 15 sec; 95°C for 1 sec; then
cooling to 40°C. Relative miRNA expression of miR-132-3p was
normalized against the endogenous control, U6, using the Δ-Δ Cq
method (18).
Transient transfection
A total of 6×105 cells were equally
seeded in the 6-well plates with 2 ml RPMI-1640 culture medium
containing serum and antibiotics. At the same time, miR-132-3p
mimic, inhibitor, miR negative control or siRNA targeting MUC13
(CCA GCU UGU UGA GGU AGA AGU AGU A) or non-target control siRNA
(Shanghai GenePharma Co., Ltd., Shanghai, China) were mixed with
HiperFect transfection reagent (Qiagen GmbH, Hilden, Germany) and
incubated at room temperature for 10 min. The complex was then
transfected into MKN28 cells for 48 h.
Cell viability analysis
To examine cell viability, MKN28 cells were seeded
in 96-well plates at a density of 1.0×104 cells/per
well. miR-132 mimics, inhibitors or a scramble/non-targeting oligo
negative control (NC) were transfected into cells at 24, 48, 72 h
after seeding of cells. MTT assay was performed as previously
described (19).
Adenovirus vector construction and
transfection
The adenovirus vector (Ad)-MUC13 and Ad-control
(Ad-con) were purchased from the Chinese National Human Genome
Center (Beijing, China). In brief, 6×105 cells were
equally seeded in the 6-well plates with 2 ml RPMI-1640 culture
medium containing serum and antibiotics. Following 24 h, the cells
were transfected with Ad-MUC13 or Ad-con at the density of 100
multiplicity of infection (MOI) for 48 h. The transfection
efficiency was calculated as the green fluorescent protein-positive
cells/all cells in each field ×100%.
Luciferase target assay
The 3′untranslated region (UTR) of MUC13 containing
the predicted target site for miR-20a-5p, was cloned into the
pmirGLO (Promega Corporation, Madison, WI, USA) luciferase reporter
vector which had been cleaved at the SacI and XhoI sites. Details
of PCR procedures are described as follows: a heated initial
denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C
for 15 sec, 55°C for 45 sec and 72°C for 30 sec. Prior to
conducting the luciferase reporter assay, 5×104 cells
per well were seeded in 24-well plates in a 500 µl medium and
cultured for 18 h. The cells were transfected with the modified
firefly luciferase vector (500 ng/µl) mixed with Vigofect
transfection reagent, according to the manufacturer's protocol.
Following a 48 h continuous exposure, the luciferase activities
from firefly and renilla were measured with the Dual-luciferase
reporter assay system (Promega Corporation). Renilla activity was
used as the normalized parameter.
Establishment of MUC13-expressing
MKN28 stable cell line
MKN28 cells were transfected with
pmirGLO-MUC13–3′UTR or empty vector (pmirGLO) using VigoFect
transfection reagent (Vigorus Biotechnology, Beijing, China).
Individual G418 resistant clones (1 mg/ml; Invitrogen; Thermo
Fisher Scientific, Inc.) were selected and applied for further
study.
Western blotting analyses
Tissue or MKN28 cell protein was extracted using
RIPA buffer (Solarbio Science & Technology Co., Ltd., Beijing,
China). A bicinchoninic protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.) was used to determine the protein concentration.
Equal quantities of protein (15 µg) were resolved by 10% SDS-PAGE
and transferred onto a PVDF membrane. The protein was detected with
primary antibodies, MUC13 (catalog no. ab124654; Abcam), HER2
(catalog no. 4290; 1:1,000), p-ERK (catalog no. 1150; 1:1,000), ERK
(catalog no. 9194; 1:1,000), p-Akt (catalog no. 8200; 1:1,000), Akt
(catalog no. 9840; 1:1,000) and GAPDH (catalog no. 2118; 1:5,000)
all obtained from Cell Signaling Technology, Inc., (Danvers, MA,
USA) overnight at 4°C. Nonspecific binding was blocked using 8%
(w/v) milk in Tris-buffered saline with 1% Tween-20 (TBST; Beijing
SolarBio Science & Technology Co., Ltd.) for 2 h at room
temperature. Following several washes with TBST, the membranes were
incubated with horseradish-peroxidase (HRP)-conjugated goat
anti-rabbit and anti-mouse IgG or HRP-conjugated mouse anti-goat
IgG (all 1:5,000; Origene Technologies, Inc.) for 2 h at room
temperature and then washed. GAPDH was used as the internal
control. Signals were detected with enhanced chemiluminescence
according to the manufacturer's protocol (EMD Millipore, Billerica,
MA, USA). ImageJ software (National Institutes of Health, Bethesda,
MD, USA) was used for density analysis.
