Introduction
MicroRNAs (miRs) are small (20–25 nucleotides)
endogenous regulatory non-coding RNAs present in eukaryotes.
Previous studies have revealed an important role for miRNAs in
oncogenesis and tumor metabolism (1–3).
Expression of specific miRNAs (including miR-15a, miR-16-1,
miR-155, miR-17-92, miR-0372, miR-373 and Let-7) has been
demonstrated in various tumor tissues, and ~50% of these miRNAs are
positioned in tumor-related fragile sites of the genome (1–4).
Therefore, miRNAs are crucial in the occurrence and development of
tumors.
Various tumor-associated specific molecular markers,
such as miRs, are important in the early diagnosis of cancer and
other diseases (5). miR expression
in cancer tissues is dysregulated (2,3,6), and
their expression pattern exhibits a certain degree of tissue
specificity (7,8). miR-205 is a multi-functional gene
located in the 1q32.2 locus of the human genome, and is involved in
various physiological and pathological processes, including
tumorigenesis, inflammation and immunity (8). miR-205 is abnormally expressed in a
variety of malignant tumors and its expression is closely related
to the incidence and development of tumors, such as head and neck
cancer, ovarian cancer and breast cancer (8–10).
Previously, Lebanony et al (11) and the authors (12) both reported that miR-205 expression
is significantly increased in non-small cell lung cancer (NSCLC)
tissues, and that it is associated with tumor differentiation grade
(12). However, the function of
miR-205-5p in lung cancer remains poorly understood.
In the present study, the aim was to explore the
effects of miR-205-5p on proliferation, apoptosis, and invasion of
A549 lung cancer cells. The results of the present study may
provide important information about the role of miR-205-5p in the
biological functions of lung cancer cells and might provide new
potential targets for lung cancer treatment.
Materials and methods
Cell culture and transfection
A549 human lung carcinoma cells were cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific,
Inc.), 100 IU/ml penicillin, and 1 µg/ml streptomycin in 5%
CO2 at 37°C. Cells were passaged every 2–3 days. Prior
to transfection, cells were seeded in 24-well plates at
1×105 cells/well. Cells were cultured until 90%
confluence. hsa-miR-205-5p inhibitor small interfering RNA
(5′-AATTCCAGACTCCGGTGGAATGAAGGACGATCAGACTCCGGTGGAATGAAGGAACCGGTCAGACTCCGGTGGAATGAAGGATCACCAGACTCCGGTGGAATGAAGGATTTTTTACCGG-3′;
Hibio Technologies Co., Ltd., Hangzhou, China) or miRNA inhibitor
scramble negative control (cat. no. A06001; Shanghai GenePharma
Co., Ltd., Shanghai, China) were diluted in 100 µl Opti-MEM (Thermo
Fisher Scientific, Inc.) and mixed with 1 µl Lipofectamine 2000
(Thermo Fisher Scientific, Inc.) diluted in 100 µl Opti-MEM. The
transfection solution (200 µl/well) was added to the 24-well plate.
Following 4–6 h, the solution was replaced by only RPMI-1640
medium.
Cell viability
The cells were divided into three groups: control
check group (CK; untransfected cells), negative control group (NC;
cells transfected with scramble negative control siRNA) and
hsa-miR-205-5p siRNA group (siRNA; cells transfected with
has-miR-205-5p inhibitor siRNA). At one day prior to transfection,
A549 cells, in the logarithmic phase of growth, were diluted to
1×104 cells/ml with RPMI-1640 complete medium and were
seeded into 96-cell plates (100 µl/well). The cells were cultured
at 37°C with 5% CO2 to obtain a confluence of ~30% at 24
h. The cells were then transfected as described above and cultured
for an additional 72 h. At 24, 48 and 72 h post-transfection, 10 µl
Cell Counting kit-8 solution (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was added to each well, and the culture was
continued for 1 h. Absorbance was then measured in a ELx800
spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA) at
450 nm. Cell survival rate (%) was calculated as follows: (OD of
each experimental group/OD of the CK group) ×100.
Apoptosis
Following transfection for 24, 48 and 72 h, cells
were dissociated using EDTA-free trypsin, centrifuged at 252 × g
rpm for 5 min at 4°C, and the medium was discarded. Cells were
rinsed twice with cooled PBS and suspended in 400 µl 1x binding
buffer, as per the kit's instructions (Alexa Fluor 488 Annexin
V/Dead Cell Apoptosis kit; Thermo Fisher Scientific, Inc.). Annexin
V-Alexa Fluor 488 (5 µl) and propidium iodide (1 µl) were then
added and cells were analyzed using a BD FACSVerse flow cytometer
(BD Biosciences) and BD FACSuite software (version 1.0.0.1477; BD
Biosciences, Franklin Lakes, NJ, USA).
Cell invasion assay
Matrigel was liquefied at 4°C and diluted with
cooled serum-free RPMI-1640 medium. Then, 100 µl diluted gel was
added to the upper chamber of a 24-well Transwell plate, and
incubated at 37°C overnight in order to solidify. Following
incubation, the gel was gently washed with serum-free RPMI-1640
medium. Cells were dissociated following transfection for 24, 48
and 72 h, and diluted to 1×105 cells/ml in 1%
FBS/RPMI-1640. The cells (200 µl) were then added to the upper
chamber of the Transwell and 10% FBS/RPMI-1640 medium was added to
the lower chamber as a chemoattractant. Following 24 h, the
non-invaded cells on the upper side of the chamber filters were
removed with cotton swabs, and filters were air-dried. Finally,
filters were stained with 0.1% crystal violet. Invaded cells were
calculated as cells/field by imaging four random fields
(magnification, ×200).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
Purity was determined using the A260/A280 and A260/A230 ratios. RNA
quality with A260/A280 of 1.8–2.0 was determined adequate for
RT-qPCR. Reverse transcription was performed using 10 µl total RNA,
4 µl 5X reaction buffer, 1 µl RiboLock RNase inhibitor (Thermo
Fisher Scientific, Inc.), 2 µl 10 mM dNTP, 1 µl RevertAid reverse
transcriptase (Thermo Fisher Scientific, Inc.), and 1 µl 10 µM
primers. cDNA was stored at −70°C. RT-qPCR was performed using 12.5
µl iQ SYBR-Green Supermix (Bio-Rad Laboratories, Inc., Hercules,
CA, USA), 1 µl 10 µM primers, and 10.5 µl cDNA in a Real-time PCR
ABI Prism 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). PCR parameters were: 50°C for 3 min and 95°C for
3 min, followed by 40 cycles of 95°C for 10 sec, 65°C for 20 sec,
72°C for 15 sec, and a final step of 76°C for 5 sec. Primer
sequences were: miR-205-5p stem-loop,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCAATA-3′;
miR-205-5p (accession no. MIMAT0000069; product size 66 bp),
forward 5′-TATCCAGTGCAGGGTCCGAGGTAT-3′ and reverse
5′-CGGCGGTAGCAGCACGTAAATAT-3′; RNU6B (internal reference), forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. For
erb-B2 receptor kinase 3 (erbB3), zinc finger E-box binding
homeobox 2 (ZEB2), clathrin heavy chain (CLTC) and mediator complex
subunit 1 (MED1), the following primers were used: GAPDH (internal
reference; Genbank ID 2597; product size 258 bp), forward
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse 5′-AGGGGCCATCCACAGTCTTC-3′;
erbB-3 (Genbank ID 2065; product size 225 bp), forward
5′-TGCTGAGAACCAATACCAGACA-3′ and reverse
5′-GCAAACTTCCCATCGTAGACC-3′; ZEB2 (Genbank ID 9839; product size
283 bp), forward 5′-CGTACTCGCAGCACATGAATC-3′ and reverse
5′-TCCTCCTCGAACTCCTCGTC-3′; CLTC (Genbank ID 1213; product size 111
bp), forward 5′-TCCAGAACCTGGGTATCAACC-3′ and reverse
5′-TTACCACCTGGGCCTGCTC-3′; and MED1 (Genbank ID 5469; product size
123 bp), forward 5′-GTGGCTCTTCCATGTCATCCT-3′ and reverse
5′-TGGTGACAACCCCATGCTTC-3′.
