Introduction
Gastric cancer is the second most common malignancy
in the world; however, the outcome of early gastric cancer (EGC)
has markedly improved (1). An
increasing number of patients with EGC receive endoscopic treatment
and benefit from an improvement in their prognosis; consequently,
laparotomy is avoided and an improved quality of life is maintained
in several patients suffering from EGC (2). Endoscopic resection as the standard
therapy for EGC is widely accepted and regularly used
worldwide.
Previously, Ono et al (3) developed an innovational technique,
endoscopic submucosal dissection (ESD), which has become a
well-accepted technique for the treatment of EGC, and pathological
examination provided by the ESD method enables accurate diagnosis
(4). ESD is associated with a good
prognosis of EGC (5,6), however, multiple EGCs may occur
following curative resection, with reports demonstrating that 6–14%
of patients with EGC deteriorate (7–9).
Consequently, it is difficult to determine the optimal follow-up
strategy, which aims at identifying recurrences of EGC following
ESD, as there is no suitable biomarker to predict recurrence.
MicroRNAs (miRNAs) are a group of hairpin-derived
RNAs consisting of 20–24 nucleotides, which bind to the 3′
untranslated region (3′UTR) of messenger RNA (mRNA), resulting in
post-transcriptional repression of the expression of target genes
in disease and normal physiological contexts in a broad range of
organisms (10,11). Typically, nuclear RNase III
(Drosha) cleaves the long primary precursors (pri-miRNAs) into
pre-miRNAs of ~70 nt in length following transcription mediated by
RNA polymerase II. A stem-loop structure is present in the
resulting transcripts, which are then exported to the cytoplasm,
followed by processing by RNase III (Dicer), generating mature
double-stranded ~22 nt miRNAs (12). The miRNA-target interaction may be
altered or the maturation process of miRNA may be affected when
genetic variation occurs in the pre-miRNA sequence. Several studies
have indicated that the risk of cancer or pre-cancer may be
increased by the presence of miRNA polymorphisms (13); however, few have examined the
association between cancer susceptibility and miRNA polymorphisms.
The arm selection of miR-499a or the target selection of miRNA may
be altered directly by the rs3746444 in the seed region of the
miR-499a (14).
The present study aimed to investigate the molecular
mechanism, including the potential regulatory and signaling
pathways, of platelet-derived growth factor receptor β (PDGFRB),
which underlies the recurrence of EGC. PDGFRB was identified as a
target of miR-499a. As the expression of PDGFRB has been reported
to be associated with EGC recurrence following ESD (15), and the rs3746444 polymorphism may
alter the expression of miR-499a, it was hypothesized that the
presence of rs3746444 may be associated with the recurrence of EGC
following ESD via targeting PDGFRB.
Materials and methods
Patients and specimens
A total of 128 patients (84 males and 44 females;
mean age, 57 years; range, 26–80 years), who received ESD for EGC
in the Affiliated Hospital of Shandong Academy of Medical Sciences
(Jinan, China) between September 2013 and September 2014, were
included in the present study. The EGC was defined as
adenocarcinoma confined to the submucosa or mucosa of the stomach.
Patients who had received ESD or multiple ESDs previously were
excluded from the investigation. Peripheral blood was collected
from all 128 patients, tissue samples (~0.5 cm) were available from
32 patients for RNA extraction and western blot analysis. The
duration between the first dose of ESD and the final dose was
defined as the follow-up period. Following the first dose of ESD,
patients received follow-up ESDs up to 42 months. Patient clinical
information, including age, gender, smoking history, body mass
index, tumor location, grade of gastric atrophy, tumor
differentiation and Helicobacter pylori infection status at
the time of first dose of ESD, were obtained based on medical
records. All subjects signed written informed consent prior to
enrollment, and the study protocol was approved by the Ethics
Committee of Shandong Academy of Medical Sciences. The principles
of the declaration of Helsinki were complied with in the present
study.
Polymorphism genotyping
Genomic DNA was extracted and purified from the
whole-blood samples with a QIAamp DNA midi kit (Qiagen Sciences,
Inc., Germantown, MD, USA). The TaqMan real-time polymerase chain
reaction (PCR) method and the ABI PRISM 7500 sequence detection
system (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) were used to analyze the genotypes of miR-499a
T>C (rs3746444) in a 96-well format primer used was
5′-AAACAUCACUGCAAGUCUUAA-3′. The PCR analysis was performed using
10 µl reaction mixture of ddH2O, 0.25 µl primer/probe
mixture, 5 µl TaqMan Universal PCR Master mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and 20 ng DNA using the following
thermocycling conditions: 94°C or 10 min, followed by 40 cycles at
94°C for 20 sec and 60°C for 45 sec. In order to determine the
genotype of samples, SDS v1.3.1 software (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used to analyze the
allele-specific fluorescence data in each plate.
RNA extraction and stem-loop reverse
transcription-PCR (RT-PCR) analysis
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from the 32 tissue
samples. In brief, the tissue samples were homogenized with 1 ml
TRIzol reagent, and 0.2 ml chloroform was added to extract
proteins. Isopropanol (0.5 ml) was added to precipitate the RNA. A
Nanodrop 1000 spectrophotometer (Nanodrop Technologies, Inc.;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA) was used to
determine the quantity, purity and concentration of total RNA. A
reaction mixture comprising a stem-loop RT, SuperScript III Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.), and RT
primers for miR-499a and PDGFRB mRNA were used for reverse
transcription of the total RNA. The following primers were used:
β-actin forward, 5′-GTCATTCCAAATATGAGATGCGT-3′ and reverse,
5′-GCATTACATAATTTACACGAAAGCA-3′, RNU6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The reaction conditions were as follows: 16°C for 30 min, followed
by 30 sec at 20°C, 30 sec at 42°C, 1 sec at 50°C for 50 cycles.