Bioinformatic predictions
To determine the potential miRNAs that target MUC13,
bioinformatic prediction was performed using TargetScan (http://www.targetscan.org).
Cell invasion assay
Invasion of cells were examined using a Transwell
system (Invitrogen; Thermo Fisher Scientific, Inc.). The MKN28
cells transfected with miR-132 inhibitors or ad-MUC13 were cultured
in the lower chamber with fresh medium containing 10% FBS. After
incubation for 24 h at 37°C, the cells on the upper chamber was
stained with 0.5% crystal violet and dissolved in 10% acetic acid
for measurement of absorbance at 560 nm.
Cell migration assay
The in vitro wound healing assay was
performed as previously described (20). Briefly, MKN28 cells were seeded in
6-well plates to form a confluent monolayer. The monolayer was
scratched with a sterile 10 µl pipette tip, and the floating cells
were carefully removed by washing with PBS. Then, the cells were
cultured in RPMI-1640 medium without FBS at 37°C in a 5%
CO2 atmosphere. The wound scratches were photographed at
0 and 12 h then scraped to collect cells.
Statistical analysis
The data are expressed as the mean ± standard error.
The number of independent experiments was represented by ‘n’. Data
were analyzed using SPSS software, version 13.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulation of MUC13 in gastric
cancer tissues
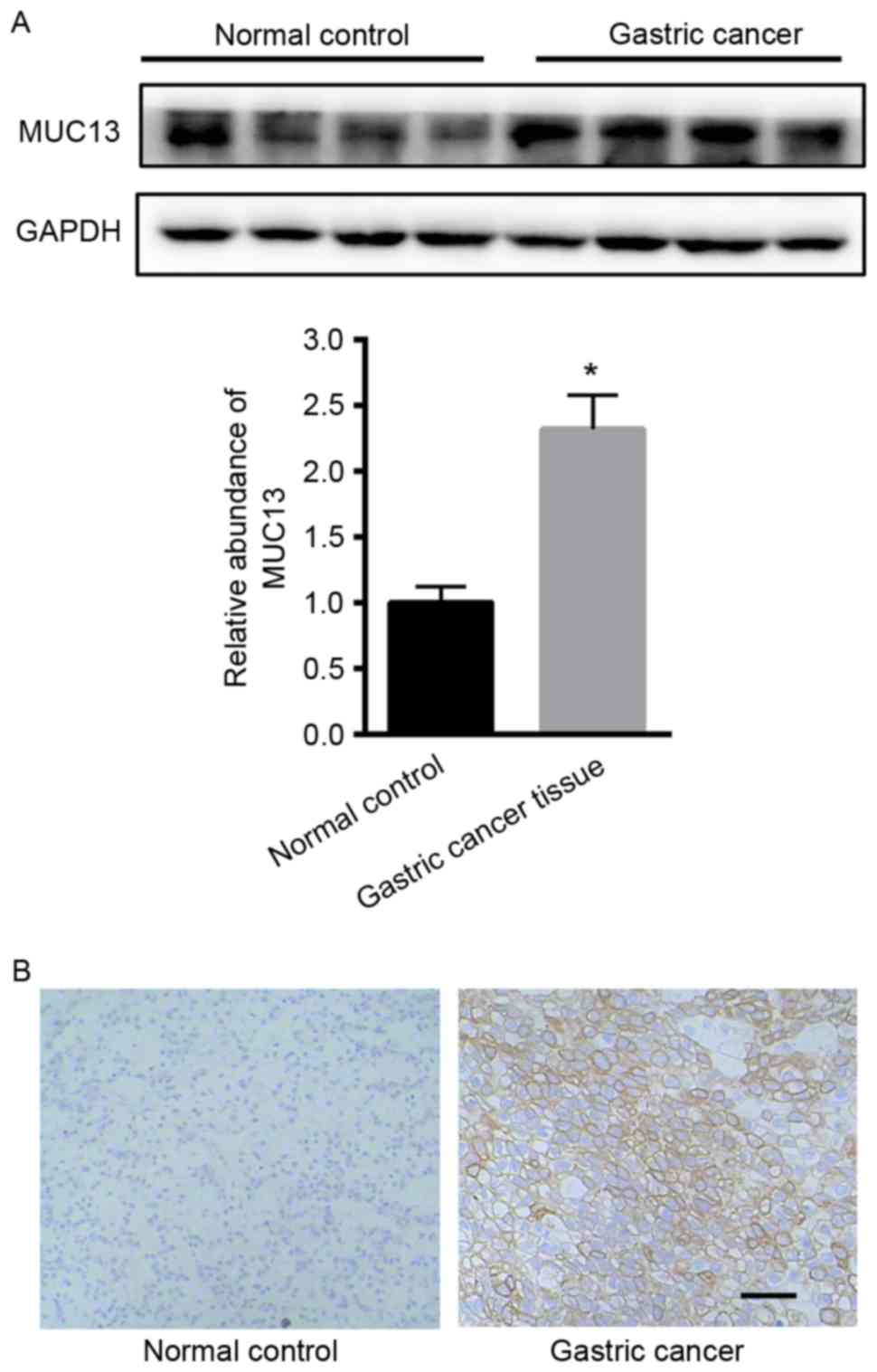
Initially, the expression of MUC13 in gastric cancer
tissues was examined. Western blot analysis demonstrated that MUC13
was significantly upregulated in gastric cancer tissues compared
with adjacent normal tissues (Fig.