Prediction of hsa-miR-205-5p target
genes
The hsa-miR-205-5 sequences were acquired from
miRBase (http://www.mirbase.org; hsa-miR-205-5p;
MIMAT0000 266) (13). The
hsa-miR-205-5p target genes were predicted using three network
platforms: TargetScan (http://www.targetscan.org/vert_61/) (14), 416 loci of conservative target
sequences; PicTar (http://pictar.mdc-berlin.de/) (15), 274 loci of conservative target
sequences; and miRDB (http://mirdb.org/miRDB/) (16), 421 loci of conservative target
sequences.
Using miRDB and TargetScan2, there were 157 common
sequences. Using TargetScan2 and PicTar, there were 70 common
sequences. Using TargetScan2, PicTar, and miRDB, there were 33
common sequences. The predicted target genes were analyzed using
the Gene Ontology Consortium (17–18)
and Mas 3.0 Molecule Annotation System (http://bioinfo.capitalbio.com/mas3/) (18). Finally, 16 genes were analyzed, and
the miRDB and TargetScan2 common sequences were considered as
references.
Western blot analysis
Cells were dissociated with trypsin and centrifuged
at 600 × g at 4°C for 5 min. Cells were resuspended in PBS and
centrifuged a second time. RIPA lysis buffer (Applygen
Technologies, Inc., Beijing, China) was then added. Following
lysis, samples were centrifuged at 16, 099 × g at 4°C for 10 min.
The supernatant was transferred to a new centrifuge tube and stored
at −70°C. Protein concentration was determined using the
bicinchoninic acid assay method (Applygen Technologies, Inc.).
Protein samples (20 µg) were diluted with sample buffer and
denatured at 95°C for 5 min. Proteins were separated using 10%
SDS-PAGE at 25 mA for 30 min and then at 30 mA for 2 h. Proteins
were transferred to a polyvinylidene fluoride membrane at 200 mA
overnight. Membranes were blocked with 5% non-fat milk or bovine
serum albumin at room temperature overnight. The membranes were
incubated at room temperature for 2 h with the following primary
antibodies: TRAP220/MED1 (1:1,000 dilution; cat. no. ab64695;
Abcam, Cambridge, MA, USA), Smad Interacting Protein 1 (1:2,000
dilution; cat. no. ab25837; Abcam), HER3/ErbB3 (1:1,000 dilution;
cat. no. 4754S; Cell Signaling Technology, Inc., Danvers, MA, USA),
clathrin heavy chain (P1663) antibody (1:2,000 dilution; 2410S;
Cell Signaling Technology, Inc.) and β-actin (1:1,000 dilution;
cat. no. SC-47778; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). The membranes were rinsed thrice for 15 min each time.
Membranes were incubated with the secondary antibody (goat
anti-rabbit immunoglobulin G-horseradish peroxidase; 1:5,000
dilution; cat. no. BS13278; Bioworld Technology Inc., Louis Park,
MN, USA) for 45 min at room temperature and rinsed thrice.
Membranes were developed using an enhanced chemiluminescence kit
(Pierce; Thermo Fisher Scientific, Inc.). The relative band
intensity was acquired using the Quantity One software version 4.4
(Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS
software (version, 12.0; SPSS. Inc., Chicago, IL, USA). Data were
expressed as mean ± standard deviation and analyzed using analysis
of variance with Tukey's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-205-5P siRNA silencing in A549
cells
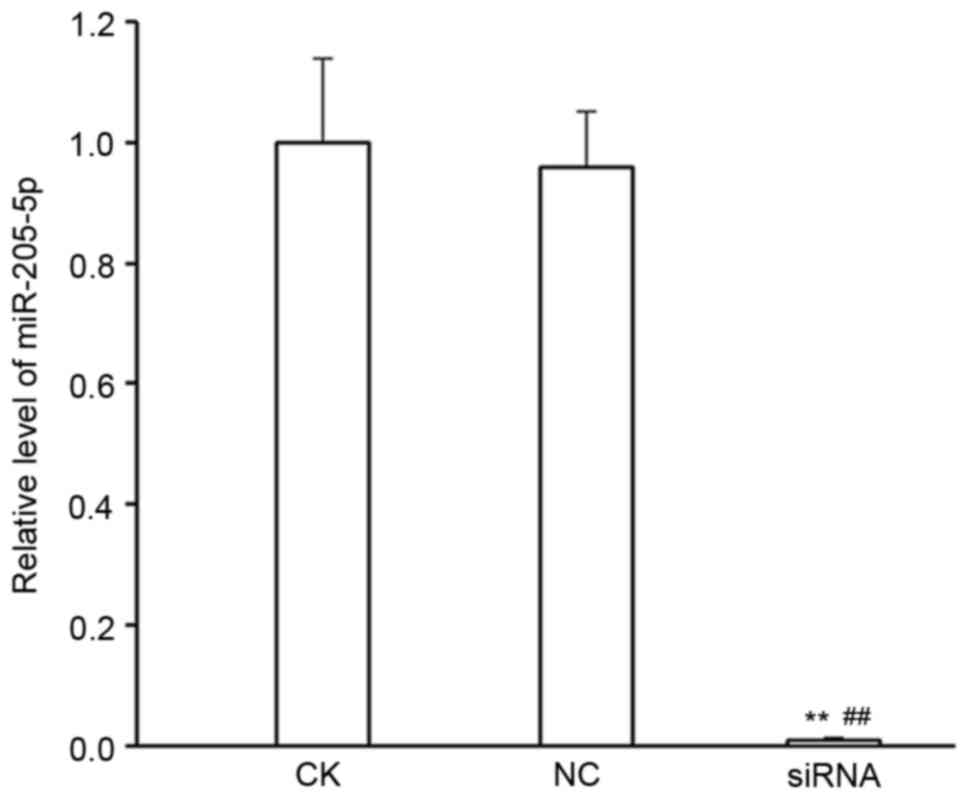
Firstly, the efficiency of silencing miR-205-5p
expression by siRNA transfection was validated. There were no
differences in miR-205-5p expression levels between the CK and NC
groups (P>0.05), while miR-205-5p expression levels were
significantly reduced in the miR-205-5p siRNA group compared with
the CK and NC group (P<0.01; Fig.