Finally, the reaction mixture was incubated at 85°C for 5 min to
inactivate the enzyme. A SYBR Green I assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using the following thermocycling
conditions: 95°C for 30 sec, 95°C at 3 sec for 40 cycles, 60°C for
15 sec to detect gene expression. The mRNA expression level of
PDGFRB and the expression level of miR-499a were normalized to that
of U6 (2−ΔΔCq) (16).
Cell culture and oligonucleotide
transfection
MKN-45 cells were obtained from American Type
Culture Collection (Manassas, VA, USA). The cells were cultured in
RPMI-1640 supplemented with streptomycin and penicillin, and FBS
(Gibco; Thermo Fisher Scientific, Inc.). The miR-499a mimics and
inhibitors were synthesized by Sangon Biotech Co., Ltd. (Shanghai,
China), and Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to perform the transfection, cells were
seeded at density of 8×106 cells/well at room
temperature for 10–15 min. The cells were collected for the
proliferation assay 24 h following transfection, and at 48 h
post-transfection, the cells were harvested for the analyses of
protein, RNA and cell viability. MiRanda (www.microrna.org) and TargetScan (www.targetscan.org) were used in the present
study.
Luciferase reporter assay
The 3′UTR luciferase assay was performed using a
Psicheck™-2 Dual-Luciferase miRNA target expression vector (Promega
Corporation, Madison, WI, USA). PCR was performed to amplify the
3′UTR sequence of the PDGFRB gene. The cDNA was then amplified by
qPCR using a SYBR green qPCR kit (Takara Bio, Inc., Tokyo, Japan)
in a 25 µl with 12.5 µl of 2xSYBR Fast qPCR Mix (Takara Bio, Inc.),
2 µl cDNA, 1 µl reverse primer (10 µm), 1 µl of forward primer (10
µm) and 8.5 µl water without nuclease. The amplification was
carried out at 98°C for 60 sec, and followed by 30 cycles (98°C for
30 sec, 58°C for 30 sec, 72°C for 120 sec) and 72°C for 5 min. The
product was cloned into a Psicheck™-2 vector to generate a
wild-type reporter. A Site-directed gene mutagenesis kit (Beyotime
Institute of Biotechnology, Shanghai, China) was used to produce
the mutant reporter, according to the manufacturer's protocol. For
the luciferase assay, the Psicheck™-2 Dual-Luciferase miRNA target
expression vector containing the mutant or wild-type target
sequences were used to co-transfect the miR-499a-overexpressing
MKN-45 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, the cells were
harvested and lyzed in passive lysis buffer (Promega Corporation).
The firefly luciferase activity was detected in a Steady Glo
Luciferase Assay system (Promega Corporation) with renilla activity
as a control.
Cell viability assays
Following transfection, the cells were incubated in
96-well plates at a density of 1×104 MKN-45 cells per
well. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium
bromide was added into each well at a final concentration of 20 µl
of 5 mg/ml to a final volume of 250 µl, and the cells were then
incubated at 37°C for 4 h. DMSO was added to dissolve the remaining
crystals following removal of the culture medium. The absorbance at
570 nm was measured using a microplate reader.
Immunoblot analysis
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was used to extract total proteins from the
transfected MKN-45 cells or tissue samples according to the
manufacturer's protocol. A bicinchoninic acid protein assay was
used to quantify whole cell protein extract. An equivalent volume
(30 µg per lane) of cell lysate was separated in a 10% SDS-PAGE gel
and then transferred onto a polyvinylidene fluoride membrane. The
membranes were blocked in TBST supplemented with 5% (w/v) skimmed
milk at 4°C for 1 h, following which primary antibodies were
incubated at 4°C for 12 h using PDGFRB (cat. no. sc-12910; 1:500;
Santa Cruz Biotechnology, Inc., Dallas, Texas) and β actin (cat.
no. sc-47778; 1:1,500; Santa Cruz Biotechnology, Inc.). The
enhanced chemiluminescence reagent (EMD Millipore, Billerica, USA)
was used to visualize the protein bands following incubation at
room temperature for 2 h with horseradish peroxidase-conjugated
secondary antibody (cat. no. sc-516086; 1:10,000; Santa Cruz
Biotechnology, Inc.). The signals on the blots were detected using
an ECL kit (Amersham ECL detection system; GE Healthcare Life
Sciences, Chalfont, UK), and the relative density of the target
bands were analyzed using Image J software (National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
The χ2-test was used to analyze the differences
between discrete variables and one-way analysis of variance was
used to analyze differences between continuous variables in each
group. The Kaplan-Meier method was used to plot recurrence-free
curves, which were compared using the log rank test. Hazard ratios
were calculated using the univariate and multivariate Cox
proportional hazard model. P<0.05 was considered to indicate a
statistically significant difference. Data are expressed as the
mean ± standard deviation. SPSS 18 (SPSS, Inc., Chicago, IL, USA)
was used to perform all statistical analyses.
Results
PDGFRB is a target of miR-499
PDGFRB has been previously reported to be a growth
factor receptor, the expression level of which affects the
recurrence of EGC following ESD treatment (17). The present study aimed to
investigate the molecular mechanism, including the potential
regulatory and signaling pathways, of PDGFRB, which underlies the
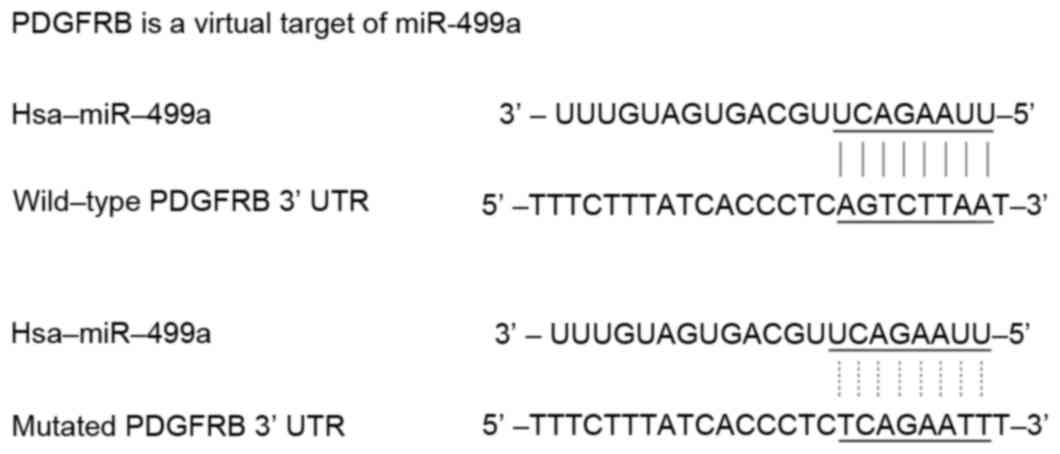
recurrence of EGC following ESD treatment. As shown in Fig. 1, online miRNA target prediction
tools were used to identify the regulatory miRNA of PDGFRB, and
consequently identified PDGFRB as the candidate target gene of
miR-499a in gastric cancer cells with the ‘seed sequence’ in the
3′UTR of PDGFRB.