1A). Immunohistochemistry analysis also demonstrated the
enhanced expression of MUC13 in gastric cancer tissues (Fig. 1B).
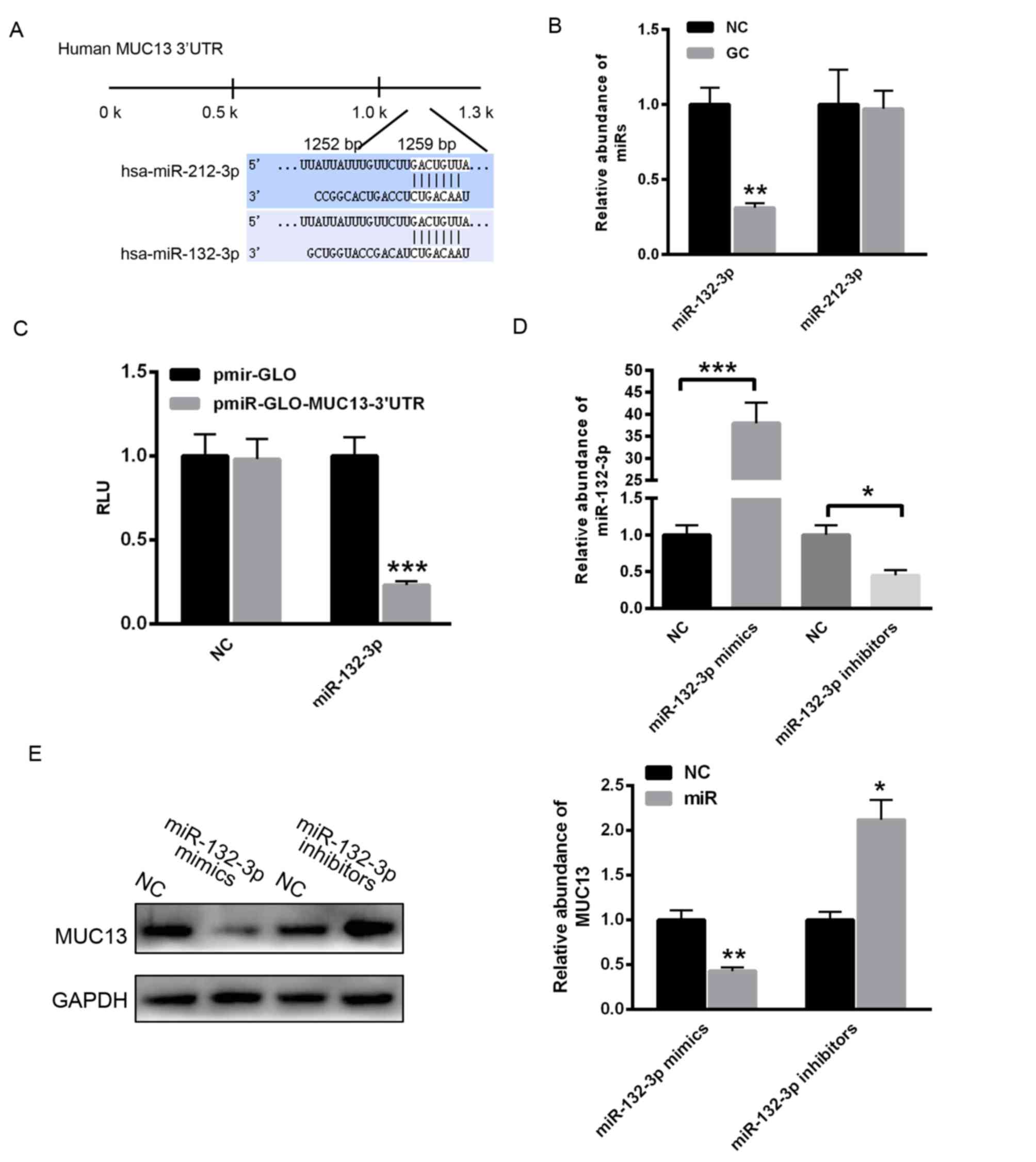
MUC13 is a target gene of miR-132-3p
in gastric cancer
To identify the potential miRNAs that regulate the
expression of MUC13, the TargetScan online prediction program was
used. As demonstrated in Fig. 2A,
two putative conserved binding miRNAs, miR-132-3p and miR-212-3p,
were identified to potentially bind the 3′untranslated region
(3′UTR) of MUC13. The present study determined that miR-132-3p
levels were reduced in gastric cancer tissues compared with
adjacent normal control tissue, whereas miR-212-3p did not
demonstrate significant a change (Fig.
2B). Subsequently, the 3′UTR of MUC13 was cloned into the
pmirGLO plasmid. A dual luciferase reporter assay demonstrated that
miR-132-3p significantly decreased the relative luciferase units of
pmirGLO-MUC13–3′UTR compared with with the empty vector pmirGLO
(Fig. 2C). Additionally,
miR-132-3p mimics or inhibitors were transfected into MKN28 cells.
RT-qPCR analysis revealed that transfection with miR-132-3p mimics
markedly enhanced the level of miR-132-3p, whereas transfection
with miR-132-3p inhibitors significantly reduced the level of
