Introduction
Preterm birth, defined as birth before 37 weeks of
gestation, accounts for 75% of infant mortality worldwide and is
the leading cause of the long-term morbidity, such as
neurodevelopmental impairments, respiratory and gastrointestinal
complications (1,2). Between 65 and 75% of preterm births
follow spontaneous preterm birth (SPTB), but the precise pathologic
mechanisms remain to be elucidated due to their complexity
(3). Previous studies analyzing
the recurrence of SPTB across generations and between siblings
suggested that genetic factors are likely contributors to
variations in the length of gestation with heritability estimates
in the range of 27–36% (4,5). Numerous candidate gene association
studies have explored the role of single nucleotide polymorphisms
(SNPs) in the development of SPTB and differences in the prevalence
across populations (6–11). Meanwhile, a publicly available
online database of genetic association data on preterm birth has
been established to keep track of the evolving evidence base of
genetic factors in preterm birth (12).
Prostaglandins (PGs) serve an important role in the
processes of term and preterm labor, contributing to uterine
contractility, membrane rupture and cervical ripening (13,14).
The rate-limiting step in PG biosynthesis is catalyzed by
phospholipase A2 (PLA2) enzymes, which comprise >20 members
(15,16). One member of PLA2 family, group IVC
PLA2 (PLA2G4C), has been reported to be expressed in uterus and
regulate uterine PG production by its expression and activity
(17). Previously, polymorphisms
in this gene were identified to be associated with the risk of
preterm birth in a case-control study with US Hispanic, White and
Black mothers (18). It remains
unclear whether these polymorphisms also confer SPTB susceptibility
in Asians.
In addition to PLA2G4C, it should be noted that
another member of PLA2 family, group IVD PLA2 (PLA2G4D) was
demonstrated to be uniquely expressed in the stratified squamous
epithelium of the cervix and involved in PG regulation (19). PLA2G4D encodes for a protein of ~90
kDa with significant homology with known cytosolic PLA2 proteins in
the catalytic domains and has been implicated to serve a role in
many diseases, such as schizophrenia, autism and psoriatic skin
(19–21). However, to date, no published data
have examined the association of the polymorphisms of PLA2G4D with
SPTB.
With these in mind, the authors hypothesized that
PLA2G4C and PLA2G4D may be potential candidates for SPTB
susceptibility. The associations of maternal PLA2G4C rs1366442 and
PLA2G4D rs4924618 polymorphisms were investigated with the risk of
SPTB in a case-control study with a Chinese population. Moreover,
bioinformatics tools were used to elucidate potential mechanism of
the SNP function.
Materials and methods
Subjects
The present study is a nested case-control study,
embedded in a prospective study from the Guangzhou Women and
Children's Medical Center (GWCMC) that was designed to investigate
the health consequences of genetic and environmental factors on
pregnancy outcomes and offspring. Pregnant women who attended the
routine oral glucose tolerance test were invited to participate in
the study at 24–28 weeks of gestation, if they met the following
inclusion criteria: ≥18 years old, Chinese and intend to deliver at
GWCMC. From April 2013 to March 2014, 3597 (69.0% of those
eligible) pregnant women were enrolled to this prospective study
and donated their blood samples.
Of those who gave singleton live birth, 174 women
had preterm deliveries (<37 weeks of gestation). Women with
medically indicated preterm delivery (n=53), in vitro
fertilization (n=5), fetal anomaly (n=1) or uterine malformation
(n=1) were excluded, resulting in 114 women having a spontaneous
preterm birth (42 preterm labor and 72 preterm premature rupture of
membranes) in the present study as the case group. In addition, 250
pregnant women, who delivered singleton term babies (38–41 weeks of
gestation) and had no preeclampsia, eclampsia, placental abruption,
placenta previa, haemolysis, elevated liver enzymes or low platelet
count syndrome, were selected and included as the control group.
Controls were frequency-matched to the cases by age within 3 years.
The study was approved by the Institutional Review Board of GWCMC.
Informed consent was obtained from all participants.
SNP selection
To select SNPs, the authors conducted a systematic
literature review, assessing the evidence for the associations of
PLA2G4C and PLA2G4D with SPTB. For the PLA2G4C gene, four SNPs had
been reported to influence preterm birth risk in the US Hispanic or
White populations (18). However,
three of them (rs8110925, rs2307276 and rs11564620) were excluded
due to the minor allele frequencies (MAF) reported in HapMap
(http://hapmap.ncbi.nlm.nih.gov/) were
<10% for Chinese subjects. Only rs1366442 in the intron region
with a MAF of 13% was chosen to ensure that an adequate number of
individuals within the population would be carriers of the minor
allele. For the PLA2G4D gene, published reports on polymorphisms of
this gene and diseases are rare. The authors chose rs4924618 based
on the following two reasons: i) a weak association between this
SNP and schizophrenia was observed in a Chinese population
(20), while schizophrenia was
related to a higher risk of preterm birth (22,23);
ii) this SNP is a non-synonymous SNP that is likely to alter the
protein folding structure, suggesting that this SNP may have a
functional effect.