Furthermore, to validate the regulatory association
between miR-449a and PDGFRB, the present study performed a
luciferase activity reporter assay in gastric cancer cells
co-transfected with wild-type PDGFRB 3′UTR constructs and different
concentrations of miR-499a mimics (25, 50 and 100 nM). To verify
PDGFRB as the direct target gene of miR-499a, another system
transfected with mutant PDGFRB 3′UTR constructs and miR-499a mimics
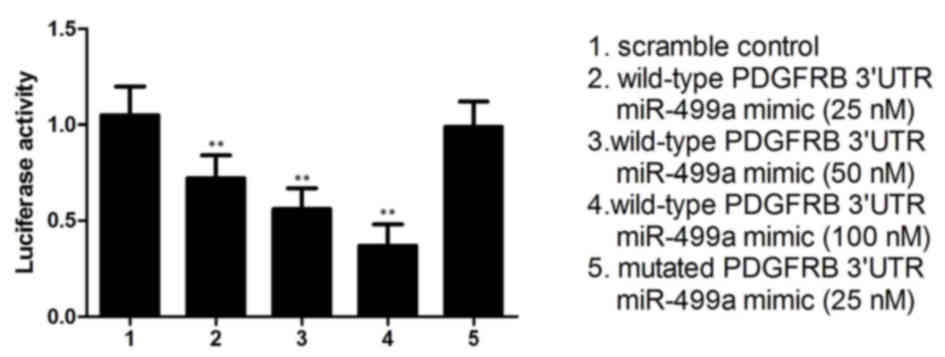
(100 nM) was examined. As shown in Fig. 2, compared with the scramble
control, the relative luciferase activity of the cells transfected
with the wild-type PDGFRB 3′UTR constructs decreased as the
concentration of miR-499a mimics increased, exhibiting a negative
regulation in a step-wise manner. By contrast, cells carrying
mutant PDGFRB 3′UTR constructs exhibited a comparable relative
luciferase activity index, compared with the scramble controls,
indicating PDGFRB as the direct target gene of miR-499a, with the
binding site located in the mutated segment.
Expression levels of miR-499a and
PDGFRB vary in different genotype groups of the rs3746444
polymorphism
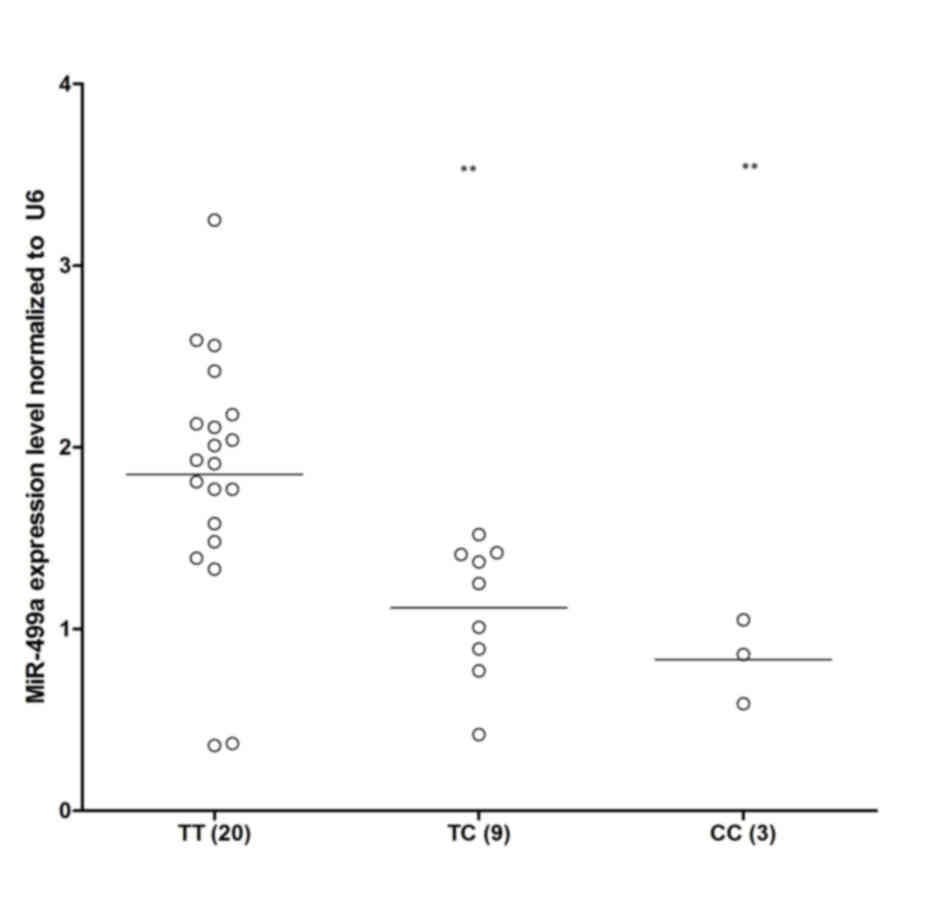
The rs3746444 polymorphism has previously been
reported to interfere with the expression of miR-499, leading to
different expression levels of miR-499a (18). Among the EGC samples collected, as
shown in Fig. 3, TT (n=20) showed
a markedly higher expression level of miR-499a, compared with TC
(n=9) and CC (n=3), indicating that the presence of minor allele
(C) of the rs3746444 polymorphism compromised the expression of
miR-499a.