miR-132-3p (Fig. 2D).
Overexpression of miR-132-3p significantly increased the level of
miR-132-3p, however decreased the protein level of MUC13 (Fig. 2E). By contrast, inhibition of
miR-132-3p reduced the level of miR-132-3p, however increased the
expression of MUC13 (Fig. 2E).
These data indicated that reduction of miR-132-3p led to enhanced
MUC13 expression in gastric cancer cells.
Overexpression of MUC13 increases the
activation of HER signaling and cell invasion and migration in
MKN28 cells
To explore the role of MUC13 on gastric cancer
progression, cell invasion and migration were analyzed. As
demonstrated in Fig. 3A, the
transfection efficiency of Ad-MUC13 or Ad-con was nearly 100%
(Fig. 3A). No alterations of MUC13
protein levels were observed following simple transfection of blank
adenovirus vectors (data not shown). A Transwell assay demonstrated
that overexpression of MUC13 significantly enhanced cell invasion
capacity (Fig. 3B). Furthermore, a
scratch assay demonstrated that MKN28 cell migration was enhanced
by MUC13 overexpression (Fig. 3C).
The signaling downstream of MUC13 was also examined. As
demonstrated in Fig. 3D,
overexpression of MUC13 obviously enhanced the level of HER2, and
the phosphorylation of ERK and Akt (Fig. 3D).