Genotyping
Blood samples were stored at −80°C until laboratory
use. Genomic DNA was extracted from cells isolated from 2 ml of
blood samples using the Qiagen DNA Blood Mini kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's instructions.
Genotyping for PLA2G4C rs1366442 and PLA2G4D rs4924618 was
performed by using a MassARRAY system (Sequenom, San Diego, CA,
USA). A total of 20 samples (5.5% of total) were tested in
duplicates to determine the quality of genotyping, and the
concordance rates were 100%. DNA from 4 (1.1%) and 5 (1.4%) samples
failed to be genotyped for PLA2G4C rs1366442 and PLA2G4D
rs4924618.
Covariates
Information on maternal age, educational level,
smoking status, maternal height and pre-pregnancy weight, parity
and previous history of preterm delivery were collected by using
face to face questionnaire. In addition, diagnosis of gestational
diabetes mellitus (GDM), infant's gestational age, birth weight and
sex, were abstracted from medical records. Gestational age was
evaluated based on the ultrasound examination in the first or
second trimester. Pre-pregnancy body mass index (BMI;
kg/m2) was calculated as the ratio of pre-pregnancy
weight (kg) to squared height (m). Since the prevalence of previous
history of preterm delivery and smoking exposure were quite low
(≤1% in the control group), the authors did not include these
variables for further analysis.
Statistical analysis
The differences between the case and control groups
were evaluated by using the chi-squared test or Fisher's exact test
(for categorical variables) and Student's t-test (for continuous
variables). The Hardy-Weinberg equilibrium for PLA2G4C rs1366442
and PLA2G4D rs4924618 was examined by a goodness-of-fit chi-squared
test among the control group. The multivariate logistic regression
was used to investigate the relationship between genotypes and the
risk of preterm birth, adjusting for maternal age, educational
level (high school or below vs. college or above), pre-pregnancy
BMI, parity (primiparous vs. multiparous), GDM status (yes vs. no)
and infant's sex. The odds ratios (ORs) and the corresponding 95%
confidence intervals (CIs) were calculated. Stratified analysis was
performed to examine whether the effects of genotypes with the risk
of preterm birth varied across different status of maternal age
(<30 vs. ≥30), GDM status (no vs. yes) and infant's sex (female
vs. male). The multiplicative interaction between genotypes and
participants' characteristics were evaluated in the logistic
regression models.
A two-tailed P<0.05 was considered statistically
significant. All statistical analysis was performed using SAS
statistical software (version, 9.3; SAS Institute, Cary, NC,
USA).
cPLA2 structure model
The structure of cPLA2 δ catalytic domain was
constructed through homology modeling. Sequences of four cytosolic
phospholipase A2 (cPLA2) (α, β, γ and δ) were obtained from UniProt
(www.uniprot.org). To check the identity and
homology, sequences of cPLA2 catalytic domains were compared by
BLAST using Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
and ClustalW (https://www.ebi.ac.uk/Tools/msa/clustalw2/). cPLA2 δ
catalytic domain is localized between amino acids 273 and 818 and
the cPLA2 α (Protein Data Bank ID: 1CJY) was chosen as a template.
The SWISS-model workspace (https://swissmodel.expasy.org/; Swiss Institute of
Bioinformatics, Basel, Switzerland) was used to construct the
homology model of cPLA2 δ to obtain a structural insight of the
protein and to locate the SNP-related amino acid in the protein.
Manual alignment of cPLA2 δ to the template was exported in FASTA
format to the Swiss-Model Server (24). The homology model was then
validated by the Ramachandran plot plugin in VMD 1.9.2 (http://www.ks.uiuc.edu/; Theoretical and Computational
Biophysics Group, NIH Center for Macromolecular Moedling and
Bioinformatics, at the Beckman Institute, University of Illinois at
Urbana-Champaign, Champaign, IL, USA) to determine stereochemical
aspects along with main chain and side chain parameters (25).
Results
Demographic characteristics
The baseline characteristics of the studied
population, including 114 SPTB cases and 250 controls, are
presented in Table I. No
significant differences were observed in the distributions of
maternal age, educational level, history of abortion, history of
preterm delivery, pre-pregnancy BMI, smoking status and GDM status
between the cases and controls (P>0.05). As expected,
gestational age at delivery (P<0.001) and birth weight
(P<0.001) were significantly lower in the case group, when
compared with those in the control group, respectively. In
addition, cases were more likely to be multiparous (P=0.01) and
neonates from the case group had higher percentage of boys
(P=0.001).
 | Table I.Characteristics of participants. |
Table I.