1).
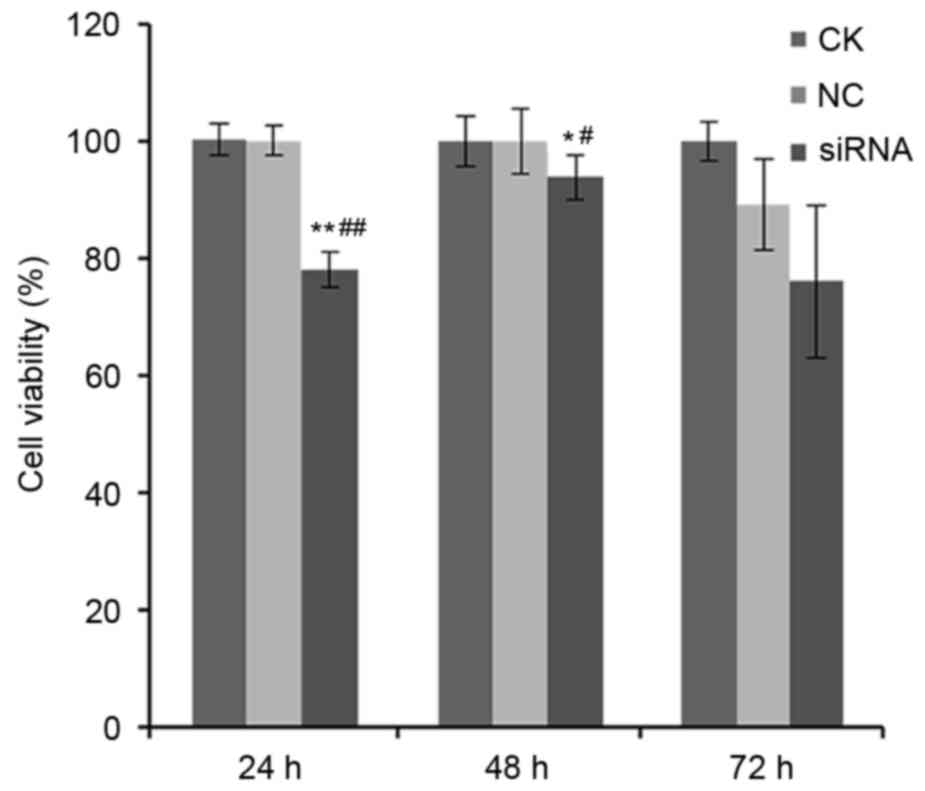
Effect of miR-205-5p silencing on cell
viability
Next, the effect of silencing miR-205-5p expression
on cell viability was assessed in A549 cells. The relative survival
rates at 24, 48 and 72 h post-siRNA transfection in the NC group
were 100.0±2.6, 100.0±5.6 and 89.2±7.7%, respectively, while those
in the miR-205-5p siRNA group were 78.1±3.0, 93.9±3.8 and
76.0±13.0%, respectively (P<0.01 for 24 h and P<0.05 for 48
h; Fig. 2). However, there were no
significant differences between the NC and CK groups, suggesting
that the negative control scramble siRNA had no effect on cell
viability (P>0.05; Fig. 2).
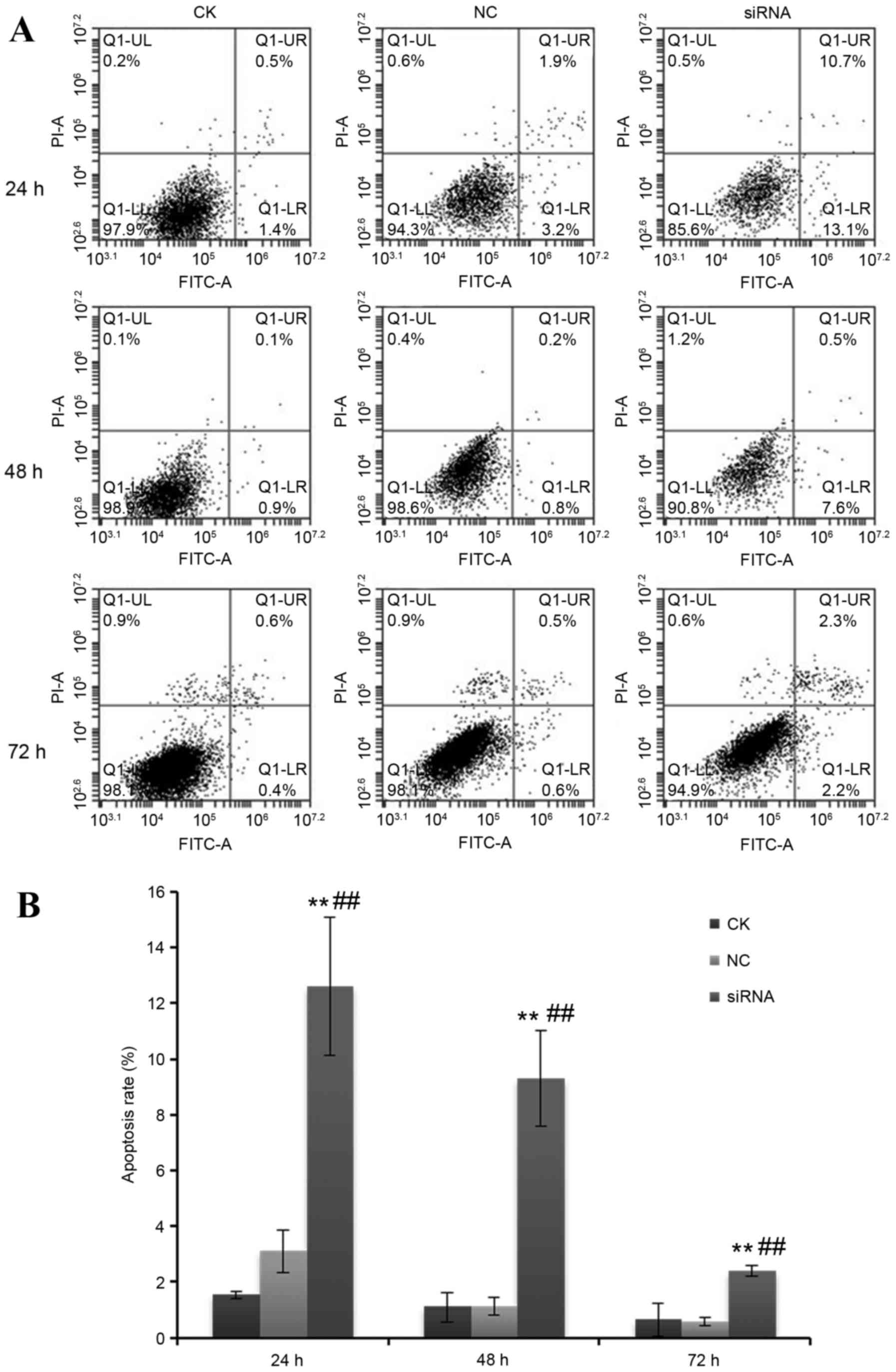
Effect of miR-2015-5p silencing on
apoptosis
The effect of silencing miR-205-5p expression was
then examined on cell apoptosis in A549 cells. At 24, 48 and 72 h
following transfection with miR-205-5p siRNA, the % of apoptotic
cells in the total cell population in the miR-205-5p siRNA group
were 12.6±2.5, 9.3±1.7 and 2.4±0.2%, respectively, while in the NC
group, these were 3.1±0.8, 1.1±0.3, and 0.6±0.2%, respectively
(P<0.01; Fig. 3).