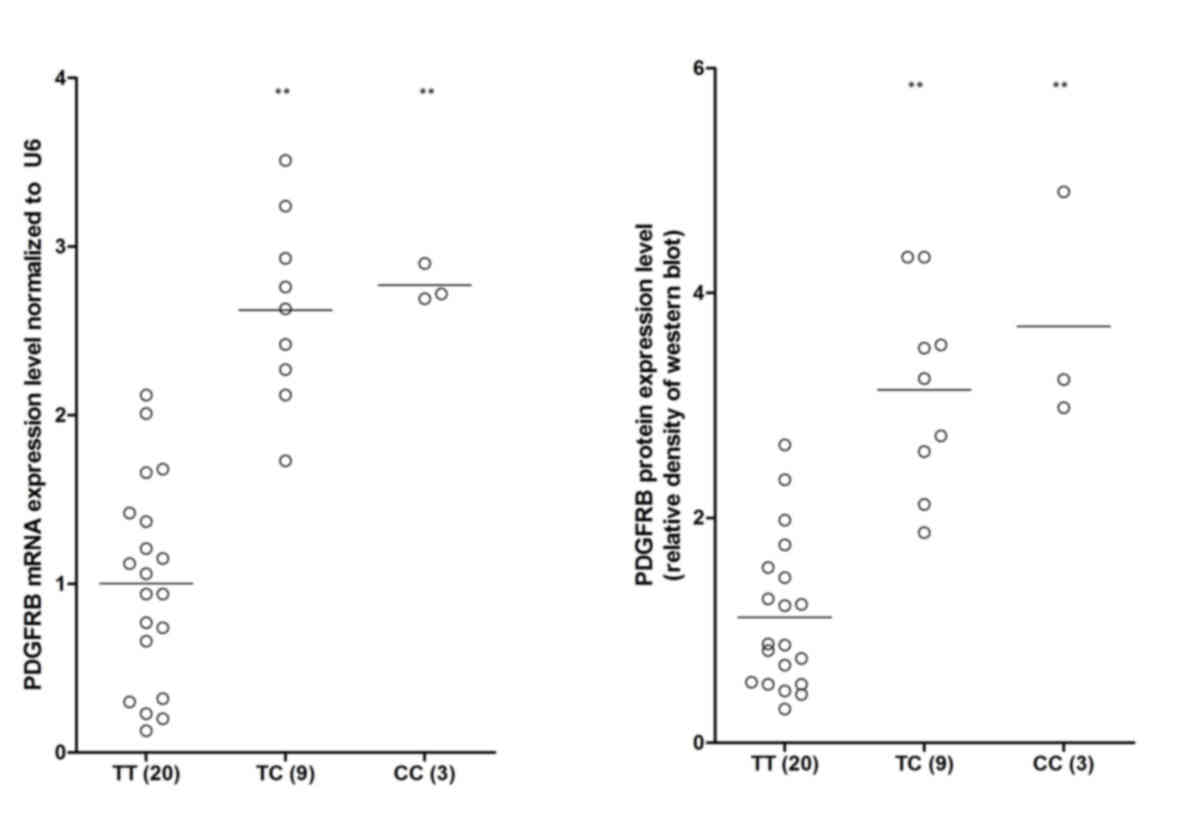
In the present study, RT-qPCR and western blot
analyses were also performed to investigate the mRNA and protein
expression levels of PFGRBR among the different genotypes. As shown
in Fig. 4, the mRNA and protein
expression levels of PFGRBR in the TT sample group were markedly
lower, compared with those in the minor allele TC and CC sample
groups, indicating the negative regulatory association between
miR-499a and PDGFRB.
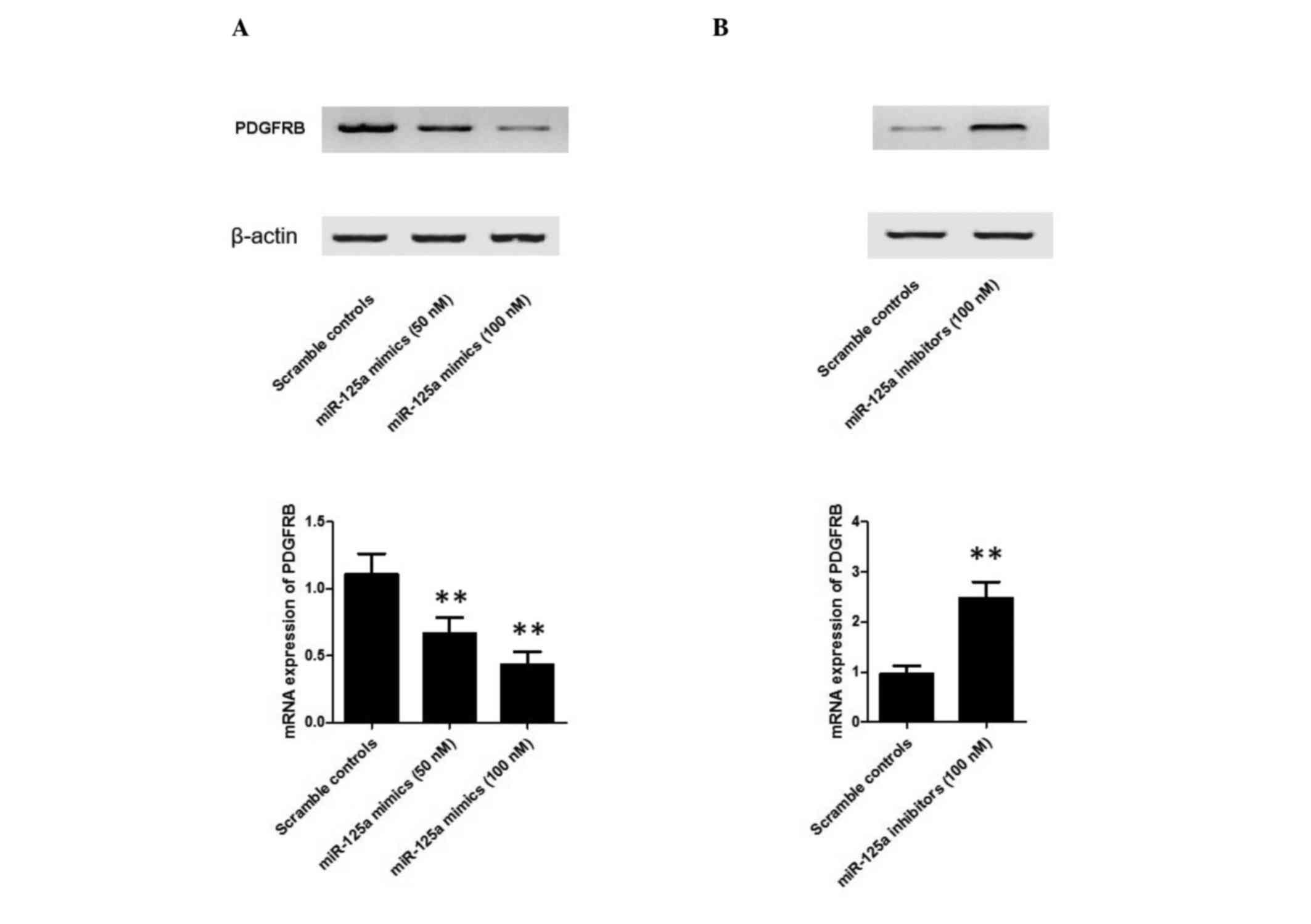
miR-499a inhibits the expression of
PDGFRB
To further confirm the hypothesis that a negative
regulatory association exists between miR-499a and PDGFRB, the
present study investigated the mRNA and protein expression levels
of PDGFRB in EGC cells treated with 50 nM miR-499a mimics, 100 nM
miR-499a mimics and 100 nM miR-499a inhibitors. As shown in
Fig. 5, the protein (upper panel)
and mRNA (lower panel) expression levels of PDGFRB in EGC cells
treated with 50 nM miR-499a mimics appeared lower, compared with
those in the scramble control, whereas those in the sample group