Characteristics of participants.
| A, Mother |
|---|
|
|---|
| Characteristics | Cases, n (%) | Controls, n (%) | P-value |
|---|
| Age (years) |
|
<30 | 55 (48.3) | 127 (50.8) |
|
| ≥30 | 59 (51.8) | 123 (49.2) | 0.65 |
| Mean
(SD) | 29.8 (3.5) | 29.8 (3.5) | 0.97 |
| Educational
level |
|
|
|
| High
middle school or below | 12 (10.5) | 36 (14.4) | 0.31 |
| College
or above | 102 (89.5) | 214 (85.6) |
|
| Marital Status |
|
|
|
|
Unmarried | 0 (0.0) | 2 (0.8) |
>0.99# |
|
Married | 114 (100) | 248 (99.2) |
|
| Parity |
|
|
|
|
Primiparous | 92 (80.7) | 225 (90.0) | 0.01 |
|
Multiparous | 22 (19.3) | 25 (10.0) |
|
| History of
abortion |
|
|
|
|
Yes | 73 (76.8) | 149 (73.0) | 0.48 |
| No | 22 (23.2) | 55 (27.0) |
|
| No
data | 19 | 46 |
|
| Previous history of
preterm delivery |
|
|
|
| No | 94 (100.0) | 205 (99.0) |
>0.99# |
|
Yes | 0 (0.0) | 2 (1.0) |
|
| No
data | 20 | 43 |
|
| Pre-pregnancy
BMI |
|
|
|
|
<20 | 74 (64.9) | 169 (67.6) | 0.61 |
|
≥20 | 40 (35.1) | 81 (32.4) |
|
| Mean
(SD) | 20.0 (2.1) | 19.8 (2.1) | 0.53 |
| Smoking |
|
|
|
| No | 67 (100.0) | 152 (99.4) |
>0.99# |
|
Yes | 0 (0.0) | 1 (0.7) |
|
| No
data | 47 | 97 |
|
| GDM |
|
|
|
| No | 88 (77.2) | 201 (80.4) | 0.48 |
|
Yes | 26 (22.8) | 49 (19.6) |
|
|
| B, Newborn |
|
|
Characteristics | Cases, n (%) | Controls, n
(%) | P-value |
|
| Sex |
|
Female | 46 (40.4) | 145 (58.0) |
|
Male | 68 (59.7) | 105 (42.0) | 0.002 |
| Birth weight (g),
median (25th, 75th) | 2660 (2300,
2880) | 3280
(3030,3540) | <0.001 |
| Gestational age
(weeks), median (25th, 75th) | 36
(35,36) | 39 (39,40) | <0.001 |
Association of maternal PLA2G4C,
PLA2G4D polymorphism with the risk of SPTB
Genotype distributions of the two selected SNPs in
cases and controls are summarized in Table II. There was no deviation from the
Hardy-Weinberg equilibrium observed among the controls (P=0.94 for
rs1366442 and P=0.58 for rs4924618). Logistic regression analysis
was performed to assess the effects of each SNP on SPTB risk. In
the multivariate analysis, compared to the homozygous genotype GG,
the TT genotypes of PLA2G4D rs4924618 was associated with a
marginally reduced risk of SPTB (adjusted OR and 95% CI, 0.50
[0.24–1.02]), and the AT/TT (dominant model) genotypes was
associated with a reduced risk of SPTB (adjusted OR, 0.61
[0.37–0.99]). No significant association was observed between the
risk of SPTB and PLA2G4C rs1366442 polymorphism (P>0.05).
 | Table II.Associations of genetic variants with
the risk of SPTB. |
Table II.
Associations of genetic variants with
the risk of SPTB.
| Genotype | Controls, n
(%) | Cases, n (%) | OR (95%
CI)a | OR (95%
CI)b |
|---|
| PLA2G4C
rs1366442 |
|
|
|
|
| CC | 159 (64.4) | 76 (67.3) | 1.00
(reference) | 1.00
(reference) |
| CA | 80 (32.4) | 34 (30.1) | 0.89 (0.55,
1.45) | 0.90 (0.54,
1.49) |
| AA | 8 (3.2) | 3 (2.7) | 0.79 (0.2,
3.04) | 1.06 (0.26,
4.24) |
|
CA/AA | 88 (35.6) | 37 (32.7) | 0.88 (0.55,
1.41) | 0.91 (0.56,
1.49) |
| PLA2G4D
rs4924618 |
|
|
|
|
| AA | 66 (26.9) | 42 (36.8) | 1.00
(reference) | 1.00
(reference) |
| AT | 133 (54.3) | 57 (50.0) | 0.67 (0.41,
1.11) | 0.65 (0.38,
1.09) |
| TT | 46 (18.8) | 15 (13.2) | 0.51 (0.26,
1.03) | 0.50 (0.24,
1.02) |
|
AT/TT | 179 (73.1) | 72 (63.2) | 0.63 (0.39,
1.02) | 0.61 (0.37,
0.99) |
Stratified analysis
Further stratified analysis was performed to assess
whether the association between these SNPs and the risk of SPTB
would be modified with specific participant's characteristics, such
as maternal age, GDM status and neonatal sex (Table III). All associations of SNPs
were independent of maternal age, GDM status and neonatal sex (all
P>0.05). There were no significant interactions between PLA2G4D
rs4924618 and participant's characteristics on the risk of SPTB
(P>0.05).