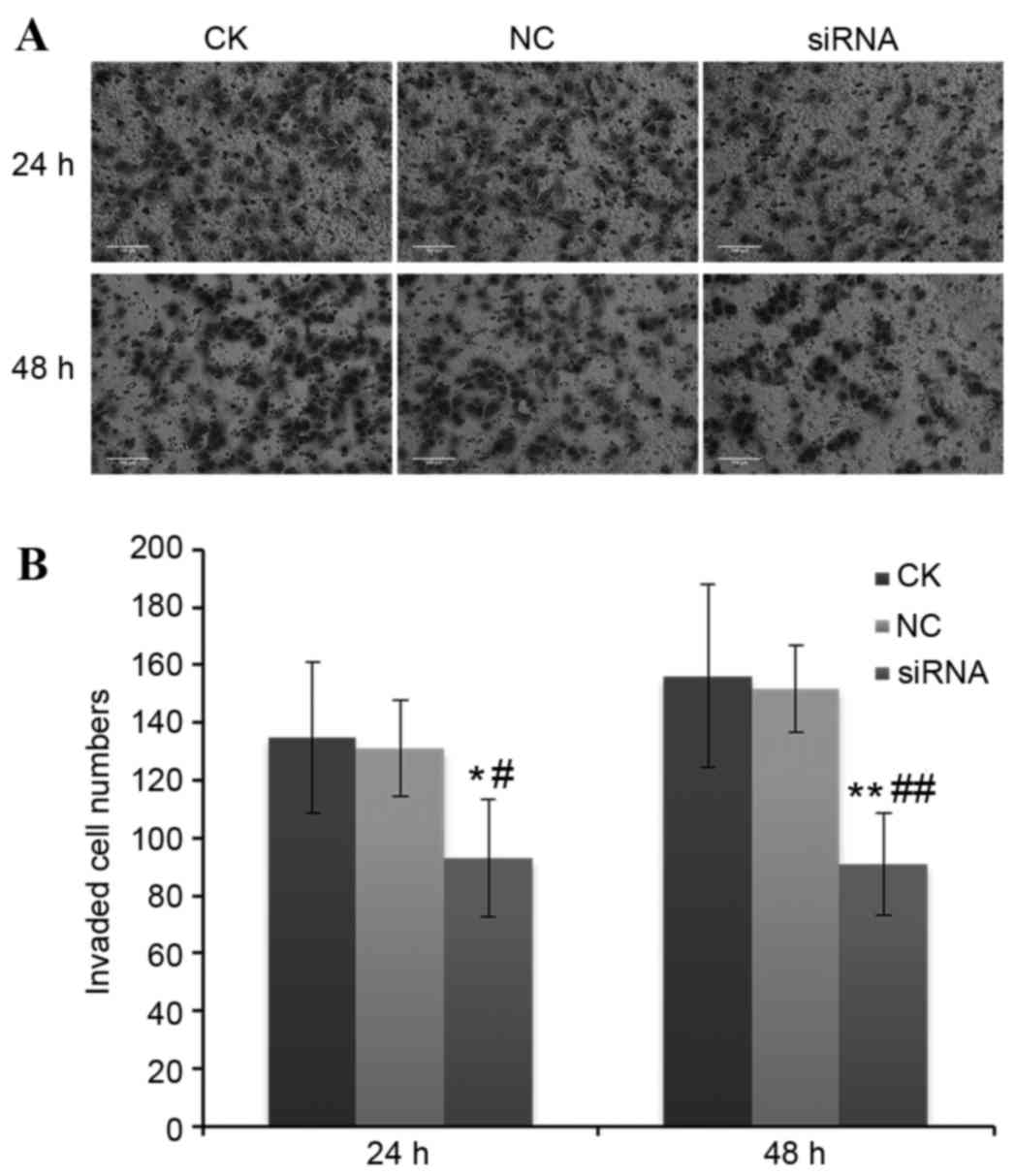
Effect of miR-2015-5p silencing on
cell invasion
The effect of silencing miR-205-5p expression on
cell invasion of A549 cells was examined. The average number of
invaded cells per field at 24 and 48 h following transfection with
miR-205-5p siRNA were 93±20 and 91±18 in miR-205-5p siRNA group,
respectively, while those in the NC group were 131±17 and 135±26,
respectively (P<0.05; Fig.
4).