treated with 100 nM miR-499a mimics were lower, compared with those
in the 50 nM treatment group (Fig.
5A). This confirmed the effect of different miR-499a
concentrations on the interaction between miR-499a and PDGFRB. In
addition, the miR-499a inhibitor treatment group showed markedly
higher protein (upper panel) and mRNA (lower panel) expression
levels of PDGFRB, compared with the scramble controls and the
miR-499a mimic treatment groups (Fig.
5B), confirming the negative regulatory association between
miR-499a and PDGFRB.
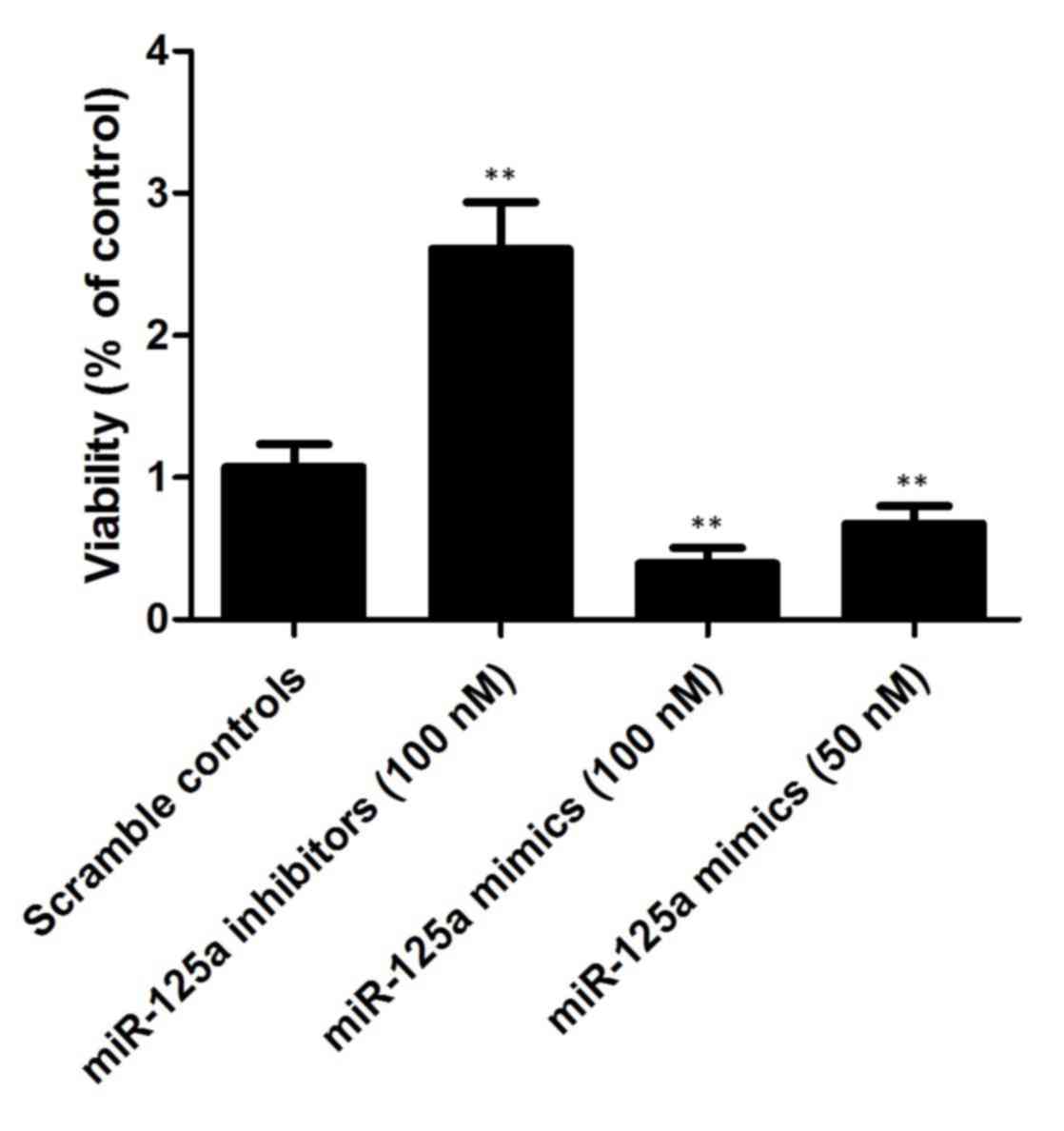
miR-499a interferes with the viability
of EGC cells
As shown in Fig. 6,
the present study also investigated the relative viability of EGC
cells when transfected with miR-499a mimics (50 and 100 nM) and
miR-499a inhibitors (100 nM). The cells transfected with 100 nM
miR-499a inhibitors showed increased viability, compared with the
scramble controls, whereas the cells transfected with 50/100 nM
miR-499a mimics showed comparably lower viability, indicating that
miR-499a negatively interfered with the viability of the EGC
cells.