 | Table III.Stratified analysis for associations
between PLA2G4C and PLA2G4D variants and the risk of SPTB. |
Table III.
Stratified analysis for associations
between PLA2G4C and PLA2G4D variants and the risk of SPTB.
|
| PLA2G4C
rs1366442 | PLA2G4D
rs4924618 |
|---|
|
|
|
|
|---|
| Stratified
variables | Genotype | Case/Control,
n | OR (95%
CI)a | Genotype | Case/Control,
n | OR (95%
CI)a |
|---|
| Age |
|
|
|
|
|
|
|
<30 | CC | 34/83 | 1.00
(reference) | AA | 16/35 | 1.00
(reference) |
|
| CA/AA | 20/43 | 1.13 (0.56,
2.28) | AT/TT | 39/89 | 0.83 (0.39,
1.8) |
|
≥30 | CC | 42/76 | 1.00
(reference) | AA | 26/31 | 1.00
(reference) |
|
| CA/AA | 17/45 | 0.75 (0.37,
1.51) | AT/TT | 33/90 | 0.42 (0.21,
0.83) |
| P-value for
interaction |
|
| 0.33 |
|
| 0.10 |
| GDM |
| No | CC | 60/132 | 1.00
(reference) | AA | 32/52 | 1.00
(reference) |
|
| CA/AA | 27/66 | 1.02 (0.58,
1.79) | AT/TT | 56/144 | 0.59 (0.34,
1.04) |
|
Yes | CC | 16/27 | 1.00
(reference) | AA | 10/14 | 1.00
(reference) |
|
| CA/AA | 10/22 | 0.50 (0.17,
1.52) | AT/TT | 16/35 | 0.68 (0.22,
2.08) |
| P-value for
interaction |
|
| 0.43 |