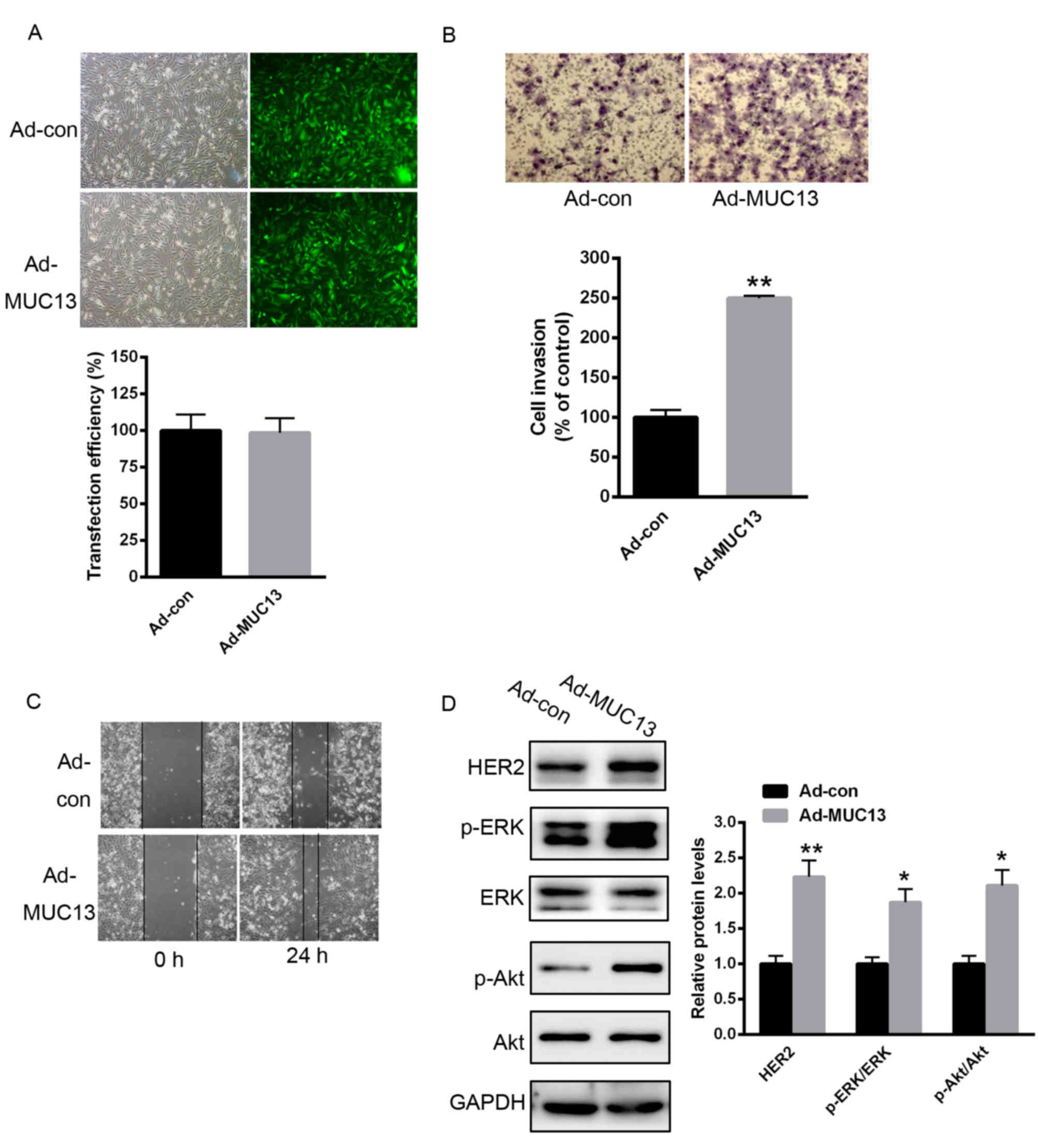 | Figure 3.Overexpression of MUC13 prompted MKN28
cell invasion and migration through activation of HER signaling.
(A) Transfection efficiency of Ad-MUC13 or Ad-con. (B) Transwell
assay demonstrated that overexpression of MUC13 significantly
enhanced MKN28 cell invasion capacity. (C) Scratch assay
demonstrated that MKN28 cell migration was significantly enhanced
when MUC13 was overexpressed. (D) Overexpression of MUC13 obviously
enhanced the expression of HER2, and the phosphorylation of ERK and
Akt. n=3 independent experiments, *P<0.05, **P<0.01 vs.
control. Ad, adenovirus; con, control; MUC13, mucin 13; HER2, human
epidermal growth factor receptor 2; p-, phosphorylated; ERK,
extracellular signal-regulated kinase; Akt, Akt serine/threonine
kinase. |
Knockdown of MUC13 partially reverses
miR-132-3p inhibition-induced MKN28 cell invasion and
migration
To explore whether miR-132-3p exerts its role
through MUC13, miR-132-3p inhibitors were transfected into MKN28
cells. As demonstrated in Fig. 4A,
inhibition of miR-132-3p significantly enhanced the protein level
of MUC13, and enhanced the level of HER2, and ERK and Akt
phosphorylation. Notably, an siRNA targeting MUC13 was selected to
suppress the expression of MUC13. miR-132-3p inhibition reduced the
effects of MUC13 siRNA on HER2 expression, and ERK and Akt
activation (Fig. 4B).
Additionally, the effect was on cell invasion and migration was
also determined. miR-132-3p inhibitors reduced the effect of MUC13
knockdown on cell invasion and migration (Fig. 4C and D).