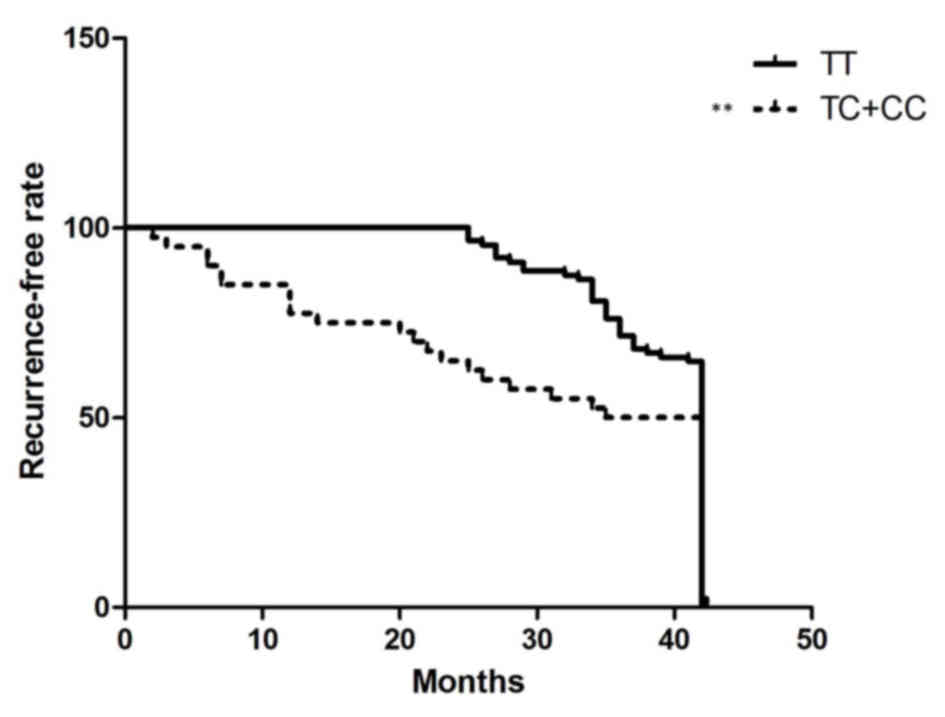
miR-499a rs3746444 polymorphism as a
biomarker to predict recurrence following ESD in patients with
EGC
To investigate the association between the rs3746444
polymorphism and the risk of recurrence in patients with EGC who
have received ESD treatment, the present study enrolled 100
patients with EGC to observe the time period between ESD treatment
and EGC recurrence using Cox proportional hazard model analysis. As
shown in Fig. 7, when the
recurrence rate was analyzed using the log rank test, the
recurrence-free duration was significantly longer in the TT
individuals, compared with the TC/CC individuals, indicating that
the miR-499a rs3746444 polymorphisms may function as a biomarker to
predict recurrence following ESD in patients with EGC.
Discussion
For patients with EGC, endoscopic treatment has been
used as curative therapy, and the prognoses of patients with EGC
has been improved by endoscopic tumor resection (19–21).
Although ESD provides accurate histological diagnosis, it also
enables the endoscopic curative resection of EGCs, however, it has
strict criteria for small, well-differentiated adenocarcinomas with
minimal submucosal invasion (22,23).
Only gastric cancer tissues are excised using endoscopic treatment,
indicating that gastric carcinoma may recur in the patients
following ESD. Therefore, a molecular diagnosis, which enables the
prediction of patients at high risk for recurrence of EGC is
urgently required (24). It has
been demonstrated that the severity of gastric atrophy may be
predictive in multiple types of gastric cancer in previous reports
(25,26). Although the development of EGC has
previously been delineated by infection with H. pylori by
comparing H. pylori-positive patients and H.
pylori-naive-negative patients, the risk of EGC recurrence was
only reduced to one-third when H. pylori was eradicated
(27).
In the present study, it was found that the miR-499a
rs3746444 polymorphism may function as a biomarker to predict
recurrence following ESD in patients with EGC via analyzing the
recurrence-free rate of patients with EGC with different
genotypes.
Several investigations have examined the function of
single nucleotide polymorphisms (SNPs) located in mature or
precursor miRNAs, and how they affect progression and
susceptibility in different human diseases. SNPs, which are
associated with miRNA can have indirect or direct effects. SNP
miRNA promoters may have indirect effects on transcription, and
SNPs in mRNA may destroy or create target sites (28). SNPs may also have direct effects on
mature miRNA, pre-miRNA or pri-miRNA, which may enhance or impair
the function or processing of miRNA (28). It has been previous reported that
the rs3746444 polymorphism, situated in the seed region of
pri-miR-499a, may interfere with arm selection during the
production and processing of mature miRNA, and reduce the
expression of the miRNA (14).