|
| 0.06 |
| Sex |
|
|
|
|
|
|
|
Female | CC | 46/71 | 1.00
(reference) | AA | 27/27 | 1.00
(reference) |
|
| CA/AA | 21/31 | 1.10 (0.55,
2.21) | AT/TT | 41/75 | 0.47 (0.23,
0.94) |
|
Male | CC | 30/88 | 1.00
(reference) | AA | 15/39 | 1.00
(reference) |
|
| CA/AA | 16/57 | 0.81 (0.40,
1.64) | AT/TT | 31/104 | 0.76 (0.36,
1.59) |
| P-value for
interaction |
|
| 0.67 |
|
| 0.31 |
Three-dimensional structure of cPLA2
δ
The SNP rs4924618 introduced a Ser-to-Thr point
mutation at position 434 of cPLA2 δ, which encodes a polypeptide of
818 amino acids that display 30% identity in its catalytic domain
(amino acids 273 to 818) with three other lipases, cPLA2 α, β and
γ. In order to elucidate the possible structural effect of this
substitution, homology modeling was performed from the crystal
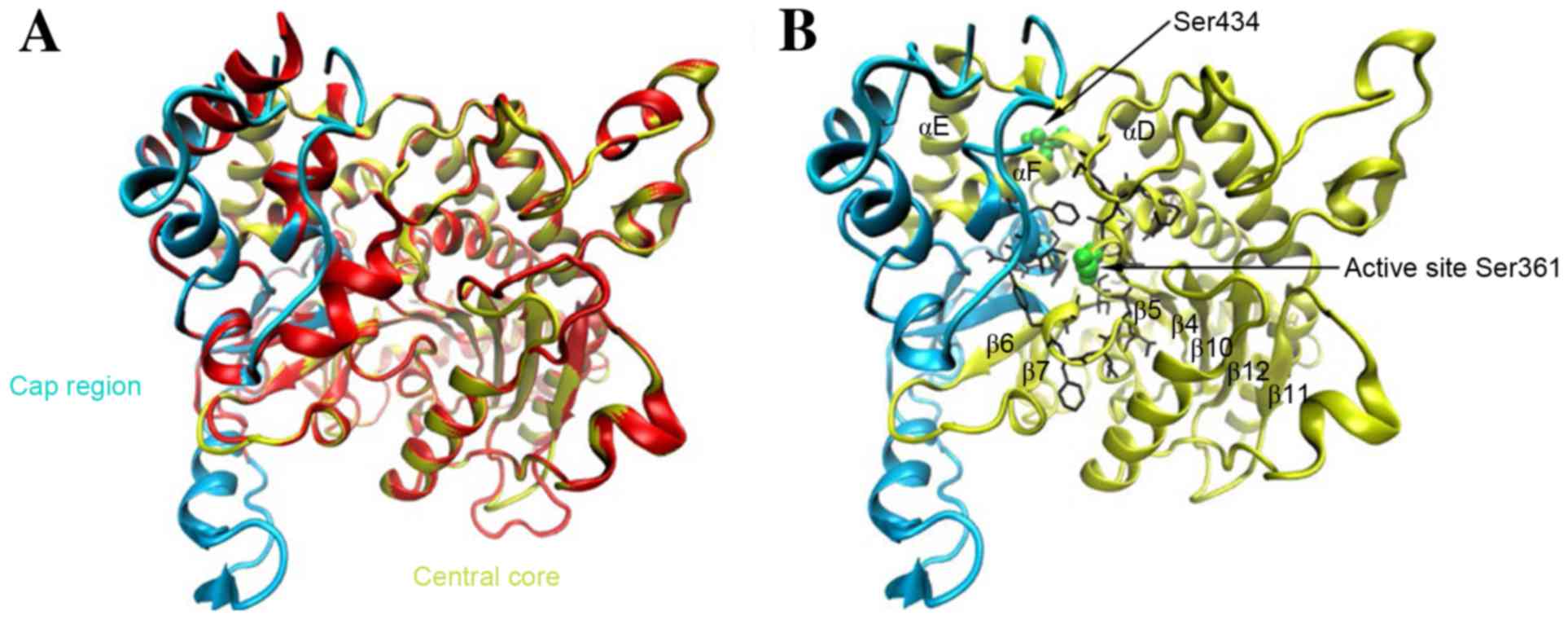
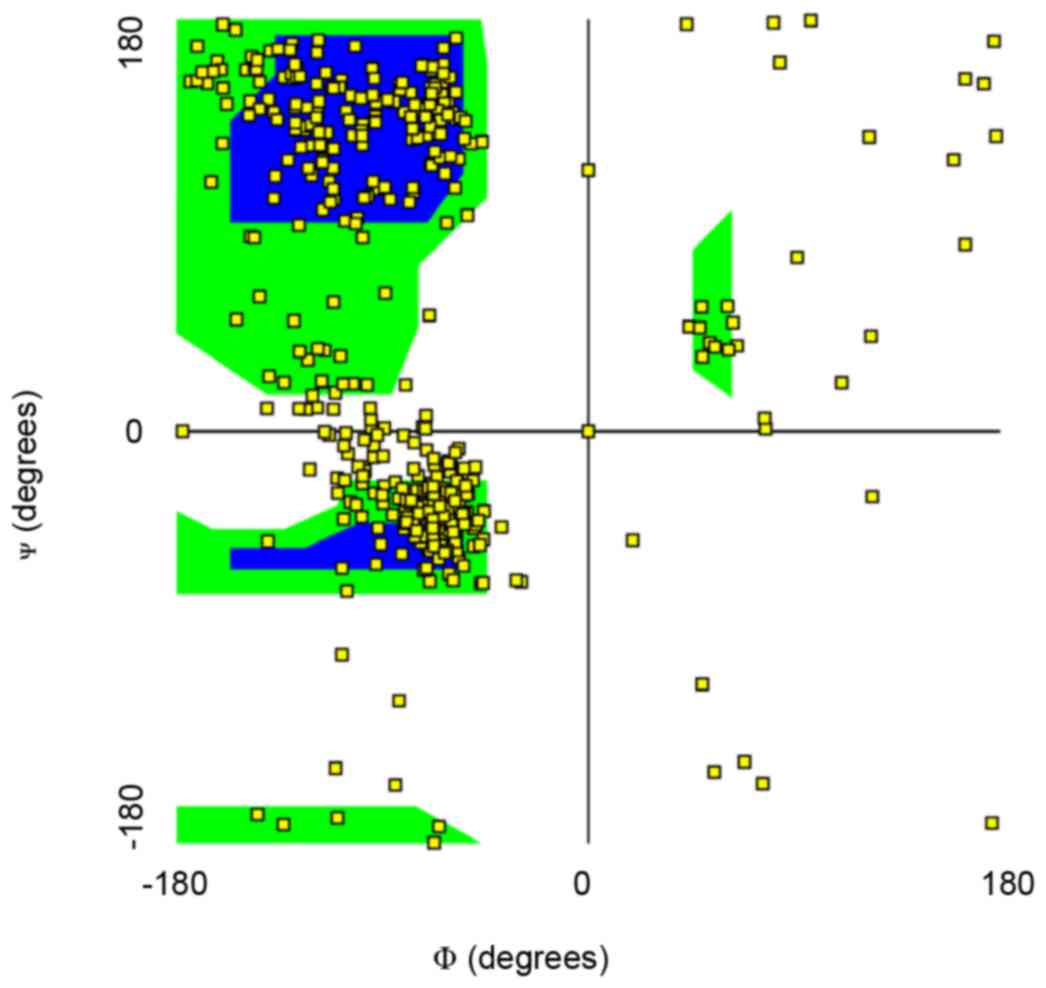
structure of cPLA2 α to locate the mutation site (Fig. 1). A Ramachandran plot of main chain
torsion angles indicated that most residues have a reasonable
conformation (Fig. 2),
demonstrating that the model is of good quality.