Prediction of miR-205-5p target genes
by bioinformatics analysis
In order to explore the pathways involved in the
effects of miR-205-5p, the potential target genes of miR-205-5p
were predicted by bioinformatics analysis, using miRDB, TargetScan
and PicTar web-based software (data not shown). Bioinformatics
analysis revealed 16 candidate miR-205-5p target genes (Table I). Based on to the annotated
functions for these genes, erbB3 (involved in cell proliferation),
ZEB2 (promoter of metastasis), CLTC (promoter of tumor growth and
angiogenesis) and MED1 (involved in proliferation, differentiation
and metabolism) were speculated to be the major potential target
genes of miR-205-5p.
 | Table I.miR-205-5p candidate target
genes. |
Table I.
miR-205-5p candidate target
genes.
| Target rank | Target score | miRNA name | Gene symbol | Gene
description |
|---|
| 260 | 59 | hsa-miR-205-5p | SBF2 | SET binding factor
2 |
| 17 | 90 | hsa-miR-205-5p | RAB11FIP1 | RAB11 family
interacting protein 1 (class I) |
| 70 | 79 | hsa-miR-205-5p | ERBB3 | Erb-b2 receptor
tyrosine kinase 3 |
| 28 | 86 | hsa-miR-205-5p | PLCB1 | Phospholipase C,
beta 1 |
| 404 | 51 | hsa-miR-205-5p | PHC2 | Polyhomeotic
homolog 2 |
| 4 | 98 | hsa-miR-205-5p | MED1 | Mediator complex
subunit 1 |
| 37 | 84 | hsa-miR-205-5p | LRP1 | Low density
lipoprotein receptor-related protein 1 |
| 278 | 58 | hsa-miR-205-5p | LAMC1 | Laminin subunit
gamma 1 |
| 26 | 86 | hsa-miR-205-5p | KLF12 | Kruppel-like factor
12 |
| 19 | 89 | hsa-miR-205-5p | ZEB2 | Zinc finger E-box
binding homeobox 2 |
| 169 | 66 | hsa-miR-205-5p | HS3ST1 | Heparan
sulfate-glucosamine 3-O-sulfotransferase 1 |
| 202 | 64 | hsa-miR-205-5p | FRK | Fyn-related
kinase |
| 87 | 75 | hsa-miR-205-5p | CLTC | Clathrin heavy
chain |
| 64 | 79 | hsa-miR-205-5p | BTBD3 | BTB domain
containing 3 |
| 307 | 56 | hsa-miR-205-5p | ADAMTS9 | ADAM
metallopeptidase with thrombospondin type 1 motif 9 |
| 21 | 88 | hsa-miR-205-5p | ACSL1 | Acyl-CoA synthetase
long-chain family member 1 |
Effect of miR-205-5p silencing on mRNA
expression of erbB3, ZEB2, CLTC, and MED1 in A549 cells
In order to confirm that the predicted target genes
were indeed regulated by miR-205-5p, the mRNA expression levels of
erbB3, ZEB2, CTLC and MED1 were assessed in A549 cells using
RT-qPCR following miR-205-5p silencing. The results demonstrated
that erbB3 mRNA expression in NC and miR-205-5p siRNA groups was
0.594±0.185 and 0.831±0.269, respectively (P>0.05; Fig. 5). ZEB2 mRNA expression in the NC
and siRNA groups was 0.616±0.046 and 1.337±0.372, respectively
(P<0.05; Fig. 5). CLTC mRNA
expression in the NC and siRNA groups was 0.421±0.117 and
0.470±0.105, respectively (P>0.05; Fig. 5). MED1 mRNA expression in the NC
and siRNA groups was 0.461±0.052 and 0.524±0.298, respectively
(P>0.05; Fig. 5). Therefore,
out of the four genes tested, only ZEB2 exhibited a significant
increase in mRNA expression following miR-205-5p silencing
(Fig. 5).
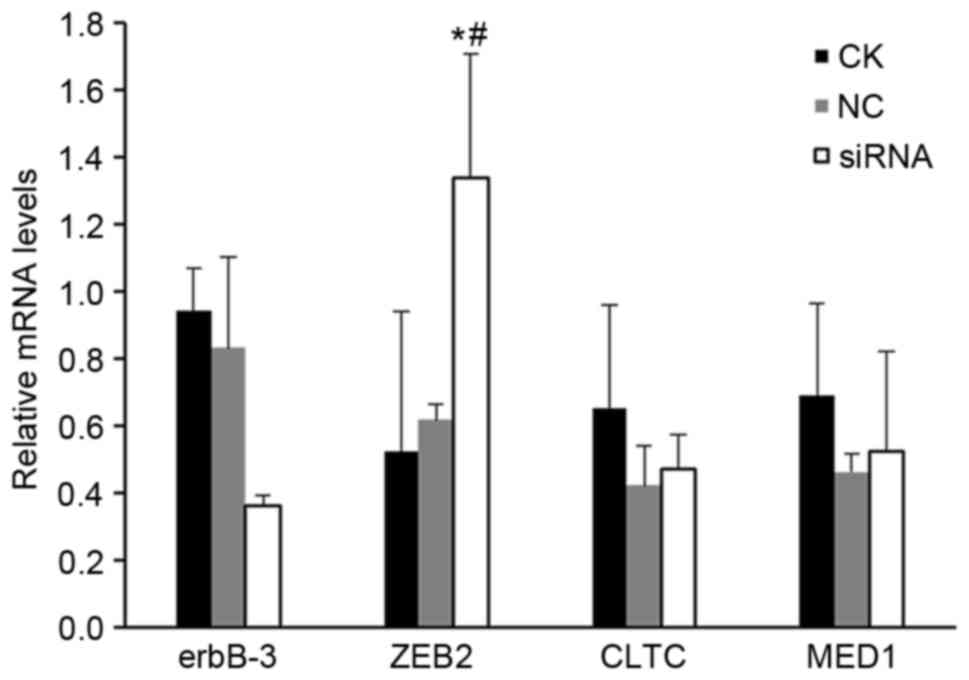 | Figure 5.Effect of miR-205-5p silencing on
mRNA expression of predicted target genes. The mRNA expression
levels of predicted target genes erbB3, ZEB2, CLTC and MED1 were
assessed in A549 cells following transfection with hsa-miR-205-5p
siRNA by reverse transcription-quantitative polymerase chain
reaction. Each experiment was performed 3 times. *P<0.05 vs. CK
group; #P<0.05 vs. NC group. erbB3, Erb-B2 receptor
kinase 3; ZEB2, zinc finger E-box binding homeobox 2; CLTC,
clathrin heavy chain; MED1, mediator complex subunit 1; siRNA,
small interfering RNA; CK, control check group; NC, scramble
negative control group; siRNA, miR-205-5p siRNA group. |
Effect of miR-205-5p silencing on
protein expression of erbB3, ZEB2, CLTC and MED1 in A549 cells
In order to confirm the results obtained for the
mRNA expression levels, protein expression levels were also
examined by western blotting (Fig.
6). The results demonstrated that erbB3 protein expression in
the NC and siRNA groups was 0.283±0.042 and 0.130±0.009,
respectively (P<0.01; Fig. 6).
ZEB2 protein expression in the NC and siRNA groups was 0.335±0.025
and 0.465±0.031, respectively (P<0.01; Fig. 6). CLTC and MED1 protein expressions
in the NC and siRNA groups were 0.905±0.090 and 0.914±0.052, and
1.029±0.025 and 0.913±0.198, respectively (P>0.05; Fig. 6). Therefore, miR-205-5p silencing
resulted in a significant decrease of erbB3 and a significant
increase of ZEB2 protein expression levels compared with control
cells (Fig. 6).