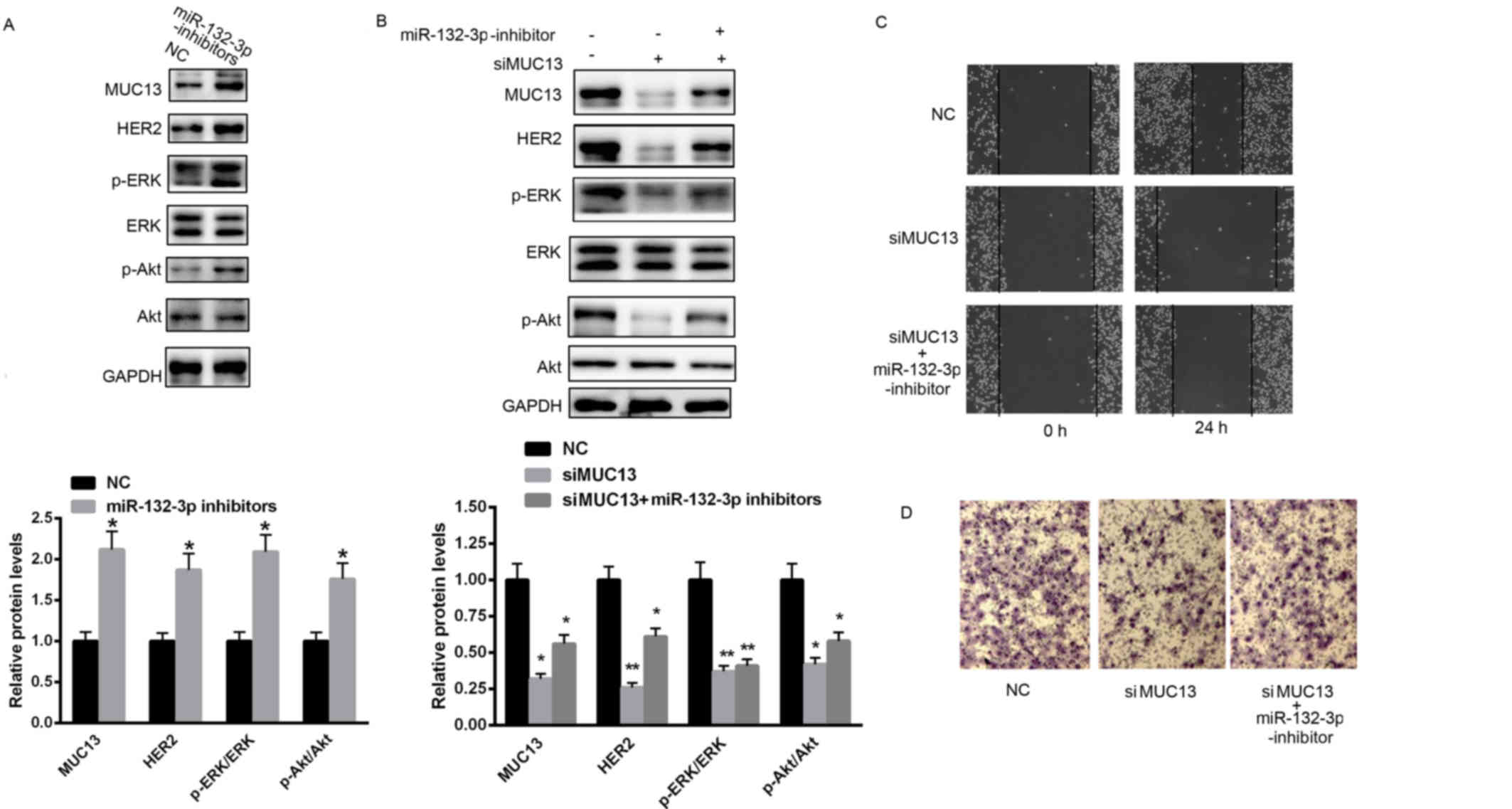 | Figure 4.Knockdown of MUC13 partially reverses
miR-132-3p inhibition-induced effects on MKN28 cell invasion and
migration. (A) Inhibition of miR-132-3p significantly enhanced the
expression of MUC13 and the activation of downstream signaling. (B)
Knockdown of MUC13 reversed the effects of miR-132-3p inhibition on
HER2, ERK and Akt. (C) Cell invasion and (D) migration were
monitored in cells transfected with miR-132-3p inhibitors or/and
si-MUC13 or NC. n=3 independent experiments, *P<0.05,
**P<0.01 vs. control. NC, negative control; miR, microRNA;
MUC13, mucin 13; HER2, human epidermal growth factor receptor 2;
p-, phosphorylated; ERK, extracellular signal-regulated kinase;
Akt, Akt serine/threonine kinase; siMUC13, MUC13-targeting small
interfering RNA. |
Discussion
Mucins are considered as potential oncogenes and
possible therapeutic targets in various malignancies (21–23).