Fig. 1A
demonstrated the constructed model superposed with the template. As
expected, the tertiary structure of cPLA2 δ displayed considerable
agreement with that of cPLA2 α, especially in the α/β central core
(yellow region in Figs. 1A and B),
which consists of a 10-stranded mixed β sheet surrounded by 9 α
helices. This result was further verified by the multiple amino
acid sequence alignment of all the four cPLA2 isozymes (Fig. 3), where the conserved residues are
mainly concentrated within the core (26). The active site necessary for
catalysis is conserved in cPLA2 δ at Ser361, following β5 (Fig. 3). Similarly to the active site of
cPLA2 α, Ser361 is placed at the bottom of a deep, narrow active
site funnel, which penetrates one-third of the way into the
catalytic domain (Fig. 1B).
The mutation site Ser434 lies in the C-terminus of α
helix F of the conserved central core, which is outside but in
close to the bottom of the active site funnel spatially (14.92 Å).
Moreover, helix F, together with helices D, E and the interwoven
loops, connect the region encompassing the catalytic site (the
interconnecting loop between β5 and helix C) with the further four
β strands of the core (β6-β9).
Discussion
In the present study, the authors investigated
whether maternal PLA2G4C and PLA2G4D gene polymorphisms modified
the risk of SPTB in a Chinese population. The results indicated
that the maternal AT/TT genotype (dominant model) of PLA2G4D
rs4924618, but not PLA2G4C rs1366442 was associated with a reduced
risk of SPTB. To the best of the authors' knowledge, the protective
effect of PLA2G4D polymorphism on SPTB has not been reported before
in any population.
PLA2G4D encodes the δ form of cPLA2, which is one of
the key proteins involved in PGs synthesis (19,27).
PGs are well-known mediators in the inflammatory signaling pathways
to accompany the switch of the myometrium from a quiescent to a
contractile state (14). Increased
expression and bioavailability of PGs have been observed in
pregnant compared to non-pregnant women, in late compared to early
gestation, suggesting a link between PGs and initial steps in
parturition (28,29). Indeed, fetal PG infusion was
illustrated to provoke premature delivery of the ovine fetus
(30). Clinically, PGs are used as
a pharmacological labor inducer, and drugs inhibiting PG synthesis
are effective in preventing preterm labor (14).
Based on the studies mentioned above, the authors
concluded that the missense SNP rs4924618 may result in
disadvantage for cPLA2 δ with regard to the expression,
localization and/or activity. The homology modeling result
indicated that rs4924618-associated position (Ser434) located at
the conserved central core of the catalytic domain and was exposed
on the protein surface (Fig. 1),
demonstrating that even small changes at this site may not be
tolerated. In addition, the proximity of Ser434 to the active site
Ser361 prompted us to predict that the Ser434Thr mutation would
cause perturbation in the physical conformation of the active site
funnel to decrease the catalytic activity of cPLA2 δ, downregulate
the synthesis of PGs and, therefore, prolong the process of labor.
The facts that human cPLA2 δ was tissue-specifically expressed in
cervix (19) and murine cPLA2 δ
was exclusively expressed in placenta (31) support the present speculation
indirectly.
A case-control study involving diverse clinical
populations showed minor A allele of PLA2G4C rs1366442 was
detrimentally associated with PTB in the US Hispanic population
(18). However, for this PLA2G4C
SNP analyzed in the present study, a null association was observed,
which reminded us of considering the heterogeneity and
population-specific effects among different ethnic cohorts.
Possibly, this provided more evidence that differences in the
genetic background of ethnic groups contribute to the variation in
the rate of preterm birth among regions and countries.