This effect of the polymorphism in its host miRNA may lead to
altered risks of certain human diseases via the downstream
signaling pathway or mediators. In the present study, the negative
regulatory association between miR-499a and PDGFRB was established
via examining the relative luciferase activity in the presence of
different concentrations of miR-499a mimics. Furthermore, as the
rs3746444 polymorphism has previously been reported to interfere
with the expression of miR-499a, the present study investigated the
expression level of different genotypes, including TT (n=20), TC
(n=9) and CC (n=3), which supported the hypothesis that the
presence of minor allele (C) of the rs3746444 polymorphism
compromised the expression of miR-499a. The present study also
performed RT-qPCR and western blot analyses to investigate the mRNA
and protein expression levels of PFGRBR among different genotypes
or between cells treated with different concentrations of miR-499a
mimics/inhibitors. The results indicated the negative regulatory
association between miR-499a and PDGFRB.
PDGFR and its ligand, PDGFm are involved in
carcinogenesis and tumor development. Guo et al (15) found that the overexpression of
PDGF-B and PDGFR-β correlates with cancer progression and the
lymphogenous metastasis of gastric carcinoma (15). The overexpression of PDGFR-α has
been observed in metastatic medulloblastoma patient samples,
compared with non-metastatic patient samples, in which the
disruption of PDGFR-α function inhibited the metastatic potential
of medulloblastoma cells in vitro (29). Mathey et al (30) also found that PDGFR-β may be
important as an anti-angiogenic agent and has now become a
component of the standard treatment of ovarian cancer. Furthermore,
the expression levels of PDGFR are associated with the
angiogenesis, invasion and metastasis of colon cancer (31–33).
A previous study showed significantly increased mRNA levels of
PDGFR-β in locally advanced rectal tumors, compared with the
corresponding normal mucosa (34).
PDGFR is considered to provide a favorable microenvironment for the
growth and survival of cancer cells (35,36).
In a previous study, Gialeli et al (37) found that the PDGF/PDGFR axis is of
paramount importance in the tumor microenvironment, and the
inhibition of PDGF receptor activation represents a major target
for future anticancer therapies. Therefore, the present study
hypothesized that attenuating the expression of PDGFR may inhibit
the growth, invasion and metastasis of tumors. The present study
investigated the relative viabilities of GC cells when transfected
with miR-499a mimics (50 and 100 nM) and miR-499a inhibitors (100
nM), and found that cells transfected with 100 nM miR-499a
inhibitors showed upregulated viability, compared with the scramble
control group, whereas cells transfected with 50/100 nM miR-499a
mimics showed comparably lower viabilities, indicating that
miR-499a negatively interfered with the viability of the EGC
cells.
Taken together, the present study demonstrated that
patients carrying at least one minor allele of the rs3746444
polymorphism had a higher recurrence rate, compared with those with
wild-type following ESD. The rs3746444 polymorphism, which may
downregulate the expression of miR-499a and thereby upregulate the
expression of PDGFRB in primary EGC, may become a useful biomarker
for predicting patients at high risk of recurrence following
ESD.
References
|
1
|
Correa P: International cancer
epidemiology meetings. Cancer Epidemiol Biomarkers Prev. 1:245–247.
1992.PubMed/NCBI
|
|
2
|
Nagano H, Ohyama S, Fukunaga T, Seto Y,
Fujisaki J, Yamaguchi T, Yamamoto N, Kato Y and Yamaguchi A:
Indications for gastrectomy after incomplete EMR for early gastric
cancer. Gastric Cancer. 8:149–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ono H, Kondo H, Gotoda T, Shirao K,
Yamaguchi H, Saito D, Hosokawa K, Shimoda T and Yoshida S:
Endoscopic mucosal resection for treatment of early gastric cancer.
Gut. 48:225–229. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Toyonaga T, Nishino E, Hirooka T, Ueda C
and Noda K: Intraoperative bleeding in endoscopic submucosal
dissection in the stomach and strategy for prevention and
treatment. Digestive Endoscopy. 18:(Suppl). S123–S127. 2006.
View Article : Google Scholar
|
|
5
|
Imaeda H, Iwao Y, Ogata H, Ichikawa H,
Mori M, Hosoe N, Masaoka T, Nakashita M, Suzuki H, Inoue N, et al:
A new technique for endoscopic submucosal dissection for early
gastric cancer using an external grasping forceps. Endoscopy.
38:1007–1010. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gotoda T: Endoscopic resection of early
gastric cancer. Gastric Cancer. 10:1–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang JQ, Sridhar S, Chen Y and Hunt RH:
Meta-analysis of the relationship between Helicobacter
pylori seropositivity and gastric cancer. Gastroenterology.
114:1169–1179. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parsonnet J, Friedman GD, Vandersteen DP,
Chang Y, Vogelman JH, Orentreich N and Sibley RK:
Helicobacter-pylori infection and the risk of
gastric-carcinoma. N Engl J Med. 325:1127–1131. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nomura A, Stemmermann GN, Chyou PH, Kato
I, Perezperez GI and Blaser MJ: Helicobacter-pylori
infection and gastric-carcinoma among Japanese-Americans in Hawaii.
N Engl J Med. 325:1132–1136. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: Approaches and considerations. Nat Rev Genet.
13:358–369. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang JS and Lai EC: Alternative miRNA
biogenesis pathways and the interpretation of core miRNA pathway
mutants. Mol Cell. 43:892–903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:398–402. 2010. View Article : Google Scholar
|
|
14
|
Hou YY, Lee JH, Chen HC, Yang CM, Huang
SJ, Liou HH, Chi CC, Tsai KW and Ger LP: The association between
miR-499a polymorphism and oral squamous cell carcinoma progression.