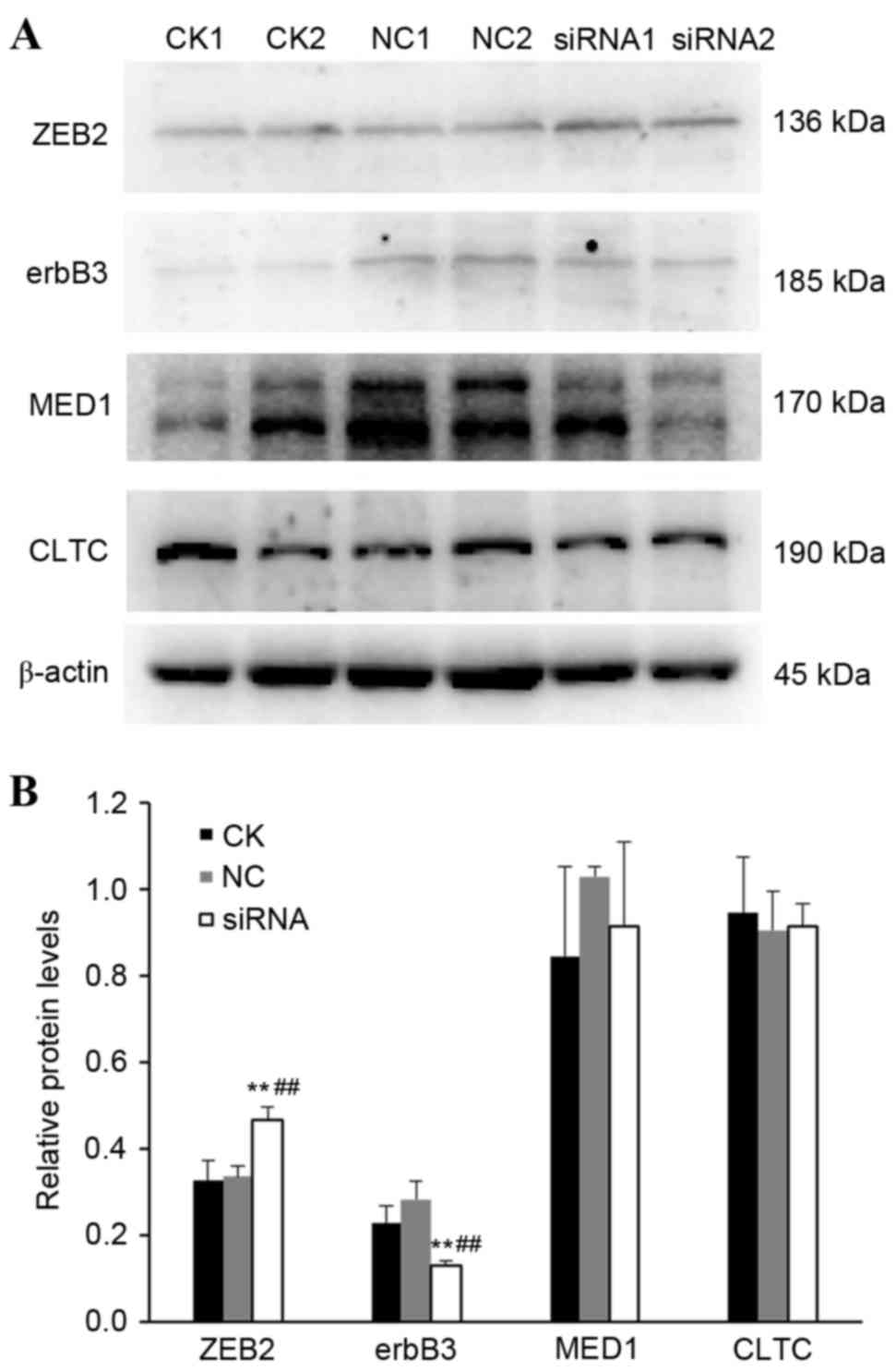 | Figure 6.Effect of miR-205-5p silencing on
protein expression of predicted target genes. The protein
expression levels of target genes erbB3, ZEB2, CLTC and MED1 were
assessed in A549 cells following transfection with hsa-miR-205-5p
siRNA, by western blotting. (A) Representative immunoblotting
results from a duplicate experiment. (B) Densitometric analysis and
quantification from three independent experiments. Each experiment
was performed 3 times. **P<0.01 vs. CK group;
##P<0.01 vs. NC group. erbB3, Erb-B2 receptor kinase
3; ZEB2, zinc finger E-box binding homeobox 2; CLTC, clathrin heavy
chain; MED1, mediator complex subunit 1; siRNA, small interfering
RNA; CK, control check group; NC, scramble negative control group;
siRNA, miR-205-5p siRNA group. |
Discussion
The aim of present study was to explore the effects
of miR-205-5p on viability, apoptosis and invasion of lung cancer
A549 cells. Results demonstrated that cell viability and cell
invasion were significantly decreased, while apoptosis was
significantly increased in miR-205-5p siRNA group compared with the
control group. Bioinformatics analysis speculated that erbB3, ZEB2,
CLTC and MED1 may be potential target genes of miR-205-5p. Reduced
expression of miR-205-5p significantly increased the expression
levels of ZEB2 mRNA and protein, while it decreased the expression
of erbB3 protein, suggesting that ZEB2 and erbB3 are indeed target
genes of miR-205-5p in A549 cells. No significant effect was
observed on the expression of CLTC and MED1 following miR-205-5p
silencing.
Proliferation, abnormal apoptosis, invasion and
metastasis are important features of malignant tumors, and
metastasis is the primary cause of death in cancer patients
(19,20). In the present study, miR205-5p
silencing led to decreased A549 cell invasion, suggesting that
knocking down expression of miR-205-5p in A549 cells may reduce the
invasive properties of tumor cells. Similar results were previously
observed in breast cancer cells following miR-205-5p silencing
(21). TargetScan is a web-based
bioinformatics tool used to predict target genes of miRNAs
(14). PicTar is a bioinformatics
tool mainly focused on identifying target genes containing single
miRNA binding sites (15,22,23).
Target gene predictions in nematodes using the PicTar software have
been confirmed to correspond to several verified miRNA target genes
(23). miRDB is a software used
for prediction and functional annotation of miRNA targets, which
performs target predictions for mature miRNA using a support vector
machine algorithm (16,24). Since each prediction software has
its limitations, and since there is a lack of high complementarity
between miRNAs and their target mRNAs, a miRNA is often associated
with hundreds of target genes, leading to a number of false
positive predictions (25).