Admittedly, the current study has several
limitations. First, only a very limited number of SNPs from the
PLA2G4C and PLA2G4D genes were analyzed. In addition, the sample
size is relatively small, which may not be typical for the general
population and has not enough power to examine the association of
SNPs in early PTB (<34 weeks of gestation). Although sample size
is a limitation to study design, given the patterns of associations
observed within genes and the functional analysis with the
bioinformatics methods, it is clear that several of the findings
are consistent with established literature. These results need to
be confirmed by larger replication studies, particularly in another
ethnic cohorts. More research, especially experiments using
appropriate cell and animal models, are required to explain how the
SNP rs4924618 influence the protein's structure and function during
parturition progress.
In conclusion, the authors have presented that the
polymorphism of PLA2G4D rs4924618 was associated with a reduced
risk of SPTB in a Chinese population. Meanwhile, bioinformatics
tools were used to investigate the potential SNP function. These
results provide novel insights into the genetic contribution to the
etiology of SPTB.
Acknowledgments
The authors would like to thank the pregnant women
who have participated in the present study and all obstetric care
providers who have assisted the authors in the implementation of
the study. This work was supported by the grants from the Guangzhou
Science and Technology Bureau (grant nos. 2011Y2-00025 and
2012J5100038), Guangdong Provincial Department of Science and
Technology (grant no. 2014A020213022) and National Natural Science
Foundation of China (grant no. 81673181).
Glossary
Abbreviations
Abbreviations:
|
SNP
|
single nucleotide polymorphism
|
|
SPTB
|
spontaneous preterm birth
|
|
PGs
|
prostaglandins
|
|
cPLA2
|
cytosolic phospholipase A2
|
|
GDM
|
gestational diabetes mellitus
|
|
GWCMC
|
Guangzhou Women and Children's Medical
Center
|
References
|
1
|
Howson CP, Kinney MV, McDougall L and Lawn
JE: Born Too Soon Preterm Birth Action Group: Born too soon:
Preterm birth matters. Reprod Health. 10 Suppl 1:S12013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldenberg RL, Culhane JF, Iams JD and
Romero R: Epidemiology and causes of preterm birth. Lancet.
371:75–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moutquin JM: Classification and
heterogeneity of preterm birth. BJOG. 110 Suppl 20:30–33. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhattacharya S, Raja EA, Mirazo ER,
Campbell DM, Lee AJ, Norman JE and Bhattacharya S: Inherited
predisposition to spontaneous preterm delivery. Obstet Gynecol.
115:1125–1133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clausson B, Lichtenstein P and Cnattingius
S: Genetic influence on birthweight and gestational length
determined by studies in offspring of twins. BJOG. 107:375–381.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Romero R, Edwards DR Velez, Kusanovic JP,
Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Chaiworapongsa T,
Pearce BD, Friel LA, et al: Identification of fetal and maternal
single nucleotide polymorphisms in candidate genes that predispose
to spontaneous preterm labor with intact membranes. Am J Obstet
Gynecol. 202:431. e1–e34. 2010. View Article : Google Scholar
|
|
7
|
Velez DR, Fortunato SJ, Thorsen P,
Lombardi SJ, Williams SM and Menon R: Preterm birth in Caucasians
is associated with coagulation and inflammation pathway gene
variants. PLoS One. 3:e32832008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Engel SA, Erichsen HC, Savitz DA, Thorp J,
Chanock SJ and Olshan AF: Risk of spontaneous preterm birth is
associated with common proinflammatory cytokine polymorphisms.
Epidemiology. 16:469–477. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Simhan HN, Krohn MA, Roberts JM, Zeevi A
and Caritis SN: Interleukin-6 promoter-174 polymorphism and
spontaneous preterm birth. Am J Obstet Gynecol. 189:915–918. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai HJ, Liu X, Mestan K, Yu Y, Zhang S,
Fang Y, Pearson C, Ortiz K, Zuckerman B, Bauchner H, et al:
Maternal cigarette smoking, metabolic gene polymorphisms, and
preterm delivery: New insights on GxE interactions and pathogenic
pathways. Hum Genet. 123:359–369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mann PC, Cooper ME, Ryckman KK, Comas B,
Gili J, Crumley S, Bream EN, Byers HM, Piester T, Schaefer A, et
al: Polymorphisms in the fetal progesterone receptor and a
calcium-activated potassium channel isoform are associated with
preterm birth in an Argentinian population. J Perinatol.