Oral Dis. 21:195–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Y, Yin J, Zha L and Wang Z:
Clinicopathological significance of platelet-derived growth factor
B, platelet-derived growth factor receptor-β and E-cadherin
expression in gastric carcinoma. Contemp Oncol (Pozn). 17:150–155.
2013.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sachinidis A, Skach RA, Seul C, Ko Y,
Hescheler J, Ahn HY and Fingerle J: Inhibition of the PDGF
beta-receptor tyrosine phosphorylation and its downstream
intracellular signal transduction pathway in rat and human vascular
smooth muscle cells by different catechins. FASEB J. 16:893–895.
2002.PubMed/NCBI
|
|
18
|
Liu J, Xie B, Chen S, Jiang F and Meng W:
Association study of two inflammation-related polymorphisms with
susceptibility to hepatocellular carcinoma: A meta-analysis. BMC
Med Genet. 15:922014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amano Y, Ishihara S, Amano K, Hirakawa K,
Adachi K, Fukuda R, Watanabe M, Fukumoto S, Fujishiro H and Imaoka
T: An assessment of local curability of endoscopic surgery in early
gastric cancer without satisfaction of current therapeutic
indications. Endoscopy. 30:548–552. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sano T, Kobori O and Muto T: Lymph node
metastasis from early gastric cancer: Endoscopic resection of
tumour. Br J Surg. 79:241–244. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haruma K, Sumii K, Inoue K, Teshima H and
Kajiyama G: Endoscopic therapy in patients with inoperable early
gastric cancer. Am J Gastroenterol. 85:522–526. 1990.PubMed/NCBI
|
|
22
|
Kato M: Endoscopic submucosal dissection
(ESD is being accepted as a new procedure of endoscopic treatment
of early gastric cancer. Intern Med. 44:85–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hirasaki S, Tanimizu M, Nasu J, Shinji T
and Koide N: Treatment of elderly patients with early gastric
cancer by endoscopic submucosal dissection using an insulated-tip
diathermic knife. Intern Med. 44:1033–1038. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyoshi E, Haruma K, Hiyama T, Tanaka S,
Yoshihara M, Shimamoto F and Chayama K: Microsatellite instability
is a genetic marker for the development of multiple gastric
cancers. Int J Cancer. 95:350–353. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hanaoka N, Uedo N, Shiotani A, Inoue T,
Takeuchi Y, Higashino K, Ishihara R, Iishi H, Haruma K and Tatsuta
M: Autofluorescence imaging for predicting development of
metachronous gastric cancer after Helicobacter pylori
eradication. J Gastroenterol Hepatol. 25:1844–1849. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shiotani A, Uedo N, Iishi H, Yoshiyuki Y,
Ishii M, Manabe N, Kamada T, Kusunoki H, Hata J and Haruma K:
Predictive factors for metachronous gastric cancer in high-risk
patients after successful Helicobacter pylori eradication.
Digestion. 78:113–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fukase K, Kato M, Kikuchi S, Inoue K,
Uemura N, Okamoto S, Terao S, Amagai K, Hayashi S and Asaka M:
Japan Gast Study Group: Effect of eradication of Helicobacter
pylori on incidence of metachronous gastric carcinoma after
endoscopic resection of early gastric cancer: An open-label,
randomised controlled trial. Lancet. 372:392–397. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salzman DW and Weidhaas JB: SNPing cancer
in the bud: microRNA and microRNA-target site polymorphisms as
diagnostic and prognostic biomarkers in cancer. Pharmacol Ther.
137:55–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
MacDonald TJ, Brown KM, LaFleur B,
Peterson K, Lawlor C, Chen Y, Packer RJ, Cogen P and Stephan DA:
Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK
pathway as therapeutic targets for metastatic disease. Nat Genet.
29:143–152. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mathey S, Graeser MK, Eulenburg Zu C,
Woelber L, Trillsch F, Jaenicke F, Müller V, Milde-Langosch K and
Mahner S: Platelet-derived growth factor receptor beta serum
concentrations during first-line therapy in ovarian cancer.
Oncology. 85:69–77. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kitadai Y, Sasaki T, Kuwai T, Nakamura T,
Bucana CD, Hamilton SR and Fidler IJ: Expression of activated
platelet-derived growth factor receptor in stromal cells of human
colon carcinomas is associated with metastatic potential. Int J
Cancer. 119:2567–2574. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mancuso MR, Davis R, Norberg SM, O'Brien
S, Sennino B, Nakahara T, Yao VJ, Inai T, Brooks P, Freimark B, et
al: Rapid vascular regrowth in tumors after reversal of VEGF
inhibition. J Clin Invest. 116:2610–2621. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song S, Ewald AJ, Stallcup W, Werb Z and
Bergers G: PDGFRbeta+ perivascular progenitor cells in tumours
regulate pericyte differentiation and vascular survival. Nat Cell
Biol. 7:870–879. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Erben P, Horisberger K, Muessle B, Müller
MC, Treschl A, Ernst T, Kähler G, Ströbel P, Wenz F, Kienle P, et
al: mRNA expression of platelet-derived growth factor receptor-beta
and C-KIT: Correlation with pathologic response to cetuximab-based
chemoradiotherapy in patients with rectal cancer. Int J Radiat
Oncol Biol Phys. 72:1544–1550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liotta LA and Kohn EC: The
microenvironment of the tumour-host interface. Nature. 411:375–379.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Radinsky R and Fidler IJ: Regulation of
tumor cell growth at organ-specific metastases. In Vivo. 6:325–331.
1992.PubMed/NCBI
|
|
37
|
Gialeli C, Nikitovic D, Kletsas D,
Theocharis AD, Tzanakakis GN and Karamanos NK: PDGF/PDGFR signaling
and targeting in cancer growth and progression: Focus on tumor
microenvironment and cancer-associated fibroblasts. Curr Pharm Des.
20:2843–2848. 2014. View Article : Google Scholar : PubMed/NCBI
|