Therefore, a cross prediction of miRNA target genes using several
prediction programs is likely to greatly improve accuracy. In the
present study, prediction of miR-205-5p target genes was performed
using three different prediction bioinformatics tools: TargetScan2,
PicTar and miRDB, and 16 miR-205-5p candidate target genes were
identified (SBF2, RAB11FIP1, erbB3, PLCB1, PHC2, MED1, LRP1, LAMC1,
KLF12, ZEB2, HS3ST1, FRK, CLTC, BTBD3, ADAMTS9 and ACSL1; Table I). Among these, four genes were
selected to be examined further in A549 cells: MED1 as a verified
target of miR-205-5p in human trophoblasts (26), erbB3 as a major regulator of cell
proliferation (27), ZEB2 as a
promoter of metastasis (28) and
CLTC as a promoter of tumor growth and angiogenesis (29). RT-qPCR and western blot analyses
were used to determine the expression of these four genes at the
mRNA and protein level respectively, following transfection with
miR-205-5p-targeting siRNA. The results demonstrated that ZEB2 and
erbB3 expression were significantly affected by miR-205-5p
silencing.
ERBB family members are involved in cell
proliferation and differentiation. Members of the ERBB family are
dysregulated in breast, lung, ovarian, prostate and
gastrointestinal tumors (30). The
present study demonstrated that, although silencing miR-205-5p
expression in A549 cells did not significantly change the mRNA
expression of erbB3, expression of the erbB3 protein levels were
significantly reduced compared with control cells. This result
indicated that high miR-205-5p expression in NSCLC might enhance
the activation of downstream signal pathways through increasing
erbB3 expression to promote cell proliferation and malignant
transformation.
Epithelial mesenchymal transition (EMT) serves an
important role in invasion and metastasis of tumor cells (31,32).
E-cadherin maintains the epithelial cell integrity and epithelial
cell polarity, and its downregulation is an important feature of
EMT. Neural cadherin is another member of the cadherin family, and
its upregulation leads to downregulation of E-cadherin and
increased cell invasion (32,33).
ZEB2 inhibits E-cadherin expression and induces EMT (34). ZEB2 has been confirmed to promote
ovarian cancer, breast cancer and gastric cancer cell metastasis,
and its upregulation is significantly correlated with poor
prognosis in head and neck cancer (35–38).
However, there is little information concerning the role of ZEB2 in
lung cancer. Oztas et al (39) reported that ZEB2 is not expressed
in normal lung tissue, but it is upregulated in up to 70% of lung
cancer tissues. Miura et al (34) observed ZEB2 overexpression in up to
58.4% of lung cancer tissues. Previous studies have demonstrated
that ZEB2 upregulation is negatively correlated with E-cadherin
expression and positively correlated with N-cadherin expression in
NSCLC, suggesting that ZEB2 may promote EMT in NSCLC (37,38,40).
The present study demonstrated that silencing miR-205-5p expression
in A549 cells significantly increased mRNA and protein expression
of ZEB2, suggesting that miR-205-5p likely regulates expression of
ZEB2 through degrading its mRNA. In addition, this result indicated
that high miR-205-5p expression may lead to decreased mRNA and
protein expression of ZEB2, as previously reported by Liu et
al (41) in melanoma
cells.
CLTC is involved in the cell cycle and spread of
viruses (42,43). CLTC promotes tumor growth and
angiogenesis by regulating hypoxia-inducible factor 1-α and
vascular endothelial growth factor expression (29). CLTC may be associated with lymphoma
(44). In the present study,
bioinformatics analysis revealed that CLTC may be a potential
target gene of miR-205-5p. However, the results did not support
that CLTC is a target of miR-205-5p, as its expression was not
affected by miR-205-5p silencing. MED1 binds to various nuclear
receptors (including peroxisome proliferator-activated receptor-α,
peroxisome proliferator-activated receptor-γ and retinoid X
receptor) and transcription factors (including p53, GATA and
CCAAT/enhancer binding protein β), serving an important regulatory
role in proliferation, differentiation and metabolism of cells
(45–47). In the present study, bioinformatics
analysis predicted that MED1 may be a potential target gene of
miR-205-5p, but miR-205-5p silencing did not alter MED1 mRNA or
protein expression levels in A549 cells.
In summary, the present study demonstrated that
reduced expression of miR-205-5p promoted apoptosis, and inhibited
viability and invasion in lung cancer A549 cells through
upregulation of ZEB2 and downregulation of erbB3. The present
results suggested that increased miR-205-5p expression in NSCLC
tissues may promote proliferation and invasion of lung cancer cells
and thus disease progression. Further studies are necessary in
order to determine the exact role of miR-205-5p in lung cancer and
to assess whether it may be a useful target for the development of
novel NSCLC treatments.
Acknowledgements
The study was supported by the Natural Science
Foundation of Jiangxi Province (grant no. 20142BAB205012) and the
555 project of Jiangxi Province Gan Po Excellence and Post-Graduate
Innovation Project of Nanchang University (grant nos. cx2015176 and
cx2016355).
References
|
1
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu XL, Wen SY, Ai ZH, Wang J, Xu YL and
Teng YC: Screening for characteristic microRNAs between
pre-invasive and invasive stages of cervical cancer. Mol Med Rep.
12:55–62. 2015.PubMed/NCBI
|
|
5
|
Hui A, How C, Ito E and Liu FF: Micro-RNAs
as diagnostic or prognostic markers in human epithelial
malignancies. BMC Cancer. 11:5002011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HY, Han SS, Rhee H, Park JH, Lee JS,
Oh YM, Choi SS, Shin SH and Kim WJ: Differential expression of
microRNAs and their target genes in non-small-cell lung cancer. Mol
Med Rep. 11:2034–2040. 2015.PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu H and Mo YY: Targeting miR-205 in
breast cancer. Expert Opin Ther Targets. 13:1439–1448. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tran N, McLean T, Zhang X, Zhao CJ,
Thomson JM, O'Brien C and Rose B: MicroRNA expression profiles in
head and neck cancer cell lines. Biochem Biophys Res Commun.
358:12–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lebanony D, Benjamin H, Gilad S, Ezagouri
M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, et
al: Diagnostic assay based on hsa-miR-205 expression distinguishes
squamous from nonsquamous non-small-cell lung carcinoma. J Clin
Oncol. 27:2030–2037. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang M, Zhang P, Hu G, Xiao Z, Xu F,
Zhong T, Huang F, Kuang H and Zhang W: Relative expressions of
miR-205-5p, miR-205-3p, and miR-21 in tissues and serum of
non-small cell lung cancer patients. Mol Cell Biochem. 383:67–75.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: MicroRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:(Database issue).
D140–D144. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:(Database issue). D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:(Database issue).