33:336–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dolan SM, Hollegaard MV, Merialdi M,
Betran AP, Allen T, Abelow C, Nace J, Lin BK, Khoury MJ, Ioannidis
JP, et al: Synopsis of preterm birth genetic association studies:
The preterm birth genetics knowledge base (PTBGene). Public Health
Genomics. 13:514–523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sykes L, MacIntyre DA, Teoh TG and Bennett
PR: Anti-inflammatory prostaglandins for the prevention of preterm
labour. Reproduction. 148:R29–R40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan AH, Carson RJ and Nelson SM:
Prostaglandins in labor-a translational approach. Front Biosci.
13:5794–5809. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Capper EA and Marshall LA: Mammalian
phospholipases A(2): Mediators of inflammation, proliferation and
apoptosis. Prog Lipid Res. 40:167–197. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kita Y, Ohto T, Uozumi N and Shimizu T:
Biochemical properties and pathophysiological roles of cytosolic
phospholipase A2s. Biochim Biophys Acta. 1761:1317–1322. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tithof PK, Roberts MP, Guan W, Elgayyar M
and Godkin JD: Distinct phospholipase A2 enzymes regulate
prostaglandin E2 and F2alpha production by bovine endometrial
epithelial cells. Reprod Biol Endocrinol. 5:162007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Plunkett J, Doniger S, Morgan T, Haataja
R, Hallman M, Puttonen H, Menon R, Kuczynski E, Norwitz E,
Snegovskikh V, et al: Primate-specific evolution of noncoding
element insertion into PLA2G4C and human preterm birth. BMC Med
Genomics. 3:622010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiba H, Michibata H, Wakimoto K, Seishima
M, Kawasaki S, Okubo K, Mitsui H, Torii H and Imai Y: Cloning of a
gene for a novel epithelium-specific cytosolic phospholipase A2,
cPLA2delta, induced in psoriatic skin. J Biol Chem.
279:12890–12897. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tao R, Yu Y, Zhang X, Shi J, Guo Y, Wang
C, Han B, Xu Q, Shang H, Zhang X, et al: A family based study of
the genetic association between the PLA2G4D gene and schizophrenia.
Prostaglandins Leukot Essent Fatty Acids. 73:419–422. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ning LF, Yu YQ, GuoJi ET, Kou CG, Wu YH,
Shi JP, Ai LZ and Yu Q: Meta-analysis of differentially expressed
genes in autism based on gene expression data. Genet Mol Res.
14:2146–2155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vigod SN, Kurdyak PA, Dennis CL, Gruneir
A, Newman A, Seeman MV, Rochon PA, Anderson GM, Grigoriadis S and
Ray JG: Maternal and newborn outcomes among women with
schizophrenia: A retrospective population-based cohort study. BJOG.
121:566–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Männistö T, Mendola P, Kiely M, O'Loughlin
J, Werder E, Chen Z, Ehrenthal DB and Grantz KL: Maternal
psychiatric disorders and risk of preterm birth. Ann Epidemiol.
26:14–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Biasini M, Bienert S, Waterhouse A, Arnold
K, Studer G, Schmidt T, Kiefer F, Cassarino T Gallo, Bertoni M,
Bordoli L and Schwede T: SWISS-MODEL: Modelling protein tertiary
and quaternary structure using evolutionary information. Nucleic
Acids Res. 42:W252–W258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Humphrey W, Dalke A and Schulten K: VMD:
Visual molecular dynamics. J Mol Graph. 14:33–38. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dessen A, Tang J, Schmidt H, Stahl M,
Clark JD, Seehra J and Somers WS: Crystal structure of human
cytosolic phospholipase A2 reveals a novel topology and catalytic
mechanism. Cell. 97:349–360. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ghosh M, Tucker DE, Burchett SA and Leslie
CC: Properties of the Group IV phospholipase A2 family. Prog Lipid
Res. 45:487–510. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hayashi M, Inoue T, Hoshimoto K,
Hirabayashi H, Negishi H and Ohkura T: The levels of five markers
of hemostasis and endothelial status at different stages of
normotensive pregnancy. Acta Obstet Gynecol Scand. 81:208–213.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Zuckerman B, Kaufman G, Wise P,
Hill M, Niu T, Ryan L, Wu D and Xu X: Molecular epidemiology of
preterm delivery: Methodology and challenges. Paediatr Perinat
Epidemiol. 15 Suppl 2:63–77. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Young IR, Deayton JM, Hollingworth SA and
Thorburn GD: Continuous intrafetal infusion of prostaglandin E2
prematurely activates the hypothalamo-pituitary-adrenal axis and
induces parturition in sheep. Endocrinology. 137:2424–2431. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ohto T, Uozumi N, Hirabayashi T and
Shimizu T: Identification of novel cytosolic phospholipase A(2)s,
murine cPLA(2){delta}, {epsilon}, and {zeta}, which form a gene
cluster with cPLA(2){beta}. J Biol Chem. 280:24576–24583. 2005.
View Article : Google Scholar : PubMed/NCBI
|