As a high-molecular-weight transmembrane glycoprotein, MUC13 is
reported to be frequently overexpressed in various epithelial
carcinomas, including gastric, colorectal and ovarian cancers
(24). MUC13 includes three
EGF-like domains and a cytoplasmic domain with phosphorylation
sites, which trigger the activation of HER2 signaling (24). The current study examined MUC13
expression in gastric cancer tissues and detected that MUC13
protein levels were significantly increased compared with adjacent
normal tissues.
A previous study reported that overexpression of
MUC13 significantly enhanced the activation of HER2, ERK and Akt
(13). To validate the role of
MUC13 in gastric cancer progression, MUC13 was exogenously
expressed in MKN28 cells. Overexpression of MUC13 observably
enhanced gastric cancer cell invasion and migration. By contrast,
knockdown of MUC13 decreased MKN28 cell invasion and migration. Due
to the three EGF domains, MUC13 is considered to stabilize the
protein level of EGF receptors, particularly HER2, thus enhancing
the activation of ERK and Akt. Activation of phosphoinositide
3-kinase/Akt and mitogen-activated protein kinase signaling through
HER2 enhances tumorigenesis in various types of cancer (25,26).
miRNAs are increasingly reported to be
differentially expressed in various tumors either as oncogenes or
tumor suppressors (27–29). In a previous study, miR-145 was
identified to target MUC13 inhibiting the tumor growth and invasion
in pancreatic tissues. The present study aimed to elucidate novel
miRNAs that regulate the expression of MUC13 in gastric cancer
(12). Bioinformatic predictions
suggested that miR-132-3p and miR-212-3p may bind to the 3′UTR of
MUC13. A luciferase reporter assay and western blot analysis
demonstrated that MUC13 is the target gene of miR-132-3p. Further
study demonstrated that miR-132-3p was obviously decreased in
gastric cancer tissues compared with normal adjacent tissues. In
line with MUC13 overexpression, reduced miR-132-3p contributed to
gastric cancer cell invasion and migration. Notably, inhibition of
miR-132-3p reduced the activation of ERK and Akt even in cells
transfected the specific siRNA targeting MUC13, suggesting the
tumor suppressor role of miR-132-3p in gastric cancer through
MUC13.
To conclude, miR-132-3p may function as a tumor
suppressor in gastric cancer tissues by targeting MUC13 and
subsequently prompting the activation of HER2 signaling.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kato M and Asaka M: Recent knowledge of
the relationship between Helicobacter pylori and gastric
cancer and recent progress of gastroendoscopic diagnosis and
treatment for gastric cancer. Jpn J Clin Oncol. 40:828–837. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li L, Ying XJ, Sun TT, Yi K, Tian HL, Sun
R, Tian JH and Yang KH: Overview of methodological quality of
systematic reviews about gastric cancer risk and protective
factors. Asian Pac J Cancer Prev. 13:2069–2079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng Z, Ma S, Zhou H, Zang A, Fang Y, Li
T, Shi H, Liu M, Du M, Taylor PR, et al: Tyrosine phosphatase SHP-2
mediates C-type lectin receptor-induced activation of the kinase
Syk and anti-fungal TH17 responses. Nat Immunol. 16:642–652. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chung C: Tyrosine kinase inhibitors for
epidermal growth factor receptor gene mutation-positive non-small
cell lung cancers: An update for recent advances in therapeutics. J
Oncol Pharm Pract. 22:461–476. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shabani M, Naseri J and Shokri F: Receptor
tyrosine kinase-like orphan receptor 1: A novel target for cancer
immunotherapy. Expert Opin Ther Targets. 19:941–955. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hatanpaa KJ, Burma S, Zhao D and Habib AA:
Epidermal growth factor receptor in glioma: Signal transduction,
neuropathology, imaging, and radioresistance. Neoplasia.