D1049–D1056. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geiger TR and Peeper DS: Metastasis
mechanisms. Biochim Biophys Acta. 1796:293–308. 2009.PubMed/NCBI
|
|
20
|
Hedley BD and Chambers AF: Tumor dormancy
and metastasis. Adv Cancer Res. 102:67–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elgamal OA, Park JK, Gusev Y,
Azevedo-Pouly AC, Jiang J, Roopra A and Schmittgen TD: Tumor
suppressive function of mir-205 in breast cancer is linked to HMGB3
regulation. PLoS One. 8:e764022013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sahoo S and Albrecht AA: Ranking of
microRNA target prediction scores by Pareto front analysis. Comput
Biol Chem. 34:284–292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hofacker IL: How microRNAs choose their
targets. Nat Genet. 39:1191–1192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mouillet JF, Chu T, Nelson DM, Mishima T
and Sadovsky Y: MiR-205 silences MED1 in hypoxic primary human
trophoblasts. FASEB J. 24:2030–2039. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jullien N, Dieudonné FX, Habel N, Marty C,
Modrowski D, Patino A, Lecanda F, Sévère N and Marie PJ: ErbB3
silencing reduces osteosarcoma cell proliferation and tumor growth
in vivo. Gene. 521:55–61. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dai YH, Tang YP, Zhu HY, Lv L, Chu Y, Zhou
YQ and Huo JR: ZEB2 promotes the metastasis of gastric cancer and
modulates epithelial mesenchymal transition of gastric cancer
cells. Dig Dis Sci. 57:1253–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tung KH, Lin CW, Kuo CC, Li LT, Kuo YH,
Lin CW and Wu HC: CHC promotes tumor growth and angiogenesis
through regulation of HIF-1α and VEGF signaling. Cancer Lett.
331:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yano T, Doi T, Ohtsu A, Boku N, Hashizume
K, Nakanishi M and Ochiai A: Comparison of HER2 gene amplification
assessed by fluorescence in situ hybridization and HER2 protein
expression assessed by immunohistochemistry in gastric cancer.
Oncol Rep. 15:65–71. 2006.PubMed/NCBI
|
|
31
|
Chang ZG, Wei JM, Qin CF, Hao K, Tian XD,
Xie K, Xie XH and Yang YM: Suppression of the epidermal growth
factor receptor inhibits epithelial-mesenchymal transition in human
pancreatic cancer PANC-1 cells. Dig Dis Sci. 57:1181–1189. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rhodes LV, Tate CR, Segar HC, Burks HE,
Phamduy TB, Hoang V, Elliott S, Gilliam D, Pounder FN, Anbalagan M,
et al: Suppression of triple-negative breast cancer metastasis by
pan-DAC inhibitor panobinostat via inhibition of ZEB family of EMT
master regulators. Breast Cancer Res Treat. 145:593–604. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gheldof A and Berx G: Cadherins and
epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci.
116:317–336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miura N, Yano T, Shoji F, Kawano D,
Takenaka T, Ito K, Morodomi Y, Yoshino I and Maehara Y:
Clinicopathological significance of Sip1-associated epithelial
mesenchymal transition in non-small cell lung cancer progression.
Anticancer Res. 29:4099–4106. 2009.PubMed/NCBI
|
|
35
|
Elloul S, Elstrand MB, Nesland JM, Tropé
CG, Kvalheim G, Goldberg I, Reich R and Davidson B: Snail, Slug,
and Smad-interacting protein 1 as novel parameters of disease
aggressiveness in metastatic ovarian and breast carcinoma. Cancer.
103:1631–1643. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rosivatz E, Becker I, Specht K, Fricke E,
Luber B, Busch R, Höfler H and Becker K: Differential expression of
the epithelial-mesenchymal transition regulators snail, SIP1, and
twist in gastric cancer. Am J Pathol. 161:1881–1891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou C, Liu J, Tang Y, Zhu G, Zheng M,
Jiang J, Yang J and Liang X: Coexpression of hypoxia-inducible
factor-2α, TWIST2, and SIP1 may correlate with invasion and
metastasis of salivary adenoid cystic carcinoma. J Oral Pathol Med.
41:424–431. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sakamoto K, Imanishi Y, Tomita T, Shimoda
M, Kameyama K, Shibata K, Sakai N, Ozawa H, Shigetomi S, Fujii R,
et al: Overexpression of SIP1 and downregulation of E-cadherin
predict delayed neck metastasis in stage I/II oral tongue squamous
cell carcinoma after partial glossectomy. Ann Surg Oncol.
19:612–619. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Oztas E, Avci ME, Ozcan A, Sayan AE,
Tulchinsky E and Yagci T: Novel monoclonal antibodies detect
Smad-interacting protein 1 (SIP1) in the cytoplasm of human cells
from multiple tumor tissue arrays. Exp Mol Pathol. 89:182–189.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xia M, Hu M, Wang J, Xu Y, Chen X, Ma Y
and Su L: Identification of the role of Smad interacting protein 1
(SIP1) in glioma. J Neurooncol. 97:225–232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu S, Tetzlaff MT, Liu A, Liegl-Atzwanger
B, Guo J and Xu X: Loss of microRNA-205 expression is associated
with melanoma progression. Lab Invest. 92:1084–1096. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dodge GR, Kovalszky I, McBride OW, Yi HF,
Chu ML, Saitta B, Stokes DG and Iozzo RV: Human clathrin heavy
chain (CLTC): Partial molecular cloning, expression, and mapping of
the gene to human chromosome 17q11-qter. Genomics. 11:174–178.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Humphries AC, Dodding MP, Barry DJ,
Collinson LM, Durkin CH and Way M: Clathrin potentiates
vaccinia-induced actin polymerization to facilitate viral spread.
Cell Host Microbe. 12:346–359. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
De Paepe P, Baens M, van Krieken H,
Verhasselt B, Stul M, Simons A, Poppe B, Laureys G, Brons P,
Vandenberghe P, et al: ALK activation by the CLTC-ALK fusion is a
recurrent event in large B-cell lymphoma. Blood. 102:2638–2641.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhu Y, Qi C, Jain S, Rao MS and Reddy JK:
Isolation and characterization of PBP, a protein that interacts
with peroxisome proliferator-activated receptor. J Biol Chem.
272:25500–25506. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Drané P, Barel M, Balbo M and Frade R:
Identification of RB18A, a 205 kDa new p53 regulatory protein which
shares antigenic and functional properties with p53. Oncogene.
15:3013–3024. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Crawford SE, Qi C, Misra P, Stellmach V,
Rao MS, Engel JD, Zhu Y and Reddy JK: Defects of the heart, eye,
and megakaryocytes in peroxisome proliferator activator
receptor-binding protein (PBP) null embryos implicate GATA family
of transcription factors. J Biol Chem. 277:3585–3592. 2002.
View Article : Google Scholar : PubMed/NCBI
|