12:675–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mellinghoff IK, Wang MY, Vivanco I,
Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute
DJ, et al: Molecular determinants of the response of glioblastomas
to EGFR kinase inhibitors. N Engl J Med. 353:2012–2024. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mizukami T, Togashi Y, Sogabe S, Banno E,
Terashima M, De Velasco MA, Sakai K, Fujita Y, Tomida S, Nakajima
TE, et al: EGFR and HER2 signals play a salvage role in
MEK1-mutated gastric cancer after MEK inhibition. Int J Oncol.
47:499–505. 2015.PubMed/NCBI
|
|
10
|
Yk W, Cf G, Xw TYZCZ, Xx L, Nl M and Wz Z:
Assessment of ERBB2 and EGFR gene amplification and protein
expression in gastric carcinoma by immunohistochemistry and
fluorescence in situ hybridization. Mol Cytogenet. 4:142011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Smet A, Blaecher C, Flahou B,
Ducatelle R, Linden S and Haesebrouck F: Gastric de novo Muc13
expression and spasmolytic polypeptide-expressing metaplasia during
Helicobacter heilmannii infection. Infect Immun.
82:3227–3239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khan S, Ebeling MC, Zaman MS, Sikander M,
Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D,
et al: MicroRNA-145 targets MUC13 and suppresses growth and
invasion of pancreatic cancer. Oncotarget. 5:7599–7609. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chauhan SC, Ebeling MC, Maher DM, Koch MD,
Watanabe A, Aburatani H, Lio Y and Jaggi M: MUC13 mucin augments
pancreatic tumorigenesis. Mol Cancer Ther. 11:24–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu G, Jiang C, Li D, Wang R and Wang W:
MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in
gastric cancer. Tumour Biol. 35:9801–9806. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu D, Xia P, Diao D, Cheng Y, Zhang H,
Yuan D, Huang C and Dang C: MiRNA-429 suppresses the growth of
gastric cancer cells in vitro. J Biomed Res. 26:389–393. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY
and Zhu ZG: miRNA-199a-3p in plasma as a potential diagnostic
biomarker for gastric cancer. Ann Surg Oncol. 20:(Suppl 3).
S397–S405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nardone RM: Curbing rampant
cross-contamination and misidentification of cell lines.
Biotechniques. 45:221–227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang F, Wang H, Jiang Z, Hu A, Chu L, Sun
Y and Han J: MicroRNA19a mediates gastric carcinoma cell
proliferation through the activation of nuclear factor-kB. Mol Med
Rep. 12:5780–5786. 2015.PubMed/NCBI
|
|
20
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khan S, Ansarullah D Kumar, Jaggi M and
Chauhan SC: Targeting microRNAs in pancreatic cancer: Microplayers
in the big game. Cancer Res. 73:6541–6547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo H, Guo W, Wang F, You Y, Wang J, Chen
X, Wang J, Wang Y, Du Y, Chen X, et al: miR-1291 targets mucin 1
inhibiting cell proliferation and invasion to promote cell
apoptosis in esophageal squamous cell carcinoma. Oncol Rep.
34:2665–2673. 2015.PubMed/NCBI
|
|
23
|
Wakata K, Tsuchiya T, Tomoshige K, Takagi
K, Yamasaki N, Matsumoto K, Miyazaki T, Nanashima A, Whitsett JA,
Maeda Y and Nagayasu T: A favourable prognostic marker for EGFR
mutant non-small cell lung cancer: Immunohistochemical analysis of
MUC5B. BMJ Open. 5:e0083662015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maher DM, Gupta BK, Nagata S, Jaggi M and
Chauhan SC: Mucin 13: Structure, function, and potential roles in
cancer pathogenesis. Mol Cancer Res. 9:531–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arias-Romero LE and Chernoff J:
p21-activated kinases in Erbb2-positive breast cancer: A new
therapeutic target? Small GTPases. 1:124–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moasser MM: The oncogene HER2: Its
signaling and transforming functions and its role in human cancer
pathogenesis. Oncogene. 26:6469–6487. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han HS, Son SM, Yun J, Jo YN and Lee OJ:
MicroRNA-29a suppresses the growth, migration, and invasion of lung
adenocarcinoma cells by targeting carcinoembryonic antigen-related
cell adhesion molecule 6. FEBS Lett. 588:3744–3750. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun S, Sun P, Wang C and Sun T:
Downregulation of microRNA-155 accelerates cell growth and invasion
by targeting c-myc in human gastric carcinoma cells. Oncol Rep.
32:951–956. 2014.PubMed/NCBI
|
|
29
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|